Abstract
Ras guanine nucleotide exchange factor (GEF) Q, a nucleotide exchange factor from Dictyostelium discoideum, is a 143-kD protein containing RasGEF domains and a DEP domain. We show that RasGEF Q can bind to F-actin, has the potential to form complexes with myosin heavy chain kinase (MHCK) A that contain active RasB, and is the predominant exchange factor for RasB. Overexpression of the RasGEF Q GEF domain activates RasB, causes enhanced recruitment of MHCK A to the cortex, and leads to cytokinesis defects in suspension, phenocopying cells expressing constitutively active RasB, and myosin-null mutants. RasGEF Q− mutants have defects in cell sorting and slug migration during later stages of development, in addition to cell polarity defects. Furthermore, RasGEF Q− mutants have increased levels of unphosphorylated myosin II, resulting in myosin II overassembly. Collectively, our results suggest that starvation signals through RasGEF Q to activate RasB, which then regulates processes requiring myosin II.
Introduction
The actin cytoskeleton is composed of actin, myosin, and several proteins that bind to them and play crucial roles in cell motility, cytokinesis, phagocytosis, and intracellular transport processes (Cooper, 1991; Matsumura, 2005). Coordinated cell movement requires protrusive forces generated by polymerization of actin filaments at the leading edge and contractile forces via myosin motors at the rear of a cell. Myosin II, the conventional two-headed myosin, is also the primary motor protein required for cytokinesis in eukaryotes. Dictyostelium discoideum cells lacking MhcA show a plethora of defects, which include cytokinesis defect in suspension but undergoing cytokinesis when grown on a solid support by a “traction-mediated” mechanism (De Lozanne and Spudich, 1987). These correspond to the localization of myosin at the contractile ring (cleavage furrow) during cytokinesis. MhcA− cells have a decreased chemotactic efficiency caused by reduction in cell polarity and an inability to suppress lateral pseudopods and retract the uropod (Wessels and Soll, 1990; Wessels et al., 1988). They also have a developmental defect, halting the developmental process shortly after cells have aggregated. The regulation of myosin II appears to differ between higher and lower eukaryotes.
In D. discoideum phosphorylation of myosin takes place at three threonine residues in the tail region by myosin II heavy chain kinases (MHCKs) (Luck-Vielmetter et al., 1990; Vaillancourt et al., 1988). Phosphorylated myosin is inactive and does not assemble into filaments, whereas unphosphorylated myosin II can spontaneously assemble into bipolar filaments. It is only these filaments that perform cellular myosin II functions (Egelhoff et al., 1993). Significant knowledge about the function of myosin II regulation has been derived from mutant myosin IIs: 3XALA myosin, where the three phosphorylatable threonines have been mutated to alanine, rendering it a poor substrate for MHCKs; and 3XASP myosin, where the three threonines were replaced by aspartate, mimicking the phosphorylated state. 3XALA myosin mutants show significant myosin overassembly in cytoskeletal fractions and form stable myosin II filaments, which accumulate in the rear cortex. Cells expressing 3XALA myosin are drastically impaired in cell migration and chemotaxis, making frequent turns and extending lateral pseudopods, which is caused by the inability to disassemble myosin filaments, and these cells have severely affected motility (Egelhoff et al., 1996; Stites et al., 1998; Heid et al., 2004). In contrast, 3XASP myosin does not assemble into bipolar filaments, is nonfunctional in vivo, and fails to complement cytokinesis and developmental defects of myosin II–null cells (Egelhoff et al., 1993).
Signaling pathways based on small GTPases of the Ras family regulate a myriad of cellular processes in eukaryotic cells. The D. discoideum genome encodes a large and varied family of Ras GTPases consisting of 15 Ras proteins. D. discoideum uses its Ras proteins to regulate several pathways controlling cell motility and polarity, cytokinesis, phagocytosis and pinocytosis, and multicellular development (Charest and Firtel, 2007).
D. discoideum expresses at least 25 Ras guanine nucleotide exchange factors (GEFs; Wilkins et al., 2005). However, it does not code for conventional receptor tyrosine kinases (RTKs), which are the major inputs for Ras signaling in higher eukaryotes (Eichinger et al., 2005). Functions of some of the RasGEFs are slowly being understood through mutant analysis. The gefA− (aimless) cells fail to aggregate upon starvation and are unable to synthesize and respond to cAMP (Insall et al., 1996; Kae et al., 2007), showing phenotypic similarities to rasC− cells, indicating that they may act to regulate activation of adenylyl cyclase. The cyclic guanosine monophosphate binding proteins GbpC and D, which have RasGEF domains, show altered myosin II localization during chemotaxis (Bosgraaf et al., 2005). Cyclic guanosine monophosphate and GbpC induce myosin II filament formation, but its corresponding Ras GTPase has not been identified. GbpD is thought to activate Rap1 and regulate cell surface adhesion and motility (Kortholt et al., 2006). RasG, the most abundant Ras in vegetative cells and the closest relative to mammalian Ras, is thought to regulate several actin cytoskeleton–based processes like cell polarity and cytokinesis (Tuxworth et al., 1997). RasGEF R appears to be required for maximal activation of RasG upon response to cAMP (Kae et al., 2007).
In this paper, we have focused on RasGEF Q. Our experiments identify RasB as a substrate for RasGEF Q. They further indicate that RasGEF Q acts upstream of RasB and regulates processes requiring myosin II, like cytokinesis, cell motility, and suppression of lateral pseudopods. Mutants lacking RasGEF Q show myosin overassembly caused by high levels of unphosphorylated myosin II and produce many random pseudopodia. Cells that overexpress the GEF domain of RasGEF Q have constitutively activated RasB, which is normally activated during aggregation upon a cAMP stimulus, and have defects in cytokinesis in suspension as do mhcA− cells. Our results also imply an involvement of MHCK A as a downstream regulator of the signaling cascade. We observe that cells that overexpress the GEF domain have higher levels of MHCK A recruited to the cytoskeletal fractions, which occurs when MHCK A is activated in response to cAMP. Furthermore, RasGEF Q is involved in cell sorting and developmental patterning and slug motility.
Results
Domain organization, expression pattern, and functional dissection of RasGEF Q
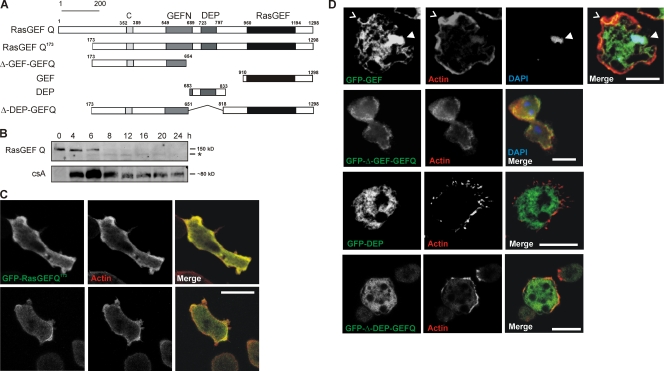
RasGEF Q, encoded by the gefQ gene, is a 1298-aa protein with a calculated molecular mass of 143,000. Apart from the RasGEF domains, it contains a DEP domain (a domain conserved among fly Dishevelled, worm Egl10, and mammalian Pleckstrin) separating the two RasGEF domains and a predicted coiled-coil region at the N terminus (Fig. 1 A). RasGEF Q mRNA as examined by RT-PCR analysis of cDNA is present throughout development with elevated levels during aggregation and the loose mound stage (6–8 h of starvation; Wilkins et al., 2005). Monoclonal antibody K-70-187-1 generated against the DEP domain of RasGEF Q recognized a protein of the expected size, which was present in vegetative cells and in early development till the aggregation stage (6 h) but could not be detected later (Fig. 1 B). Instead, a smaller and less intense band was seen at subsequent stages (Fig. 1 B, asterisk). The smaller band could arise by translation from a downstream start (ATG) site at position 565. The sequence between both start sites is highly AT rich and could function as an alternative promoter.
Figure 1.
Architecture and expression of RasGEF Q and localization of RasGEF Q domains. (A) Schematic diagram of RasGEF Q depicting its domain organization and constructs used in the study. (B) Accumulation of RasGEF Q protein during development. Total cell lysates prepared at the indicated time points were immunoblotted and probed using mAb K-70-187-1 raised against RasGEF Q. *, smaller form of the protein. Expression of the developmentally regulated cell adhesion protein csA is shown for control at the bottom. (C) Localization of GFP-RasGEF Q173 (green) in wild-type cells. In both vegetative (0 h; bottom) and aggregation-competent (6 h; top) cells, GFP-RasGEF Q173 was present throughout the cytosol and showed specific enrichment with cortical actin (arrowheads). Actin (red) was recognized by mAb Act 1–7 followed by cy3-labeled anti–mouse secondary antibody. Cells were fixed with cold methanol. (D) Localization of GFP-tagged RasGEF Q domains. The top row shows that gfp-gef localizes to the cell cortex (carat) and nucleus (arrowhead). The second row shows that GFP-Δ-GEF-GEFQ localizes throughout the cytosol but is enriched in the cell cortex. The third row shows that the DEP domain is present throughout the cytoplasm. The fourth row shows that GFP-Δ-DEP-GEFQ localizes throughout the cytosol with slight enrichment in the cell cortex. F-actin was stained by TRITC-labeled phalloidin and DNA was stained with DAPI. Cells were fixed with picric acid/formaldehyde. Bars, 10 μm.
To study the localization of RasGEF Q, we expressed RasGEF Q lacking the first 172 aa as a GFP fusion protein (GFP-RasGEF Q173). We observed that in fixed vegetative (Fig. 1 C, bottom) and aggregation-competent cells (Fig. 1 C, top), GFP-RasGEF Q173 was localized throughout the cytosol but was also enriched in the cortex, colocalizing with actin (Fig. 1 C). When living cells expressing GFP-RasGEF Q173 were observed, cells appeared more flat and adhered to the surface, having long filopodia-like protrusions, and the protein was localized throughout the cytosol (Videos 1–4, available at http://www.jcb.org/cgi/content/full/jcb.200710111/DC1). To analyze the functions of different domains of RasGEF Q, we expressed the corresponding GFP fusion proteins in AX2. In fixed cells, the RasGEF domain (GFP-GEF) was present throughout the cytosol but was enriched at the cell cortex where it colocalized with F-actin (Fig. 1 D, first row, carats) and was also present in the nucleus (Fig. 1 D, first row, arrowheads). Live cell analysis showed that the protein was present throughout the cells. It was also observed in the nucleus, and in moving cells it accumulated in extending pseudopods (Video 5). Cells expressing GFP-ΔGEF-GEFQ, which corresponds to the N-terminal domain (aa 173–654), distributed throughout the cytoplasm with enrichment in the cortex (Fig. 1 D, second row). The DEP domain was present throughout the cytoplasm but did not show a particular enrichment either in fixed or in living cells (Fig. 1 D, third row; and Video 6). The GFP fusion protein lacking the DEP domain (GFP-Δ-DEP-GEFQ) localized throughout the cytosol with slight enrichment in the cortical regions (Fig. 1 D, fourth row).
RasGEF Q association with the actin cytoskeleton
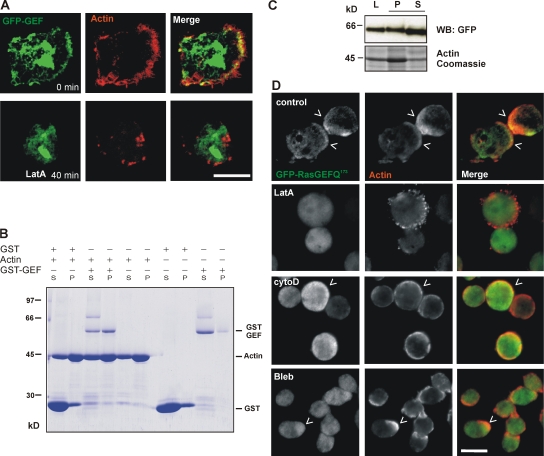
Small GTPases of the Ras superfamily and their regulatory GEFs are known to be directly involved in the regulation of the cytoskeleton. To further investigate the colocalization of GFP-GEF with F-actin in the cortex, we used latrunculin A (LatA), a drug that binds to actin monomers and prevents polymerization. Treatment with 10 μM LatA for 40 min led to a loss of cortical F-actin. In parallel, the cortical localization of GFP-GEF was lost, whereas its nuclear localization and the cytoplasmic staining were not affected (Fig. 2 A). This indicates that the GEF domain is recruited to the cortical region by F-actin. To test whether the GEF domain directly interacts with actin filaments, we performed cosedimentation assays. We used the GST-tagged GEF domain in these assays and found that it cosedimented with actin filaments by high speed centrifugation at 120,000 g, whereas the fusion protein alone stayed in the supernatant. The GST-GEF interaction with F-actin occurred even at high salt concentrations (100 mM KCl), which is known to drastically reduce the binding efficiency of several actin binding proteins like α-actinin, comitin, and plastin (Fig. 2 B; Jung et al., 1996; Prassler et al., 1997). We then used cells overexpressing GFP-GEF and prepared cytoskeletal fractions. Supernatant and pellet fractions were resolved by SDS-PAGE, and distribution of GFP-GEF in either fraction was tested with a GFP monoclonal antibody. We found that GFP-GEF was present in the cytoskeletal pellet and the supernatant fractions (Fig. 2 C).
Figure 2.
Association of the C-terminal part of RasGEF Q with F-actin. (A) The cortical localization of GFP-GEF is sensitive to LatA treatment. Cells expressing GFP-GEF (green) were incubated with LatA for 40 min, fixed with picric acid/formaldehyde, and stained for actin using TRITC-labeled phalloidin (red). Images were taken using a confocal microscope. (B) GST-GEF binds to F-actin in an in vitro cosedimentation assay using D. discoideum actin. Pellet (P) and supernatant (S) were separated by high-speed centrifugation, and proteins in the fractions were resolved by SDS-PAGE and stained with Coomassie brilliant blue. (C) Cytoskeletal fractions were prepared from AX2 cells expressing GFP-GEF using a buffer containing 2% saponin. Total cell lysates (L), pellet (P), and supernatant (S) fractions were resolved on SDS-PAGE and amounts were analyzed by Western blotting using a GFP monoclonal antibody. Coomassie-stained gel showing the actin band is used as control. (D) Effect of drugs affecting the cytoskeleton on localization of GFP-RasGEF Q173. Cells expressing GFP-RasGEF Q173 (green) were treated with DMSO (control; top row), 10 μM LatA (second row), 20 μM cytochalasin D (third row), or 100 μM Blebbistatin (fourth row) for 40 min. Cells were fixed with cold methanol. Actin (red) was recognized by mAb Act1–7 followed by cy3-labeled anti–mouse secondary antibody. Carats point to areas of colocalization. Bars, 10 μm.
We treated cells expressing GFP-RasGEF Q173 with drugs to disturb the actin cytoskeleton or myosin II. For interference with the actin cytoskeleton we used 10 μM LatA, which forms 1:1 complexes with actin monomers and inhibits actin polymerization, and 20 μM cytochalasin D, a drug that binds to the growing or barbed end (+ end) of F-actin filaments and prevents addition of G-actin at these sites. Blebbistatin (used at 100 μM) is a drug that blocks the myosin heads and lowers the affinity of myosin for actin (Kovacs et al., 2004). We found that in control cells treated with DMSO (solvent for the drugs), GFP-RasGEF Q173 showed a cytosolic staining and was present in cortical regions of the cell colocalizing with actin. Upon treatment with LatA, the cells rounded up, the typical cortical actin staining was lost, and a patchy pattern appeared. GFP-RasGEF Q173 was present in the cytosol. On treatment with cytochalasin D, we observed the cells rounding up and actin staining in the cortex was fragmented. In this case, GFP-RasGEF Q173 was also seen in the periphery of the cell together with actin. Treatment with Blebbistatin did not alter the cell morphology or the actin staining and GFP-RasGEF Q173 was still enriched in the cell periphery (Fig. 2 D). Thus, LatA that disrupts the cortical actin cytoskeleton also affects the localization of GFP-RasGEF Q172 at the cortex. Collectively, our results imply that RasGEF Q associates with F-actin in vitro and in vivo and that the C-terminal region containing the RasGEF domain may mediate this binding.
RasGEF Q–null cells have defects in developmental patterning and slug motility
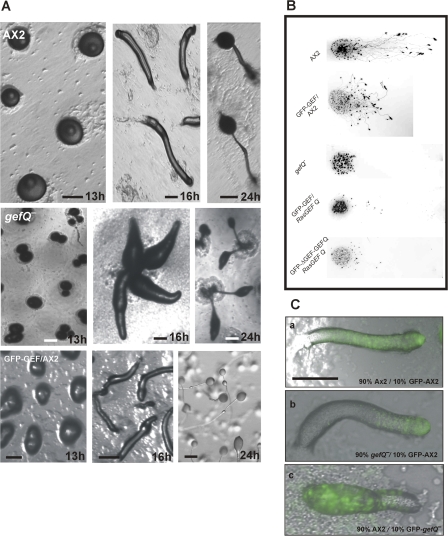
To study the functions of RasGEF Q in vivo we generated cells lacking RasGEF Q. gefQ− cells were generated by homologous recombination, screened by genomic PCR on genomic DNA, and confirmed by Southern and Western blotting and immunofluorescence analysis (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200710111/DC1). Multicellular morphogenesis can be described in its most basic form as a process where cells in a group undergo coordinated changes in cell shape and motility to form an organized structure to perform a particular function. The process of D. discoideum development is one such example of programmed multicellularization (Chisholm and Firtel, 2004). Involvement of the cytoskeleton in such processes that require continuous cellular changes is a prerequisite. When developed on nutrient-deficient agar plates, AX2 cells form aggregates by 8 h and tipped mounds by 13 h. The tip elongates to form a standing finger that falls down, becoming a migratory slug or pseudoplasmodium by 16 h. Development is completed by 20–24 h with the formation of fruiting bodies (Fig. 3 A, top; and Video 7 available at http://www.jcb.org/cgi/content/full/jcb.200710111/DC1).gefQ− cells initiate the developmental program, but in >50% of the mounds, multiple tips arise from a single mound by 13–14 h and, as a result, fruiting bodies are smaller than in AX2 (Fig. 3 A, middle; and Video 8). gefQ− cells formed slugs that were relatively smaller, their ability to move was greatly reduced, and they did not perform phototaxis toward a directed light source. In contrast, slugs from AX2 cells expressing GFP-GEF were able to move and perform phototaxis, but the migration was more disoriented compared with AX2. GFP-GEF slugs migrated at a mean angle of ∼80°, whereas AX2 slugs migrated at a mean angle of 25° toward the light. gefQ− cells expressing GFP-GEF or GFP-ΔGEF-GEFQ showed slightly improved slug motility in phototaxis assays compared with gefQ− cells. A proportion of the slugs formed could migrate toward the light source, although migration was greatly reduced, indicating that RasGEF Q is important for slug motility (Fig. 3 B).
Figure 3.
Developmental defects in RasGEF Q–null cells (gefQ−). (A) Development of gefQ− cells plated on nonnutrient agar. The top shows wild-type tipped aggregate (13 h), migratory slug (16 h), and fruiting body (24 h). The middle shows that gefQ− aggregates break up into smaller aggregates (13 h), multiple tipped structures (16 h), and fruiting bodies (24h). The bottom shows development of cells overexpressing GFP-GEF. Bars, 200 μm. (B) Altered slug motility in phototaxis assays in gefQ− and GFP-GEF–expressing cells. Wild-type AX2, GFP-GEF expressing AX2, and gefQ− cells, as well as gefQ− cells expressing GFP-GEF or GFP-Δ-GEF-GEFQ, were allowed to develop on phosphate agar plates placed in a black opaque box for 36 h with a unidirectional light source from an open slit. Migratory pattern of slugs were determined by transferring slime trails and cellular materials onto nitrocellulose membrane. Membranes were stained with 0.1% amido black. (C) Altered cell-type spatial patterning in gefQ− mutants. (a) 10% AX2 cells labeled with GFP was mixed with 90% unlabeled wild-type cells. (b) 10% labeled wild-type cells was mixed with 90% unlabeled gefQ− cells. (c) 10% labeled gefQ− cells was mixed with 90% AX2 cells and were codeveloped as a chimera. Images were taken at the slug stage using a fluorescent microscope (Leica DMR) at 5× magnification. Imaging medium was air at 22°C. Images were acquired with a camera (DC 350 FX; Leica). Bar, 200 nm.
D. discoideum serves as a model system to understand cell sorting. At the mound stage, an asymmetry is established and a fixed proportion of cell types is generated where the majority of the cells (nearly 80%) are destined to become spore cells and sort to the rear of a slug and 20% occupy the anterior tip, becoming the stalk cells that hold the spore mass in the fruiting body. Involvement of a particular protein in such cell-sorting mechanisms can be easily studied in D. discoideum by mixing strains together and allowing them to codevelop as a chimera. When 10% GFP-expressing AX2 cells was mixed with 90% unlabeled AX2 cells, GFP-labeled cells distributed evenly throughout the slug. However, when 10% GFP-labeled AX2 cells was mixed with 90% unlabeled gefQ− cells, wild-type cells localized to the anterior prestalk region and mutants were present in the posterior prespore region of the slug. In a converse experiment where 10% GFP-labeled gefQ− cells was mixed with 90% AX2 cells, mutant cells also sorted to the posterior (Fig. 3 C).
Cytokinesis defect in suspension in cells overexpressing GFP-GEF and constitutively activated RasB
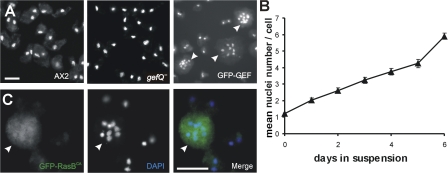
Further analysis revealed that the cells expressing GFP-GEF were defective in cytokinesis in suspension. When grown in suspension for 6 d, they attain a mean nuclei number of approximately six nuclei per cell, whereas wild-type AX2 and gefQ− cells are mostly mono- or binucleated (Fig. 4, A and B). However, when grown on a plastic surface, GFP-GEF cells are predominantly mono- or binucleated, probably dividing by traction-mediated cytofission as occurs in mhcA− cells (De Lozanne and Spudich, 1987; Neujahr et al., 1997).
Figure 4.
Cells overexpressing GFP-GEF and constitutively activated RasB have a cytokinesis defect. (A) Cytokinesis is normal in wild-type AX2 and gefQ− cells but is defective in GFP-GEF cells. Images were taken using a fluorescent microscope (DMR). Images were acquired with a camera (DC 350 FX). (B) The increase in the number of nuclei in GFP-GEF cells. GFP-GEF cells were grown in suspension, fixed at the indicated times, stained with DAPI, and the nuclei were counted. The data are the mean of three independent determinations ±SD. (C) Cytokinesis defect in cells overexpressing constitutively activated RasBG12T (GFP-RasBCA; green). Cells were fixed with methanol and nuclei are visualized with DAPI (blue). Arrowheads indicate multinucleated cells. Images were taken using a fluorescent microscope (DMR). Images were acquired with a camera (DC 350 FX). Bars, 10 μm.
Our results indicate that although karyokinesis is normal in GFP-GEF cells, the following cytokinesis is impaired. We also confirmed previous data on the localization and phenotype of cells expressing constitutively activated RasB (RasBG12T; Sutherland et al., 2001). Overexpression of GFP-tagged RasBG12T caused cells to become multinucleated in suspension. The protein was slightly enriched in the nucleus (Fig. 4 C). Because overexpressors of the GEF domain and overexpressors of RasBG12T had phenotypic similarities, we hypothesized that RasGEF Q could be an exchange factor for RasB, overexpression of the GEF domain acts as an activated form of RasGEF Q, and a regulatory mechanism must exist.
RasGEF Q activates RasB
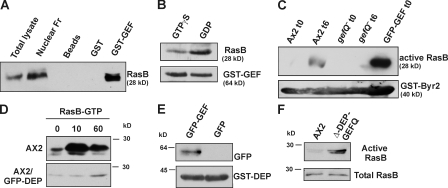
To test whether RasGEF Q interacts with and activates RasB, we performed several assays. In pulldown assays, GST-GEF bound to glutathione Sepharose beads could precipitate RasB from AX2 cell lysates (Fig. 5 A). When cell lysates were pretreated with 100 μM GDP or 100 μM GTPγS, we found that GST-GEF bound preferentially to the GDP-bound form of RasB (Fig. 5 B), which is the preferred form for a RasGEF–Ras interaction. To determine if RasGEF Q actually activates RasB, we used GST-Byr2 Ras binding domain (RBD) to pull down GTP-bound active RasB from AX2, gefQ−, or GFP-GEF cells. Schizosaccharomyces pombe Byr2 is a Ras effector (MAPK/ERK kinase homologue) and binds to Ras in its activated (GTP-bound) form (Gronwald et al., 2001; Scheffzek et al., 2001). Cell lysates of AX2 and gefQ− at the growth (0 h) and aggregation (6 h) stage and GFP-GEF at 0 h were incubated with equal amounts of GST-Byr2(RBD) bound to glutathione Sepharose beads. We found that RasB is activated at 6 h in AX2 cells, whereas gefQ− cells are unable to activate RasB at either 0 or 6 h. GFP-GEF cells at 0 h have a high level of activated RasB, indicating that RasGEF Q acts as an exchange factor for RasB (Fig. 5 C). Expression of the GEF domain alone may act as a constitutively activated form of RasGEF Q. Our results also indicate that, although RasB is expressed throughout development (Daniel et al., 1993), it is activated upon starvation, thus implying a role for cAMP in its activation process. Because RasB is not activated in gefQ−, cells we can conclude that it is the only or the predominant exchange factor for RasB.
Figure 5.
RasB activation by RasGEF Q and during development. (A) Physical association of GST-GEF and RasB. Glutathione Sepharose beads coated with either GST-GEF or GST or uncoated with any protein were incubated with AX2 cell lysates and pulldown eluates were immunoblotted with RasB antibody. (B) GST-GEF binds preferentially to GDP-bound RasB. Glutathione Sepharose beads coated with GST-GEF were incubated with AX2 cell lysates preincubated with 100 μM GTPγS or 100 μM GDP. Pulldown eluates were immunoblotted with RasB antibody. (C) RasB is activated upon starvation (6 h) and in cells overexpressing GFP-GEF (0 h), using beads coated with GST-Byr2(RBD). (D) Activation of RasB in response to cAMP. Aggregation-competent AX2 (top) cells were stimulated with 500 nM cAMP, and the amount of activated RasB bound to GST-Byr2(RBD) was determined at the indicated time points. Wild-type cells overexpressing GFP-DEP (bottom) showed reduced and delayed RasB activation upon cAMP stimulation. (E) Interaction between the DEP and the GEF domain of RasGEF Q. Glutathione Sepharose beads coated with GST-DEP and incubated with cell-free extracts from cells expressing GFP-GEF or GFP alone. (F) Deletion of DEP domain causes RasB activation. Vegetative cells overexpressing GFP-Δ-DEP-GEFQ and AX2 cells were used to pull down activated RasB using GST-Byr2–coated beads. Total cell lysates and pulldown eluates were immunoblotted with RasB antibody.
RasB is activated upon cAMP stimulation
To test whether RasB is activated upon response to cAMP, we used a GST-Byr2(RBD) to pull down activated Ras from cell lysates stimulated with cAMP. We observed an elevation in the levels of activated RasB within 10 s of stimulation with cAMP and decreasing by 60 s (Fig. 5 D, top). In a similar experiment using gefQ− cells expressing GFP-Δ-GEF-GEFQ, which also lacks the DEP domain, we saw no activation of RasB upon cAMP, further indicating that RasGEF Q is required for RasB activation (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200710111/DC1). We also observed that in cells overexpressing the DEP domain, the RasB activation was greatly reduced and delayed (Fig. 5 D, bottom). As overexpression of the GEF domain acts as a constitutively activated form of RasGEF Q, the DEP domain that resides between the RasGEF domains might serve as a possible autoregulatory domain in the activation process of RasGEF Q. Further on, in a GST pulldown experiment, GST-DEP could pull down GFP-GEF from cell lysates of cells overexpressing GFP-GEF, indicating that the two domains can physically interact (Fig. 5 E). To further test whether the DEP domain acts as an autoregulatory domain, we used AX2 cells expressing GFP-Δ-DEP-GEFQ, which is specifically lacking the DEP domain, and determined the amount of activated RasB in growing cells using GST-Byr2–coated beads. Cells overexpressing GFP-Δ-DEP-GEFQ have activated RasB even at the vegetative state, which is similar to cells expressing the GEF domain alone, further supporting that the DEP domain has autoregulatory functions in the protein (Fig. 5 F).
Role of RasGEF Q in regulating myosin II function
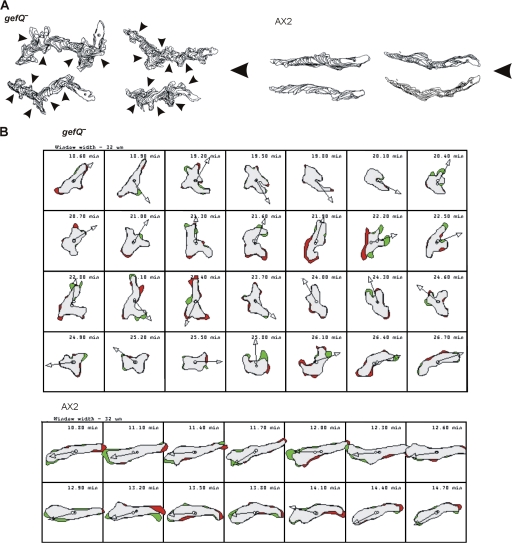
Defects in Ras pathways result in chemotaxis defects (Sasaki and Firtel, 2005, 2006; Charest and Firtel, 2006). To examine whether RasGEF Q had any effect on chemoattractant-induced cell migration, we compared the migration of aggregation-competent gefQ− and parental AX2 cells. During migration toward an exogenous cAMP source, wild-type cells are well polarized and produce pseudopodia exclusively at the leading edge and very few lateral pseudopods. In contrast, gefQ− cells produce more random pseudopodia (4.6 per cell per 10 min) than wild-type cells (1.7 per cell per 10 min; Table I) and have an increased frequency of turning (Fig. 6, A and B; and Videos 9 and 10, available at http://www.jcb.org/cgi/content/full/jcb.200710111/DC1), showing similarities to 3XALA myosin mutants. Surprisingly, the cell motility parameters (speed, persistence, and direction change) were not significantly altered (Table II).
Table I. Lateral pseudopod formation by cells crawling in buffer and in a spatial cAMP gradient.
| Cell strain | Number of cells |
0–2 lateral pseudopods per 10 min |
3–5 lateral pseudopods per 10 min |
>5 lateral pseudopods per 10 min |
Mean frequency of lateral pseudopods per cell per 10 min |
|---|---|---|---|---|---|
| % | % | % | |||
| Buffer | |||||
| AX2 | 31 | 9.6 | 45.16 | 45.16 | 5.19 |
| gefQ− | 33 | 0 | 21.21 | 78.78 | 7.03 |
| cAMP gradient | |||||
| AX2 | 35 | 82.85 | 17.14 | 0 | 1.71 |
| gefQ− | 29 | 6.9 | 65.5 | 27.50 | 4.65 |
Images were taken at a magnification of 40× every 6 s. In all cases, cells were analyzed for 10 min. χ2 test, performed between AX2 and gefQ− cells on data of the three categories of lateral pseudopods formed, showed highly significant (P > 0.001) difference between AX2 and gefQ− cells in both buffer and a cAMP gradient.
Figure 6.
Chemotactic behavior of gefQ− cells. (A) Computer-generated cell tracks of gefQ− and AX2 cells during chemotactic migration in a spatial cAMP gradient using DIAS. Arrowheads indicate turns initiated by formation of lateral pseudopods in gefQ− cells. (B) Shape changes of aggregation-competent gefQ− and AX2 cells during chemotactic migration were analyzed by DIAS. Images were taken every 6 s but only every third frame is shown. The green areas indicate new membrane protrusions, the red areas indicate retractions, and the arrows indicate the direction of migration.
Table II. Chemotactic behavior in a spatial cAMP gradient.
| AX2 | gefQ− | |
|---|---|---|
| Buffer | ||
| Speed (μm/min) | 6.04 ± 3.14 | 5.47 ± 1.41 |
| Persistence (μm/min-degree) | 1.35 ± 0.88 | 1.4 ± 0.78 |
| Directionality | 0.31 ± 0.18 | 0.34 ± 0.14 |
| Direction change (degree) | 50.9 ± 12.84 | 50.12 ± 11.74 |
| cAMP gradient | ||
| Speed (μm/min) | 10.62 ± 2.23 | 12.21 ± 3.01 |
| Persistence (μm/min-degree) | 3.76 ± 1.06 | 4.47 ± 1.28 |
| Directionality | 0.79 ± 0.12 | 0.79 ± 0.12 |
| Direction change (degree) | 21.63 ± 12 | 18.66 ± 7.16 |
Images were taken at magnification of 20× every 30 s. In all cases, cells were analyzed for at least 10 min. The DIAS software was used to trace individual cells along the image series and calculate motility parameters. Persistence is an estimation of movement in the direction of the path. Directionality is calculated as the net path length divided by the total path length and gives a value of 1 for a straight path. Directional change represents the average change of angle between frames in the direction of movement. Values are mean standard deviation of 30–90 cells from at least three independent experiments. For both strains, all parameters varied significantly between the buffer and cAMP conditions
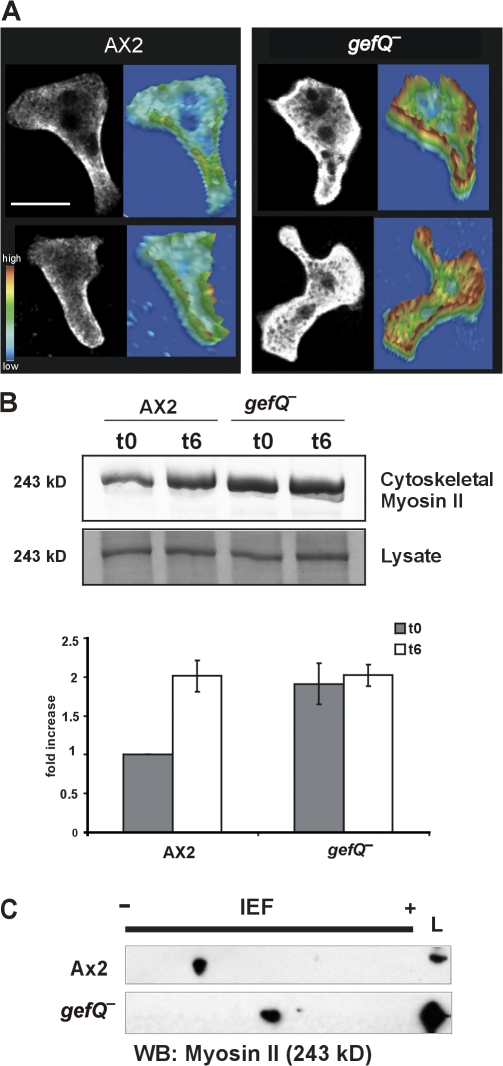
In chemotaxing cells, myosin II typically localizes to the posterior cortical regions of polarized cells, where it is required for retraction of the cell body and suppression of lateral pseudopods. When we stained gefQ− cells for myosin, we observed an increased myosin staining in the cortex that was not restricted to the rear and also an elevated myosin level throughout the cytoplasm (Fig. 7 A). To test this further, we measured the amounts of myosin II present in preparations of cytoskeletal ghosts of gefQ− and AX2 cells both from growing and aggregation-competent stages. In this assay, the amount of myosin II recovered reflects the amount of filamentous myosin associated with the actin cytoskeleton in the living cell. Myosin II filament assembly and disassembly is an extremely dynamic process and, in D. discoideum, it depends largely on the levels of myosin heavy chain phosphorylation. Also, the level of myosin II in the cytoskeletal fractions in AX2 cells is significantly higher in aggregation-competent cells than in growing cells. In contrast, gefQ− cells had a high level of myosin II in the cytoskeletal fractions obtained from growing cells, which was comparable to that in aggregating AX2 cells, and there was no significant rise upon reaching aggregation competence (Fig. 7 B, top). Total myosin II levels in both AX2 and gefQ− cells were comparable in vegetative and aggregation-competent cells (Fig. 7 B, bottom).
Figure 7.
RasGEF Q regulation of myosin II assembly. (A) Aggregation-competent AX2 and gefQ− cells were fixed and stained for myosin II. Images were taken with a confocal microscope. Every confocal section is accompanied by a pseudo-3D projection, in which the z axis represents the intensity of myosin II staining over the scanned area. Bar, 5 μm. (B) Myosin II levels in cytoskeletal ghosts from cells in vegetative and aggregation-competent stages. The graph (below) represents a mean of four independent experiments. The error bars indicate SD. (C) Lysates from vegetative wild-type AX2 and gefQ− cells were subjected to 2D SDS-PAGE analysis for determining the phosphorylation status of myosin II.
The cause for higher levels of filamentous myosin II in gefQ− cells could be because of higher levels of unphosphorylated myosin II that can spontaneously form filaments. To test this hypothesis, we determined the phosphorylation status of myosin II in vegetative AX2 and gefQ− cells. We used 2D gel electrophoresis to examine the changes in the isoelectric point (pI) of phosphorylated versus unphosphorylated myosin II. We observed myosin II as a distinct spot in a Western blot using myosin II antibodies. We found that in gefQ− cells, myosin II had a higher pI than myosin II in AX2 cells, implying that in gefQ− cells, myosin was predominantly in the unphosphorylated state (Fig. 7 C). From these results, we conclude that the higher levels of unphosphorylated myosin II in gefQ− cells lead to higher levels of filamentous myosin II and involve MHCKs in the process.
RasGEF Q regulates Myosin II through MHCK A
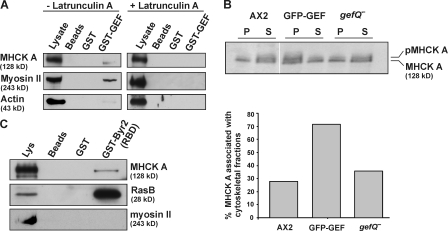
The presence of higher amounts of unphosphorylated myosin II in gefQ− cells indicated that probably myosin phosphorylation is affected. MHCK A is a major kinase regulating myosin II phosphorylation. In GST pulldown experiments, we found that GST-GEF could coprecipitate myosin II and MHCK A from cell lysates under conditions that dissociate actin–myosin complexes by the addition of 5 mM ATP. Actin was absent from the precipitate. When cell lysates were preincubated with LatA to disrupt the cytoskeleton, the association with myosin II and MHCK A was totally abolished (Fig. 8 A). These results indicate that an intact cytoskeleton is required for bringing a signaling complex together, where RasGEF Q can bind to MHCK A and myosin II in an F-actin–dependent manner.
Figure 8.
RasGEF Q regulates myosin II through MHCK A. (A) GST-GEF can associate with myosin II and MHCK A in a manner that is sensitive to Lat A treatment. (B) Cytoskeletal fractions from wild-type AX2, AX2 cells overexpressing GFP-GEF, and gefQ− cells were prepared as described in Materials and methods. Supernatant and pellet fractions were resolved by SDS-PAGE and immunoblotted using MHCK A–specific monoclonal antibody. The graph below represents the mean of two independent experiments and shows the amount of MHCK A present in the pellet fractions as the percentage of the total MHCK A content. (C) Presence of MHCK A in fractions containing active RasB. Active RasB was pulled down from GFP-GEF (6 h) cells using GST-Byr2(RBD). The precipitates were tested for the presence of MHCK A and myosin II heavy chain.
We then examined MHCK A in gefQ− cells and in cells overexpressing GFP-GEF. Under normal conditions, MHCK A is localized throughout the cytosol. Upon chemoattractant stimulation, MHCK A associates with the cytoskeleton and localizes to the cell cortex (Steimle et al., 2001). Association with the actin cytoskeleton causes a drastic increase in the autophosphorylation activity of MHCK A, which is then activated (Egelhoff et al., 2005). Using a monoclonal antibody that detects both the phosphorylated (∼145 kD) and unphosphorylated (130 kD) forms of MHCK A (Steimle et al., 2001), we found that gefQ− cells and cells overexpressing GFP-GEF have both unphosphorylated and autophosphorylated MHCK A. We then analyzed the amounts of MHCK A associated with cytoskeletal fractions, as the cytoskeleton-associated protein represents the active autophosphorylated form of MHCK A. We found that gefQ− cells and AX2 cells have similar amounts of MHCK A associated with the cytoskeletal fractions. In contrast, cells overexpressing the GEF domain had at least twofold higher levels of MHCK A associated with the cytoskeletal fractions (Fig. 8 B).
To investigate if activated Ras could transduce signals for regulation of myosin II, we used the GST-Byr2(RBD) to pull down active Ras from GFP-GEF cells at 6 h and found MHCK A in pulldown eluates with active RasB (Fig. 8 C). Pulldown eluates did not contain myosin II. Thus, MHCK A, which directly regulates myosin II phosphorylation, may either bind directly to active Ras or associate with complexes containing active Ras, suggesting that it might be regulated by Ras upon activation.
Discussion
RasGEF Q in regulation of myosin II functions
Our studies on RasGEF Q reveal that it is required for regulation of myosin II–based cellular functions, like cytokinesis and cell shape during chemotaxis, and also affects late developmental decisions. We provide evidence for RasB being a substrate for RasGEF Q and propose that regulation of myosin II functions by RasGEF Q occurs predominantly through RasB. Cells lacking RasGEF Q show myosin II overassembly associated with the inability to suppress lateral pseudopods, which is similar to that observed in 3XALA myosin II mutants. We show that the myosin overassembly is caused by higher levels of unphosphorylated myosin II in these cells, so the defect in myosin phosphorylation might arise from an inability to activate MHCKs. Cells that overexpress the GEF domain of RasGEF Q have a cytokinesis defect in suspension, and these cells also have high levels of constitutively activated RasB. We identified RasB as the substrate for RasGEF Q through in vivo and in vitro binding assays and found that RasB is activated upon cAMP, which indicates an involvement of cAMP and cAMP receptors (seven transmembrane G protein–coupled receptors [GPCRs]) in the process of activation.
We note that RasGEF Q associates with F-actin. The C-terminal region containing the catalytic GEF domain is a possible region involved in F-actin association. The cortical localization of both GFP-RasGEF Q173 and GFP-GEF are sensitive to Latrunculin A, indicating that an intact cytoskeleton is required for localization of RasGEF Q to the cortex. We also note that RasGEF Q can associate with myosin II and MHCK A, which is also sensitive to Latrunculin A. These results indicate that the F-actin cytoskeleton acts as a scaffold to recruit the signaling complex and that when the F-actin network is disassembled the signaling components do not meet. Myosin II is an F-actin cross-linking protein and, upon a cAMP stimulus, MHCK A localizes to the cell cortex with the help of F-actin. At this point when all the signaling components are assembled, it is critical for RasGEF Q to be in its active state to elicit downstream effects. Signaling components like RasGEFs are active only temporarily, the activation has to be appropriately controlled, and the downstream signals are amplified by orders of magnitude. It is likely that RasGEF Q is active for a very short time after a cAMP stimulus only.
We ruled out the possibility that RasGEF Q might activate RasG, another Ras isoform. Cells lacking RasG show a cytokinesis defect (Tuxworth et al., 1997), whereas in cells that overexpress the GEF domain of RasGEF Q resulting in constitutively high levels of active Ras, we see a cytokinesis defect and, thus, activation of RasG might be independent of RasGEF Q. We also provide a function for the less well understood function of the DEP domain in RasGEF Q, as our results point to the DEP domain acting as an autoregulatory domain of RasGEF Q. Under vegetative conditions when RasB is not activated, the DEP domain can bind to the GEF domain and RasGEF Q is in an inactive conformation. Upon starvation, GPCRs can induce a conformational change upon cAMP binding whereby the DEP domain inhibition is released and, thus, GPCRs activate RasGEF Q, which activates RasB.
We observed that myosin in gefQ− cells has a higher pI, possibly because of higher levels of unphosphorylated myosin II. A higher level of unphosphorylated myosin II is the cause of myosin overassembly in the cortex in gefQ− cells. This also implies a role of MHCK in the process. In D. discoideum, the most immediate regulators of myosin II are MHCKs. D. discoideum has four MHCKs (A–D), among which MHCK A has been most extensively studied. MhckA− cells exhibit a partial, but significant, level of myosin II overassembly because of higher levels of unphosphorylated myosin II. Overexpression of MHCK A elicits defects comparable to those observed in myosin II–null cells, namely a blocked cytokinesis in suspension and arrested development in the mound stage (Kolman et al., 1996). In chemotaxing cells, MHCK A relocalizes to the actin rich cortex in the anterior of the cell, where it presumably functions to phosphorylate and disassemble myosin II at the leading edge (Steimle et al., 2001).
The GEF domain of RasGEF Q has the ability to coprecipitate MHCK A and myosin II in an F-actin–dependent manner. In accordance with the fact that RasGEF Q regulates myosin II functions by regulating MHCK A, we observed that cells overexpressing the GEF domain have a twofold higher level of MHCK A associated with cytoskeletal fractions. This is the result of MHCK A activation by autophosphorylation, whereupon it associates with the actin cytoskeleton in aggregation-competent cells that are stimulated with cAMP (Steimle et al., 2001). Thus, overexpression of the GEF domain causes larger amounts of MHCK A to be activated. In contrast, MHCK A distribution in cytoskeletal fractions is normal in gefQ− cells, indicating that RasGEF Q plays a role in facilitating MHCK A recruitment to the cytoskeleton and that other (probably passive) processes are important for returning the kinase to the cytosol. Furthermore, the C terminus of RasGEF Q, which harbours the GEF domain, has the potential to bind to F-actin directly in F-actin cosedimentation assays. These results also suggest that the actin cytoskeleton may serve as a scaffold to recruit a signaling complex at the leading edge. RasB, which is activated by RasGEF Q, may promote myosin II phosphorylation by regulating the activity of MHCK A (Fig. 9), which leads to phosphorylation of myosin II followed by elimination of filamentous myosin II from the leading edge. In a mutant situation where RasGEF Q is absent, RasB would not be active and MHCK A activation would be limited and would result in higher levels of unphosphorylated myosin and, thus, myosin II overassembly in the cortex in these cells. In a situation where we overexpress the catalytic GEF domain of RasGEF Q, we see constitutively high levels of activated RasB, which activates MHCK A, leading to higher levels of phosphorylated myosin II. Because phosphorylated myosin II cannot assemble into filaments that are required to provide force during cytokinesis, these cells fail to divide in suspension. Such a system may be very useful for D. discoideum and the expansion of the functional roles for Ras proteins may compensate for the lack of Rho GTPases or its effector molecule ROCK, which regulate myosin II functions in higher eukaryotes (Somlyo and Somlyo, 2000)
Figure 9.
Model for a role of RasGEF Q in regulating Myosin II functions. For details see Discussion.
RasGEF Q in D. discoideum development
The process of D. discoideum development is an example of programmed multicellularization (Chisholm and Firtel, 2004), where cells that form and organize the multicellular organism undergo continuous coordinated changes in shape and motility that require the cytoskeleton. D. discoideum tip formation is an event that can serve as a model to understand similar developmental processes in other organisms. Normally, only one tip arises from a mound and this apical tip serves as an organizer to control morphogenesis of the organism by acting as a cAMP oscillator from which waves are initiated and propagated posteriorly (Siegert and Weijer, 1992; Siegert and Weijer, 1995). Mounds from gefQ− cells give rise to supernumerary tips and, as a result, smaller fruiting bodies are formed. Besides gefQ− cells, mutations in other components of signal transduction cascades lead to similar defects. A defect in Scar, a WASP-related protein identified in a mutant screen which suppresses phenotypes of cAR2− cells, also leads to a multiple-tipped phenotype (Bear et al., 1998) as well as overexpression of an activated RasDG12T (Reymond et al., 1986). Similarly, a double knockout of two isoforms of phosphatidylinositol-3-kinase (PI3K1−/2−) and overexpression of the phosphotyrosine phosphatase PTP1 also resulted in the multiple-tip phenotype (Howard et al., 1992; Zhou et al., 1995).
Multiple tips arising from gefQ− mounds may result from the formation of more than one oscillator (or embryogenic organizer). It is also possible that a single embryogenic organizer subdivides to form multiple organizers, causing the multiple-tip phenotype. The tip of the slug is also critical in sensing light and phototactic migration of the slugs. gefQ− mounds gave rise to slugs, but when analyzed for their ability to migrate in phototaxis assays they showed severe impairment. In contrast, slugs from cells overexpressing GFP-GEF formed migratory slugs but their orientation toward the light source was severely impaired. Several proteins in Ras signaling have been implicated in phototaxis, with rasD− and gefE− cells having totally impaired phototaxis (Wilkins et al., 2000; Wilkins et al., 2005).
Our data also suggests that a RasGEF Q–mediated pathway is required for proper spatial patterning in the slug. In chimeric experiments with gefQ− cells and wild-type cells, gefQ− cells are predominant in the posterior prespore region of the slug. Absence of RasGEF Q may cause defects in proper directional movement of cells within the slug necessary to maintain proper spatial patterning. The cell-autonomous nature of this defect in gefQ− mutants may suggest a role of RasGEF Q in processing external signals. Similar patterns of mutant cell distributions in chimeras have been reported for cells lacking the GPCR CrlA, G protein Gα5, or ERK1 protein kinase, suggesting that these components might function in the same or related pathways (Gaskins et al., 1996; Natarajan et al., 2000; Raisley et al., 2004). The findings that myosin heavy chain and myosin light chain are required for cell patterning during D. discoideum development may also be relevant for RasGEF Q regulating myosin II (Eliott et al., 1993; Springer et al., 1994; Chen et al., 1998). Cells lacking myosin heavy chain can aggregate but fail to proceed beyond the mound stage of development. The regulatory light chain of myosin has also been thought to be required for directional sorting of prestalk EcmAO cells at the tip of a developing mound (Clow et al., 2000). Although RasGEF Q transcripts are present throughout development, a protein of higher molecular mass is present in the first 6 h of development and a smaller isoform generated by an alternative promoter is present in low amounts during later stages of development. This smaller form of the protein might have functions in regulating processes in late development.
Materials and methods
Cell culture and development
D. discoideum cells of strain AX2 were grown either with Klebsiella aerogenes on SM agar plates or axenically in liquid nutrient medium (Claviez et al., 1982) in shaking suspension at 160 rpm at 21°C. gefQ− cells were cultivated in nutrient medium containing 7 μg/ml G418 (Invitrogen). To analyze development, cells were grown axenically to a density of 2–3 × 106/ml, washed twice in Soerensen phosphate buffer (17 mM NaK phosphate, pH 6), and 5 × 107 cells were plated on phosphate agar plates.
For phototaxis assay, 10 μl of cells at a density of 108 cells/ml were transferred to the center of a 90-mm phosphate agar plate and placed in a black opaque box with a slit to provide a unidirectional light source and cells were allowed to develop to the migratory slug stage. Slime trails and cellular material were transferred onto a nitrocellulose membrane. Membranes were stained with staining solution (0.1% amido black in 20% isopropanol and 10% acetic acid) for 10 min and destained twice with destaining solution (20% isopropanol and 10% acetic acid) for 15 min, washed with water, and air dried.
For development in chimera, gefQ− cells and wild-type cells were transfected with pBsr-GFP for expression of GFP to mark the different cell types. 90% wild-type cells was mixed with 10% gefQ− cells expressing GFP or 90% gefQ− cells was mixed with 10% GFP-expressing wild-type cells and allowed to codevelop. For control 10% GFP-expressing wild-type cells was mixed with 90% wild-type cells. Images were taken at the slug stage using a fluorescent microscope (DMR; Leica).
Generation of gefQ− cells and cells expressing RasGEF Q domains
For generation of the knockout vector, cDNA clone SLH890 (procured from the D. discoideum cDNA project, Tsukuba, Japan) encompassing nt 1798–3987 of gefQ cloned in pSPORT (Invitrogen) was used. The plasmid was digested with BsaBI to release a 55-bp fragment and the neomycin resistance cassette (Witke et al., 1987) was inserted by blunt end ligation. The resulting replacement vector was linearized by digesting with SalI and transformed into AX2 by electroporation. Transformants were selected in nutrient medium containing 7 μg/ml G418. Independent clones were screened for the disruption of the gefQ gene by PCR using genomic DNA, Southern blotting, immunofluorescence, and Western blot analysis. For Southern blot analysis, a probe encompassing nt 690–990 of the cDNA was used.
For expression of RasGEFQ173, a fragment encoding the protein from aa 173–1298 was cloned into pDex79 and expressed as a GFP fusion protein in AX2 cells. For expression of the N-terminal fragment Δ-GEF-GEFQ fused to GFP at its N terminus, a 1.45- kb fragment encoding aa residues 172–654 was cloned into pBsr (Mohrs et al., 2000). For expression of the DEP domain as a GFP-tagged protein fused at the N terminus, a 450-bp fragment encoding residues 683–833 was cloned into pMCS (Weber et al., 1999). The same fragment was recloned into pGEX-4T1 (GE Healthcare) for expression of GST-DEP in bacteria. For expression of the C-terminal domain containing the catalytic GEF domain as a GFP-GEF fusion fused at the N terminus, a 1.2-kb fragment encoding residues 910–1298 was cloned into pMCS. The same fragment was recloned into pGEX-4T1 for expression of GST-GEF in bacteria. A construct lacking the DEP domain (GFP-Δ-DEP-GEFQ) was generated by digesting pDex79/GFP-RasGEFQ173 with NsiI and SalI and ligating it with a PCR-amplified fragment encoding aa 818–1298 preserving the NsiI site. The resulting plasmid, which lacks the DEP domain, was introduced into AX2 cells.
Generation of RasGEF Q monoclonal antibodies
The procedure used to generate RasGEF Q monoclonal antibodies was as described previously (Schleicher et al., 1984). The DEP domain of RasGEF Q (residues 683–833) was used for immunization of four female BALB/c mice (Lingnau et al., 1996). mAbs K-70-187-1 and K-70-102-1 were used in this study. They recognize a protein of 143 kD in whole cell homogenates of wild-type Ax2 cells, which is absent in gefQ− cells.
Actin binding assay
All purified proteins used in the study were clarified by centrifugation at 120,000 g for 60 min at 4°C. D. discoideum actin was isolated as described previously (Haugwitz et al., 1991). Actin sedimentation assays were performed as described previously (Jung et al., 1996). In brief, 5 μM G-actin was polymerized in the presence or absence of 60 μg/∼1 μM GST-GEF or 20 μg/∼1 μM GST by addition of 0.1 vol of 10× polymerization buffer (100 mM imidazol, pH 7.6, 20 mM MgCl2, 10 mM EGTA, 1 M KCl, and 5 mM ATP) for 30 min at room temperature. The final reaction volume was 70 μl. Binding to F-actin was determined by high-speed centrifugation at 120,000 g for 1 h at 4°C. Cross-linking of filaments was analyzed by low-speed centrifugation at 12,000 g for 1 h at 4°C. Equal amounts of pellet and supernatant were resolved by SDS-PAGE (10% acrylamide) and proteins were visualized by Coomassie brilliant blue staining.
In vitro binding assays with GST fusion protein and analysis of cAMP-induced activation of RasB
For interaction with cytoskeletal proteins, 5 × 107 AX2 cells were lysed in lysis buffer (LB; 25 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, and 1 mM DTT, supplemented with protease inhibitors [Sigma-Aldrich] with 5 mM ATP added) with or without a preincubation with 10 μM Lat A (Sigma-Aldrich for 1 h) and incubated with equal amounts of GST-GEF bound to beads for 3 h at 4°C. Beads were washed with wash buffer (25 mM Tris/HCl, pH 7.5, 150 mM NaCl, and 5 mM EDTA) and the pulldown eluates were analyzed in Western blots. Monoclonal antibodies recognizing actin (Simpson et al., 1984) or myosin II (Pagh and Gerisch, 1986) and a polyclonal antibody against MHCK A (provided by T. Egelhoff, Case Western Reserve University, Cleveland, OH; Kolman et al., 1996) were used.
For interaction of GST-GEF with RasB, 5 × 107 AX2 cells were lysed by sonication in LB without Triton X-100 and membrane and nuclear-enriched pellet fraction separated by centrifugation at 100,000 g for 30 min at 4°C. The pellet fraction was resuspended in LB containing 1% Triton X-100. The pulldown reaction was done as described for the interaction of GST-GEF with cytoskeletal proteins. For analysis of binding preference of GST-GEF for GTP- or GDP-bound RasB, cell lysates from 5 × 107 AX2 cells were lysed in 1 ml LB. One 1-ml aliquot was pretreated with 100 μM GDP and 5 mM MgCl2 and another one with 100 μM GTPγS and 5 mM MgCl2 for 1 h, and the pulldown reaction was done as described for the interaction of GST-GEF with cytoskeletal proteins. Eluates were probed with RasB-specific polyclonal antibodies (provided by G. Weeks, University of British Columbia, Vancouver, Canada; Sutherland et al., 2001).
Activation of RasB was assayed by performing pulldown with beads coated with the GST-fused Ras binding domain of S. pombe Byr2 (supplied by G. Praefcke, University of Cologne, Cologne, Germany; Gronwald et al., 2001; Scheffzek et al., 2001). 5 × 107 AX2 and gefQ− cells were harvested at the vegetative (0 h) and aggregation-competent (6 h) stages and GFP-GEF–expressing cells were harvested at the vegetative stage. Cells were lysed in LB and incubated with equal amounts of GST-Byr2–bound beads. Pulldown eluates were immunoblotted and probed with RasB antibody.
cAMP-induced activation of RasB in aggregation-competent AX2 cells was done according to Kae et al. (2004). Cells were stimulated with 500 nM cAMP while shaking and immediately lysed 0, 10, or 60 s after cAMP stimulation. Lysates were incubated with equal amounts of glutathione Sepharose beads coated with GST-Byr2(RBD) to pull down active Ras and probed with RasB antibody.
Preparation of cytoskeletal ghosts
Cytoskeletal fractions were isolated as proteins insoluble in Nonidet P-40 (Chung and Firtel, 1999) or as proteins insoluble in Triton X-100 (Steimle et al., 2001). For isolation of cytoskeletal fractions for analyzing myosin II levels, 107 cells were harvested at either the vegetative or aggregation-competent stage by centrifugation and lysed in NP-40 buffer (50 mM Tris/HCl, pH 7.6, 100 mM NaCl, 10 mM NaF, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 10% glycerol, and 1 mM DTT, supplemented with protease inhibitors [Sigma-Aldrich]). After vortexing, tubes were kept on ice for 10 min followed by 10 min at room temperature. The samples were spun at 100,000 g for 4 min. Supernatants were discarded and pellet fractions washed once with NP-40 buffer. The pellet fractions were dissolved in 2× SDS-PAGE sample buffer (Laemmli, 1970) and proteins resolved by SDS-PAGE (8% acrylamide) and visualized by Coomassie brilliant blue staining. Protein bands were scanned and changes in myosin II content in the cytoskeleton quantified using Image J software (National Institutes of Health). For isolation of cytoskeletal fractions for analyzing MHCK A, 1.5 × 106 cells were lysed in buffer containing 0.1 M MES, pH 6.8, 2.5 mM EGTA, 5 mM MgCl2, 0.5 mM ATP, and0.5% Triton X-100, supplemented with protease inhibitors, briefly vortexed, and then spun at 100,000 g for 1 min. Supernatant fractions were precipitated with acetone. Pellet and supernatant fractions were resolved by SDS-PAGE and immunoblotted using MHCK A–specific monoclonal antibody (Steimle et al., 2001). The level of MHCK A in both the pellet and supernatant fractions was quantified by densitometric analysis of the scanned blot using ImageJ. The percentage of MHCK A in the pellet fraction was determined by dividing the value obtained for the band in the pellet by the total from the bands in the pellet and supernatant. Preparation of cytoskeletal fractions using saponin was done using a buffer containing 50 mM Tris/HCl, pH 7.6, 100 mM NaCl, 10 mM NaF, 1 mM EDTA, 1 mM EGTA, 2% Saponin, 10% glycerol, and 1 mM DTT, supplemented with protease inhibitors (de Curtis and Malanchini, 1997).
Analysis of cell shape and migration
Aggregation-competent AX2 and RasGEF Q− cells were plated onto glass coverslips and allowed to settle for 15 min in Soerensen phosphate buffer and chemotaxis experiments were performed with micropipettes filled with 10−4 M cAMP attached to a micromanipulator system at 22°C. The micropipette tip was carefully moved to touch the surface of the glass coverslip. Images were recorded at intervals of 6 s using an inverse microscope (40× objective; DM-IL; Leica) and a conventional charge-coupled device video camera using OPTIMAS 6.0 software (Optimas Corporation) and analyzed using Dynamic Image Analysis software (DIAS; Soll Technologies; Wessels et al., 1998).
2D SDS-PAGE
2D gel electrophoresis was performed using an adaptation of previously described protocols (Clemen et al., 2005). D. discoideum cells were lysed in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 40 mM Tris, 2% IPG buffer, 2% DTT, 1 mM PMSF, and protease inhibitors (Roche) containing bromophenol blue at room temperature and centrifuged at 16,000 g for 5 min. Samples of supernatants (250 μl) were diluted with rehydration buffer to a volume of 350 μl and applied to the IEF strips (18 cm; pH 3–10; nonlinear) via rehydration technique for 12 h at 50 V. The strips were then focused on the IPGphor system (GE Healthcare) with a current limit of 50 μA per strip at 20°C with the following program: 1 h at 200 V, 1 h at 500 V, 1 h at 1,000 V, gradient to 8,000 V in 1 h, and a final focusing step at 8,000 V for 28 kVh. After IEF, the strips were briefly rinsed with water and prepared for the second dimension by a two-step equilibration and cystine alkylation process. The strips were incubated two times in equilibration buffer (50 mM Tris/HCl, pH 8.8, 6 M urea, 30% vol/vol glycerol, and bromophenol blue) for 12 min, in which 1% (wt/vol) DTT (step one) or 4% (wt/vol) iodoacetamide were added, respectively. Subsequently, the strips were loaded on SDS gels (8% acrylamide) and resolved in the second dimension. Gels were subjected to immunoblotting using myosin II–specific monoclonal antibody mAb 56–396-2 (Pagh and Gerisch, 1986).
Miscellaneous methods
For treatment with drugs, cells were treated with 10 μM Lat A, 20 μM cytochalasin D, or 100 μM blebbistatin in nutrient medium with shaking for 40 min. Cells were fixed with 3% PFA/picric acid and stained for F-actin using TRITC-labeled phalloidin (Sigma-Aldrich) or cold methanol, and actin was detected with mAb act1–7. For staining for myosin II, aggregation-competent cells were allowed to settle on coverslips and were fixed using methanol (−20°C) and stained for myosin II using mAb 56–396-2 (Pagh and Gerisch, 1986) and images were captured using a laser scanning confocal microscope (TCS-SP; Leica). For quantitative analysis, initial scans using AX2 cells were used to optimize scanning parameters and, subsequently, gefQ− cells were scanned. Images were acquired using the accompanying Leica software. This software was also used to generate pseudo-3D projections from 2D images, in which the z axis represents intensity distribution over the scanned area. In all cases, temperature was maintained at 22°C and cells were mounted on coverslips using gelvatol. Monoclonal antibodies recognizing csA (Bertholdt et al., 1985), actin (Simpson et al., 1984), and GFP (Noegel et al., 2004) were used for Western blotting.
Online supplemental material
Fig. S1 shows the scheme for generation of gefQ− cells and verification of mutants by genomic PCR, Southern blotting, Western blotting, and immunofluorescence studies. Fig. S2 shows that gefQ− cells overexpressing GFP-Δ-GEF-GEFQ do not show RasB activation. Videos 1 and 3 show localization of GFP-RasGEF Q173 in AX2 cells and 2 and 4 show the corresponding phase-contrast images, respectively. Video 5 shows localization of GFP-GEF and 6 shows the localization of GFP-DEP in AX2 cells. Video 7 shows development of wild-type AX2 strain and 8 shows development of gefQ− cells. Video 9 shows migration of AX2 cells toward a cAMP source and 10 shows migration of gefQ− cells toward a cAMP source. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200710111/DC1.
Acknowledgments
We thank R. Müller for experimental help, Dr. M. Schleicher for discussion, Dr. T. Egelhoff for polyclonal antibodies against MHCK A, Dr. G. Weeks for RasB antibody and RasBG12T-pVE II construct, and Dr. G. Praefcke for the GST-Byr2(RBD) expression vector.
This work was supported by the Deutsche Forschungsgemeinschaft (NO 113/17-1 to A.A. Noegel and RI 1034/4 to F. Rivero), the Fonds der Chemischen Industrie and the Köln Fortune Program of the Medical Faculty, University of Cologne, and National Institutes of Health (grant 2R15GM066789-02 to P. Steimle).
D. Bakthavatsalam's present address is Department of Biochemistry and Cell Biology, Rice University, Houston, TX 77005.
Abbreviations used in this paper: DIAS, Dynamic Image Analysis software; GEF, guanine nucleotide exchange factor; GPCR, G protein–coupled receptor; LatA, latrunculin A; LB, lysis buffer; MHCK, myosin II heavy chain kinase; pI, isoelectric point; RBD, ras binding domain; RTK, receptor tyrosine kinase.
References
- Bear, J.E., J.F. Rawls, and C.L. Saxe III. 1998. SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 142:1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholdt, G., J. Stadler, S. Bozzaro, B. Fichtner, and G. Gerisch. 1985. Carbohydrate and other epitopes of the contact site A glycoprotein of Dictyostelium discoideum as characterized by monoclonal antibodies. Cell Differ. 16:187–202. [DOI] [PubMed] [Google Scholar]
- Bosgraaf, L., A. Waijer, R. Engel, A.J. Visser, D. Wessels, D. Soll, and P.J. van Haastert. 2005. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J. Cell Sci. 118:1899–1910. [DOI] [PubMed] [Google Scholar]
- Charest, P.G., and R.A. Firtel. 2006. Feedback signaling controls leading-edge formation during chemotaxis. Curr. Opin. Genet. Dev. 16:339–347. [DOI] [PubMed] [Google Scholar]
- Charest, P.G., and R.A. Firtel. 2007. Big roles for small GTPases in the control of directed cell movement. Biochem. J. 401:377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.L., W.A. Wolf, and R.L. Chisholm. 1998. Cell-type-specific rescue of myosin function during Dictyostelium development defines two distinct cell movements required for culmination. Development. 125:3895–3903. [DOI] [PubMed] [Google Scholar]
- Chisholm, R.L., and R.A. Firtel. 2004. Insights into morphogenesis from a simple developmental system. Nat. Rev. Mol. Cell Biol. 5:531–541. [DOI] [PubMed] [Google Scholar]
- Chung, C.Y., and R.A. Firtel. 1999. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 147:559–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claviez, M., K. Pagh, H. Maruta, W. Baltes, P. Fisher, and G. Gerisch. 1982. Electron microscopic mapping of monoclonal antibodies on the tail region of Dictyostelium myosin. EMBO J. 1:1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemen, C.S., D. Fischer, U. Roth, S. Simon, P. Vicart, K. Kato, A.M. Kaminska, M. Vorgerd, L.G. Goldfarb, B. Eymard, et al. 2005. Hsp27-2D-gel electrophoresis is a diagnostic tool to differentiate primary desminopathies from myofibrillar myopathies. FEBS Lett. 579:3777–3782. [DOI] [PubMed] [Google Scholar]
- Clow, P.A., T. Chen, R.L. Chisholm, and J.G. McNally. 2000. Three-dimensional in vivo analysis of Dictyostelium mounds reveals directional sorting of prestalk cells and defines a role for the myosin II regulatory light chain in prestalk cell sorting and tip protrusion. Development. 127:2715–2728. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A. 1991. The role of actin polymerization in cell motility. Annu. Rev. Physiol. 53:585–605. [DOI] [PubMed] [Google Scholar]
- Daniel, J., G.B. Spiegelman, and G. Weeks. 1993. Characterization of a third ras gene, rasB, that is expressed throughout the growth and development of Dictyostelium discoideum. Oncogene. 8:1041–1047. [PubMed] [Google Scholar]
- de Curtis, I., and B. Malanchini. 1997. Integrin-mediated tyrosine phosphorylation and redistribution of paxillin during neuronal adhesion. Exp. Cell Res. 230:233–243. [DOI] [PubMed] [Google Scholar]
- De Lozanne, A., and J.A. Spudich. 1987. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 236:1086–1091. [DOI] [PubMed] [Google Scholar]
- Egelhoff, T.T., R.J. Lee, and J.A. Spudich. 1993. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 75:363–371. [DOI] [PubMed] [Google Scholar]
- Egelhoff, T.T., T.V. Naismith, and F.V. Brozovich. 1996. Myosin-based cortical tension in Dictyostelium resolved into heavy and light chain-regulated components. J. Muscle Res. Cell Motil. 17:269–274. [DOI] [PubMed] [Google Scholar]
- Egelhoff, T.T., D. Croft, and P.A. Steimle. 2005. Actin activation of myosin heavy chain kinase A in Dictyostelium: a biochemical mechanism for the spatial regulation of myosin II filament disassembly. J. Biol. Chem. 280:2879–2887. [DOI] [PubMed] [Google Scholar]
- Eichinger, L., J.A. Pachebat, G. Glockner, M.A. Rajandream, R. Sucgang, M. Berriman, J. Song, R. Olsen, K. Szafranski, Q. Xu, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature. 435:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliott, S., G.H. Joss, A. Spudich, and K.L. Williams. 1993. Patterns in Dictyostelium discoideum: the role of myosin II in the transition from the unicellular to the multicellular phase. J. Cell Sci. 104:457–466. [DOI] [PubMed] [Google Scholar]
- Gaskins, C., A.M. Clark, L. Aubry, J.E. Segall, and R.A. Firtel. 1996. The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes Dev. 10:118–128. [DOI] [PubMed] [Google Scholar]
- Gronwald, W., F. Huber, P. Grunewald, M. Sporner, S. Wohlgemuth, C. Herrmann, and H.R. Kalbitzer. 2001. Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe. Structure. 9:1029–1041. [DOI] [PubMed] [Google Scholar]
- Haugwitz, M., A.A. Noegel, D. Rieger, F. Lottspeich, and M. Schleicher. 1991. Dictyostelium discoideum contains two profilin isoforms that differ in structure and function. J. Cell Sci. 100:481–489. [DOI] [PubMed] [Google Scholar]
- Heid, P.J., D. Wessels, K.J. Daniels, D.P. Gibson, H. Zhang, E. Voss, and D.R. Soll. 2004. The role of myosin heavy chain phosphorylation in Dictyostelium motility, chemotaxis and F-actin localization. J. Cell Sci. 117:4819–4835. [DOI] [PubMed] [Google Scholar]
- Howard, P.K., B.M. Sefton, and R.A. Firtel. 1992. Analysis of a spatially regulated phosphotyrosine phosphatase identifies tyrosine phosphorylation as a key regulatory pathway in Dictyostelium. Cell. 71:637–647. [DOI] [PubMed] [Google Scholar]
- Insall, R.H., J. Borleis, and P.N. Devreotes. 1996. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr. Biol. 6:719–729. [DOI] [PubMed] [Google Scholar]
- Jung, E., P. Fucini, M. Stewart, A.A. Noegel, and M. Schleicher. 1996. Linking microfilaments to intracellular membranes: the actin-binding and vesicle-associated protein comitin exhibits a mannose-specific lectin activity. EMBO J. 15:1238–1246. [PMC free article] [PubMed] [Google Scholar]
- Kae, H., C.J. Lim, G.B. Spiegelman, and G. Weeks. 2004. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 5:602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kae, H., A. Kortholt, H. Rehmann, R.H. Insall, P.J. Van Haastert, G.B. Spiegelman, and G. Weeks. 2007. Cyclic AMP signalling in Dictyostelium: G-proteins activate separate Ras pathways using specific RasGEFs. EMBO Rep. 8:477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolman, M.F., L.M. Futey, and T.T. Egelhoff. 1996. Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J. Cell Biol. 132:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt, A., H. Rehmann, H. Kae, L. Bosgraaf, I. Keizer-Gunnink, G. Weeks, A. Wittinghofer, and P.J. Van Haastert. 2006. Characterization of the GbpD-activated Rap1 pathway regulating adhesion and cell polarity in Dictyostelium discoideum. J. Biol. Chem. 281:23367–23376. [DOI] [PubMed] [Google Scholar]
- Kovacs, M., J. Toth, C. Hetenyi, A. Malnasi-Csizmadia, and J.R. Sellers. 2004. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279:35557–35563. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lingnau, A., T. Chakraborty, K. Niebuhr, E. Domann, and J. Wehland. 1996. Identification and purification of novel internalin-related proteins in Listeria monocytogenes and Listeria ivanovii. Infect. Immun. 64:1002–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck-Vielmetter, D., M. Schleicher, B. Grabatin, J. Wippler, and G. Gerisch. 1990. Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. FEBS Lett. 269:239–243. [DOI] [PubMed] [Google Scholar]
- Matsumura, F. 2005. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 15:371–377. [DOI] [PubMed] [Google Scholar]
- Mohrs, M.R., K.P. Janssen, T. Kreis, A.A. Noegel, and M. Schleicher. 2000. Cloning and characterization of beta-COP from Dictyostelium discoideum. Eur. J. Cell Biol. 79:350–357. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., C.A. Ashley, and J.A. Hadwiger. 2000. Related Galpha subunits play opposing roles during Dictyostelium development. Differentiation. 66:136–146. [DOI] [PubMed] [Google Scholar]
- Neujahr, R., C. Heizer, and G. Gerisch. 1997. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J. Cell Sci. 110:123–137. [DOI] [PubMed] [Google Scholar]
- Noegel, A.A., R. Blau-Wasser, H. Sultana, R. Muller, L. Israel, M. Schleicher, H. Patel, and C.J. Weijer. 2004. The cyclase-associated protein CAP as regulator of cell polarity and cAMP signaling in Dictyostelium. Mol. Biol. Cell. 15:934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagh, K., and G. Gerisch. 1986. Monoclonal antibodies binding to the tail of Dictyostelium discoideum myosin: their effects on antiparallel and parallel assembly and actin-activated ATPase activity. J. Cell Biol. 103:1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassler, J., S. Stocker, G. Marriott, M. Heidecker, J. Kellermann, and G. Gerisch. 1997. Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin-myosin complexes. Mol. Biol. Cell. 8:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisley, B., M. Zhang, D. Hereld, and J.A. Hadwiger. 2004. A cAMP receptor-like G protein-coupled receptor with roles in growth regulation and development. Dev. Biol. 265:433–445. [DOI] [PubMed] [Google Scholar]
- Reymond, C.D., R.H. Gomer, W. Nellen, A. Theibert, P. Devreotes, and R.A. Firtel. 1986. Phenotypic changes induced by a mutated ras gene during the development of Dictyostelium transformants. Nature. 323:340–343. [DOI] [PubMed] [Google Scholar]
- Sasaki, A.T., and R.A. Firtel. 2005. Finding the way: directional sensing and cell polarization through Ras signalling. Novartis Found. Symp. 269:73–87. [PubMed] [Google Scholar]
- Sasaki, A.T., and R.A. Firtel. 2006. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur. J. Cell Biol. 85:873–895. [DOI] [PubMed] [Google Scholar]
- Scheffzek, K., P. Grunewald, S. Wohlgemuth, W. Kabsch, H. Tu, M. Wigler, A. Wittinghofer, and C. Herrmann. 2001. The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast. Structure. 9:1043–1050. [DOI] [PubMed] [Google Scholar]
- Schleicher, M., G. Gerisch, and G. Isenberg. 1984. New actin-binding proteins from Dictyostelium discoideum. EMBO J. 3:2095–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert, F., and C.J. Weijer. 1992. Three-dimensional scroll waves organize Dictyostelium slugs. Proc. Natl. Acad. Sci. USA. 89:6433–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert, F., and C.J. Weijer. 1995. Spiral and concentric waves organize multicellular Dictyostelium mounds. Curr. Biol. 5:937–943. [DOI] [PubMed] [Google Scholar]
- Simpson, P.A., J.A. Spudich, and P. Parham. 1984. Monoclonal antibodies prepared against Dictyostelium actin: characterization and interactions with actin. J. Cell Biol. 99:287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlyo, A.P., and A.V. Somlyo. 2000. Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J. Physiol. 522:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, M.L., B. Patterson, and J.A. Spudich. 1994. Stage-specific requirement for myosin II during Dictyostelium development. Development. 120:2651–2660. [DOI] [PubMed] [Google Scholar]
- Steimle, P.A., S. Yumura, G.P. Cote, Q.G. Medley, M.V. Polyakov, B. Leppert, and T.T. Egelhoff. 2001. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr. Biol. 11:708–713. [DOI] [PubMed] [Google Scholar]
- Stites, J., D. Wessels, A. Uhl, T. Egelhoff, D. Shutt, and D.R. Soll. 1998. Phosphorylation of the Dictyostelium myosin II heavy chain is necessary for maintaining cellular polarity and suppressing turning during chemotaxis. Cell Motil. Cytoskeleton. 39:31–51. [DOI] [PubMed] [Google Scholar]
- Sutherland, B.W., G.B. Spiegelman, and G. Weeks. 2001. A Ras subfamily GTPase shows cell cycle-dependent nuclear localization. EMBO Rep. 2:1024–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth, R.I., J.L. Cheetham, L.M. Machesky, G.B. Spiegelmann, G. Weeks, and R.H. Insall. 1997. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138:605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt, J.P., C. Lyons, and G.P. Cote. 1988. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J. Biol. Chem. 263:10082–10087. [PubMed] [Google Scholar]
- Weber, I., G. Gerisch, C. Heizer, J. Murphy, K. Badelt, A. Stock, J.M. Schwartz, and J. Faix. 1999. Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J. 18:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, D., and D.R. Soll. 1990. Myosin II heavy chain null mutant of Dictyostelium exhibits defective intracellular particle movement. J. Cell Biol. 111:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, D., D.R. Soll, D. Knecht, W.F. Loomis, A. De Lozanne, and J. Spudich. 1988. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev. Biol. 128:164–177. [DOI] [PubMed] [Google Scholar]
- Wessels, D., E. Voss, N. Von Bergen, R. Burns, J. Stites, and D.R. Soll. 1998. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil. Cytoskeleton. 41:225–246. [DOI] [PubMed] [Google Scholar]
- Wilkins, A., M. Khosla, D.J. Fraser, G.B. Spiegelman, P.R. Fisher, G. Weeks, and R.H. Insall. 2000. Dictyostelium RasD is required for normal phototaxis, but not differentiation. Genes Dev. 14:1407–1413. [PMC free article] [PubMed] [Google Scholar]
- Wilkins, A., K. Szafranski, D.J. Fraser, D. Bakthavatsalam, R. Muller, P.R. Fisher, G. Glockner, L. Eichinger, A.A. Noegel, and R.H. Insall. 2005. The Dictyostelium genome encodes numerous RasGEFs with multiple biological roles. Genome Biol. 6:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke, W., W. Nellen, and A. Noegel. 1987. Homologous recombination in the Dictyostelium alpha-actinin gene leads to an altered mRNA and lack of the protein. EMBO J. 6:4143–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, K., K. Takegawa, S.D. Emr, and R.A. Firtel. 1995. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologs during growth and development. Mol. Cell. Biol. 15:5645–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]