Abstract
The importance of the DNA damage response (DDR) pathway in development, genomic stability, and tumor suppression is well recognized. Although 53BP1 and MDC1 have been recently identified as critical upstream mediators in the cellular response to DNA double-strand breaks, their relative hierarchy in the ataxia telangiectasia mutated (ATM) signaling cascade remains controversial. To investigate the divergent and potentially overlapping functions of MDC1 and 53BP1 in the ATM response pathway, we generated mice deficient for both genes. Unexpectedly, the loss of both MDC1 and 53BP1 neither significantly increases the severity of defects in DDR nor increases tumor incidence compared with the loss of MDC1 alone. We additionally show that MDC1 regulates 53BP1 foci formation and phosphorylation in response to DNA damage. These results suggest that MDC1 functions as an upstream regulator of 53BP1 in the DDR pathway and in tumor suppression.
Introduction
To ensure the meticulous transmission of genetic information, there exists a series of checkpoints that are activated when DNA damage is detected. At the apex of the signaling cascade initiated after the detection of DNA double-strand breaks is the serine/threonine protein kinase ataxia telangiectasia mutated (ATM; Abraham, 2001). ATM has been shown to phosphorylate a broad range of substrates upon activation, including NBS1, BRCA1, and Chk2. ATM patients show symptoms including neural degeneration, immunodeficiency, growth retardation, premature aging, cancer predisposition, and severe sensitivity to ionizing radiation (IR; Shiloh, 2003), with ATM-deficient mice displaying many of these phenotypes (Xu and Baltimore, 1996; Xu et al., 1996). Thus, understanding the ATM signaling cascade has been pivotal to discovering the mechanisms underlying genomic instability and tumorigenesis. Recently, 53BP1 and MDC1 have been recognized as critical upstream mediators in the cellular response to double-strand breaks, although the extent and nature of their interaction is not known.
53BP1 has been suggested to be the mammalian orthologue of scRad9/spCrb2, a protein critical for DNA damage response (DDR) in yeast. Like many proteins of the DDR pathways, 53BP1 contains tandem BRCA1 C-terminal (BRCT) domains. In addition, 53BP1 contains a tudor domain that binds methylated K79 of histone H3 or K20 of histone H4 (Huyen et al., 2004). 53BP1 is phosphorylated after IR in an ATM-dependent fashion and localizes to IR-induced foci (Schultz et al., 2000; Anderson et al., 2001; Rappold et al., 2001). Furthermore, moderate checkpoint defects have been reported in cells depleted of 53BP1 (Fernandez-Capetillo et al., 2002; Wang et al., 2002). Analysis of 53BP1 knockout (KO) mice confirmed an important role for 53BP1 in genomic stability (Morales et al., 2003; Ward et al., 2003), with null mice recapitulating some of the ATM-deficient phenotypes, although they were less severe (notably growth retardation, mild checkpoint defect, genomic instability, and mild tumor incidence). These moderate phenotypes challenge the notion that 53BP1 is the only orthologue of the original scRad9.
Subsequently, MDC1 was identified and described by several groups as being a critical mediator of DDR (Stucki and Jackson, 2004). MDC1 contains tandem BRCT domains as well as a forkhead-associated domain and a repeat region, which also mediate protein interactions. Through its BRCT domains, MDC1 binds γH2AX and recruits activated ATM to the sites of DNA damage, thus amplifying DNA damage signals (Stucki et al., 2005; Lou et al., 2006). siRNA against MDC1 were used to demonstrate MDC1's role in G2/M checkpoint response, IR-induced foci formation, and Chk2 signaling (Stucki and Jackson, 2004). A role of MDC1 in DNA repair has also been demonstrated (Lou et al., 2004; Zhang et al., 2005). However, it was not until MDC1 KO mice were generated that the importance of MDC1's function in ATM signaling and genomic stability became apparent (Lou et al., 2006). MDC1-null mice displayed many of the phenotypes associated with ATM deficiency: growth retardation, IR sensitivity, male infertility, gross genomic instability, and S-phase and G2/M checkpoint defects. Although it remains to be determined whether MDC1 has tumor suppressor functions in vivo, so far the phenotypes observed in MDC1−/− mice are milder than those of ATM−/− mice, suggesting that MDC1, like 53BP1, only regulates a subset of ATM signaling.
The data regarding the interactions between MDC1 and 53BP1 in the ATM signaling cascade have often been conflicting; although both 53BP1 and MDC1 are required for correct and robust DDR, there are some differences between the two proteins. Notably, 53BP1 does not seem to play a major role in checkpoint activation but is crucial in a subset of DNA repair functions, especially class-switch recombination (CSR; Manis et al., 2004; Ward et al., 2004). In contrast, MDC1 seems to have major role in checkpoint activation but a moderate role in DNA repair. Thus, it appears that MDC1 and 53BP1 may regulate separate branches of the DDR downstream of ATM. To clarify these points, we generated mice deficient of both MDC1 and 53BP1 and investigated how the loss of both MDC1 and 53BP1 affects DDR and tumorigenesis.
Results and discussion
MDC1/53BP1 deficiency and radiosensitivity
One of the hallmarks of defective DDR is hypersensitivity to IR. ATM-, MDC1-, and 53BP1-deficient mice have been shown to be hypersensitive to IR (Xu and Baltimore, 1996; Ward et al., 2003; Lou et al., 2006). Mice deficient for both MDC1 and 53BP1 (double KO [DKO]) demonstrated similar overall levels of morbidity to the MDC1−/− and 53BP1−/− mice 12 d after 7 Gy whole body irradiation (Fig. 1 A), all of whose survival were significantly lower than the survival of wild-type (WT) mice (53BP1−/−, P = 0.0164; MDC1−/−, P = 0.0494; DKO, P = 0.0455). DKO survival after IR was distinct from that of the single KOs, as the DKO onset of morbidity was earlier than that of the single KOs, although this did not correspond with a significant decrease in survival (P = 0.4672 for MDC1 KO and P = 0.7968 for 53BP1 KO). Correlating with the whole body IR results, mouse embryonic fibroblasts (MEFs) from MDC1−/− or 53BP1−/− mice show sensitivity to IR (Fig. 1 B) as previously reported (Ward et al., 2003; Lou et al., 2006). MEFs from DKO mice are only slightly more sensitive to IR than those from MDC1−/− or 53BP1−/− mice, although not statistically more so (P = 0.668 for MDC1−/− and P = 0.439 for 53BP1−/−).
Figure 1.
Radiosensitivity of DKO mice and MEFs. (A) Kaplan-Meier survival curve of WT (n = 9), 53BP1−/− (n = 8), MDC1−/− (n = 10), and DKO (n = 8) mice after 7 Gy of whole body IR. (B) Survival of WT, 53BP1−/−, MDC1−/−, and DKO MEFs 5 d after 0, 1, and 2 Gy IR as determined by Trypan blue exclusion. Error bars represent SEM from two independent experiments. (C) Survival of WT, 53BP1−/−, MDC1−/−, and DKO MEFs 48 h after continuous treatment with the indicated amounts of MMC. Error bars represent SEM from four independent experiments.
To further confirm our IR data, we chose to test the sensitivity of these cell lines to the DNA cross-linking agent mitomycin C (MMC). As shown in Fig. 1 C, 53BP1 KO MEFs showed a mild, insignificant resistance to MMC when compared with WT (P = 0.167). MMC-induced DNA damage is believed to be mainly repaired by the homologous recombination (HR) pathway (Brugmans et al., 2007), and loss of 53BP1 has been shown to enhance HR as a result of its suppressive role in HR (Xie et al., 2007), which could contribute to this mild resistance to MMC in 53BP1−/− cells. Inversely, the loss of MDC1, which has been reported to be involved in HR pathways (Zhang et al., 2005; Xie et al., 2007), resulted in mild decreased survival in comparison to WT (P = 0.071). However, the DKO line displayed a significantly higher sensitivity to MMC than WT and 53BP1 lines (P = 0.0031 and P < 0.0001, respectively) but was not significantly more sensitive to MMC than the MDC1 KO line (P = 0.2267). This result suggests that although MDC1 and 53BP1 may have distinct repair functions, the loss of MDC1 is a dominant effect in the context of DKO.
MDC1/53BP1 deficiency and genomic instability
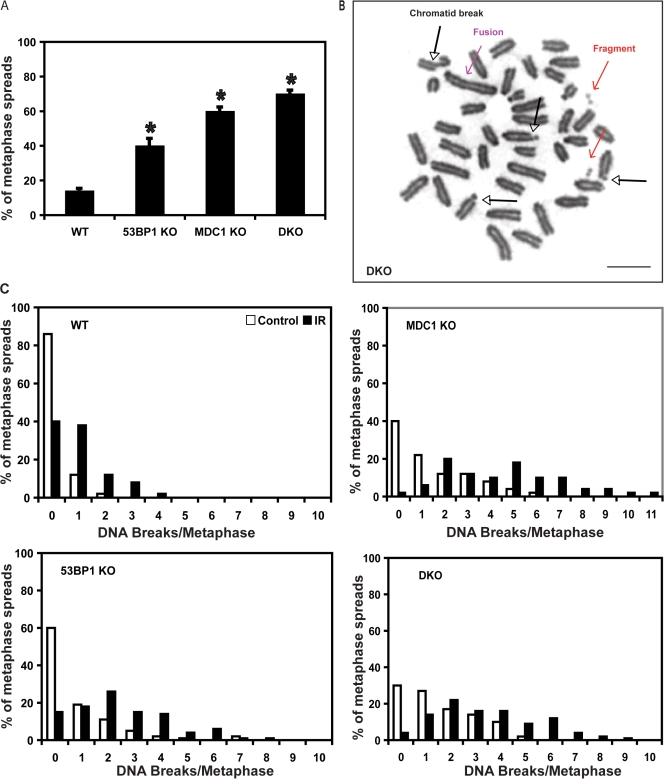
As MEFs from both MDC1−/− and 53BP1−/− mice have been shown to have higher rates of genomic instability (Ward et al., 2003; Lou et al., 2006), we next examined how the loss of both MDC1 and 53BP1 affects chromosome stability before and after IR. Mitotic spreads prepared from single and DKO passage 1 MEFs were examined by light microscopy. Although unirradiated WT MEFs had relatively few spontaneous chromosomal aberrations (14% of MEFs examined), including chromatid breaks, chromosome fusions, and fragments from chromosome breaks, 53BP1−/− and MDC1−/− MEFs had a significantly higher incidence with 40% and 60% of spreads containing chromosome aberrations, respectively (P = 0.00019 and P = 3.35 × 10−6, respectively; Fig. 2 A). The MEFs from DKO mice displayed slightly more chromosome aberrations than those of MDC1−/− MEFs (70%; Fig. 2, A and B), which is not considered a statistically significant increase (P = 0.8) but is highly significant when compared with spontaneous breaks in WT and 53BP1 KO cells (P = 1.01 × 10−11 for WT and P = 0.00016 for 53BP1 KO).
Figure 2.
Genomic instability in 53BP1−/−, MDC1−/−, and DKO MEFs. Mitotic spreads were produced as described in Materials and methods. (A) The percentage of mitotic spreads with spontaneous chromosomal abnormalities was determined by light microscopy. The asterisk denotes a significant difference from WT (53BP1−/− compared with WT, P = 0.00019; MDC1−/− compared with WT, P = 3.35 × 10−6; DKO compared with WT, P = 1.01 × 10−11). (B) Representative pictures of chromosomal aberrations from DKO MEFs. Different aberrations are indicated by colored arrows. (C) Percentage of metaphase spreads with the indicated number of DNA breaks from unirradiated or irradiated MEFs. 100 metaphase spreads/genotype were evaluated. Bar, 10 μM.
We next examined DNA breaks caused by IR. Cells were treated with 1 Gy IR and were allowed to recover for 1 h, and spreads were made and examined for chromosome aberrations. As shown in Fig. 2 C, WT MEFs had zero to four breaks per spread, with zero being the highest frequency at 40% followed by one-break frequencies of 38%. 53BP1−/− cells had between zero and eight breaks per spread, with two and then one break per spread being the most prevalent. MDC1−/− MEFs again appear more sensitive to chromosomal aberrations than WT or 53BP1−/− and had >90% of spreads containing two or more breaks. The DKOs recapitulate the MDC1−/− phenotype more closely with a larger proportion of spreads containing two or more chromosomal abnormalities after IR. The data shown in Figs. 1 and 2 relating to IR sensitivity, DNA repair, and genomic stability suggest that the loss of MDC1 contributes to most of the phenotypes observed in DKO mice and cells.
MDC1/53BP1 deficiency, development, and tumorigenesis
We also examined how the loss of 53BP1 and MDC1 affects development and tumorigenesis. Although ATM−/− mice are infertile (Xu et al., 1996), 53BP1−/− mice show normal fertility (Ward et al., 2003), as MDC1−/− mice show an intermediate phenotype with male MDC1−/− mice being infertile and females showing reduced fertility (Lou et al., 2006). The DKO mice showed the same fertility phenotype as MDC1−/− mice (unpublished data).
We next evaluated immunological phenotypes. ATM−/− mice show defects in both CSR and T cell development (Xu and Baltimore, 1996; Xu et al., 1996). Although 53BP1−/− mice show a severe defect in CSR (Manis et al., 2004; Ward et al., 2004) and a decreased number of mature T cells, overall T cell development appears to be intact (Morales et al., 2003; Ward et al., 2003). Similarly, MDC1−/− mice show a moderate defect in CSR and no apparent abnormalities in T cell development (Lou et al., 2006). To examine whether the loss of both MDC1 and 53BP1 would result in a defect in T cell development, we stained CD4 and CD8 of lymphocytes isolated from the thymus of WT, MDC1−/−, 53BP1−/−, and DKO mice. Like that of MDC1−/− and 53BP1−/− mice, no obvious defect in T cell development was observed in DKO mice (Fig. 3 A). These results suggest that unlike ATM, MDC1 and 53BP1 are not essential for T cell development.
Figure 3.
T cell development, tumor incidence, and spectrum in WT, MDC1−/−, and DKO mice. (A) Lymphocytes from WT, MDC1−/−, 53BP1−/−, and DKO mice. Numbers in each quadrant indicate percentage of the total population. (B and C) Spontaneous tumor incidence and spectrum in WT (n = 19), MDC1−/− (n = 20), and DKO (n = 27) mice for up to 21 mo were determined. The asterisk denotes significant difference from WT (tumor incidence in MDC1−/− mice compared with WT mice, P = 7.6 × 10−6; incidence in DKO mice compared with WT, P = 1.26 × 10−9). (D) Representative pictures of lymphoma and lung cancer from MDC1−/− and DKO mice.
Finally, we examined spontaneous tumor incidence in WT, MDC1−/−, and DKO mice. 10.5% of WT mice developed tumors by the age of 21 mo, whereas 25% of MDC1−/− mice developed tumors (P = 0.0001; Fig. 3 B). The majority of tumors from WT and MDC1−/− mice are lymphomas, which also metastasized to other organs, such as the lung, liver, and gastrointestinal tract (Fig. 3, C and D). These results are the first to demonstrate that MDC1 acts as a tumor suppressor, suggesting that genomic instability caused by the loss of MDC1 does contribute to tumorigenesis. However, the loss of both 53BP1 and MDC1 only resulted in a mild increase in tumor incidence in comparison to the loss of MDC1 alone (P = 0.356; Fig. 3 B).
53BP1 and MDC1 in the DNA damage signaling pathway
A previous study using siRNAs suggests that MDC1 and 53BP1 work in parallel pathways to activate ATM (Mochan et al., 2003). We used MEFs from MDC1−/−, 53BP1−/−, and DKO mice to verify this observation. In contrast to Mochan et al. (2003), we found that neither the absence of MDC1 or 53BP1 alone significantly affected the autophosphorylation at serine 1981 of ATM after IR (Fig. 4 A and Fig. S1, A and B; available at http://www.jcb.org/cgi/content/full/jcb.200801083/DC1) even at low doses of IR. Furthermore, there was no significant decrease in ATM phosphorylation in the DKO cells (Fig. 4 A and Fig. S1, A and B). Interestingly, there was some decrease in ATM phosphorylation at a later time point, supporting a possible role of MDC1 in maintaining ATM phosphorylation (Fig. 4 B). However, the loss of both MDC1 and 53BP1 did not further decrease ATM phosphorylation. These results suggest that MDC1 and 53BP1 are not critical for ATM activation.
Figure 4.
DKO MEFs do not have more severe defects in ATM activation and G2/M checkpoint activation. (A and B) ATM, Chk1, and Chk2 activation in wild-type (WT), MDC1−/−, and double knockout (DKO or MDC1/53BP1−/−) MEFs. MEFs of the indicated genotypes were irradiated at the indicated doses and harvested 1 h later (A) or irradiated (2 Gy) and harvested at different time points (B). Cell extracts were then blotted with the indicated antibodies. (C) MEFs were left untreated or irradiated (2 Gy). 1 h later, cells were stained for antiphospho-H3 antibodies, and mitotic populations were determined by FACS. Error bars represent SEM.
We also examined how the loss of MDC1 and 53BP1 affects downstream signals of the DDR. MDC1 and 53BP1 have previously been shown to regulate Chk1 and Chk2 phosphorylation (Wang et al., 2002, 2003; Peng and Chen, 2003; Stewart et al., 2003; Lou et al., 2006). MDC1−/− and 53BP1−/− MEFs showed weak activation of Chk1 and Chk2. However, the loss of both MDC1 and 53BP1 did not further decrease Chk1 and Chk2 phosphorylation (Fig. 4, A and B; and Fig. S1 B). Although we did observe a reproducible defect in Chk2 phosphorylation, the decreased level of Chk2 seen in MDC1 KO cells was observed in some, but not all, experiments. Finally, we examined checkpoint activation in MDC1−/−, 53BP1−/−, and DKO MEFs. Both MDC1 and 53BP1 have been shown to regulate the G2/M checkpoint. As has been previously reported, the loss of MDC1 resulted in a defective G2/M checkpoint, with more cells able to enter mitosis after IR than in WT cells (Fig. 4 C). Similarly, the loss of 53BP1 also resulted in some inappropriate progression into mitosis. However, in DKO cells, the observed G2/M checkpoint defect is comparable to that seen in MDC1-null cells (Fig. 4 C). These results suggest that the loss of both 53BP1 and MDC1 does not cause more severe checkpoint defects than the loss of MDC1 alone.
MDC1 regulates 53BP1 phosphorylation and localization at the sites of DNA damage
Our results with 53BP1/MDC1 DKO mice and cells suggest that MDC1 and 53BP1 likely function on the same molecular pathway downstream of ATM. Conflicting results have been reported as to whether 53BP1 foci formation requires MDC1, possibly as a result of the different siRNA used and the extent of MDC1 knockdown (Goldberg et al., 2003; Peng and Chen, 2003; Stewart et al., 2003; Bekker-Jensen et al., 2005). We found that 53BP1 failed to form foci in MDC1−/− cells in response to DNA damage (Fig. 5 A). On the other hand, MDC1 foci formation is normal in 53BP1−/− cells. These results suggest that MDC1 is an upstream regulator of 53BP1 localization after DNA damage.
Figure 5.
MDC1 regulates 53BP1 foci formation and phosphorylation. (A–C) MEFs of the indicated genotypes were irradiated (10 Gy), and foci formation of MDC1, 53BP1, RNF8, and γH2AX was determined by immunofluorescence. (D) MDC1+/+ and MDC1−/− MEF cells were irradiated (2 Gy), and, 1 h later, 53BP1 was immunoprecipitated and blotted with the indicated antibodies. (E) Proposed model of the regulation of 53BP1 phosphorylation and localization by MDC1. Bars, 10 μM.
Recent studies suggest that MDC1 might regulate the localization of DNA damage factors through the E3 ubiquitin ligase RNF8, which ubiquitinates H2AX and creates binding sites for downstream factors, such as BRCA1 and 53BP1 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). To confirm these studies, we generated RNF8−/− mice and isolated MEFs from RNF8−/− mice. Loss of RNF8 resulted in the failure of 53BP1 foci formation, whereas MDC1 foci formation remained intact (Fig. 5 B). On the other hand, RNF8 foci formation is dependent on MDC1 (Fig. 5 C), suggesting the existence of an MDC1–RNF8–53BP1 pathway for the accumulation of 53BP1 at or near the sites of DNA damage.
We further investigated whether MDC1 regulates 53BP1 phosphorylation. 53BP1 is phosphorylated at S25/29 by ATM after DNA damage, and the phosphorylation of S25 has been shown to be important for the binding of Pax transactivation domain–interacting protein and proper DDR (Munoz et al., 2007). As shown in Fig. 5 D, 53BP1 phosphorylation at S25/29 is defective in MDC1−/− cells, suggesting that MDC1 also regulates 53BP1 phosphorylation after DNA damage. Collectively, our results suggest that MDC1 and 53BP1 act in the same molecular pathway of the DDR, and MDC1 is an upstream regulator of 53BP1 by regulating 53BP1 phosphorylation and foci formation after DNA damage.
MDC1 and 53BP1 are two important mediator proteins of the DDR pathway. Our observations indicate that the phenotypes of MDC1−/− and DKO mice are substantially milder than that of ATM−/− mice, suggesting that it is unlikely that MDC1 and 53BP1 are critical for ATM activation. Concurrently, our studies of DDR (radiosensitivity, DNA repair, and genomic stability) indicate that the phenotypes of DKO mice and cells mostly resemble those of MDC1 KO mice; this is in contrast to scid/Rad54−/− mice that showed a drastic increase in radiosensitivity compared with scid or Rad54−/− mice (Essers et al., 2000). Although a role in nonhomologous end joining and HR by 53BP1 and MDC1, respectively, has been suggested (Xie et al., 2007), it is possible that 53BP1 and MDC1 only play a supporting role in these processes, which were not obvious in our in vivo studies.
We have shown that MDC1 regulates 53BP1 localization to the sites of DNA damage and 53BP1 phosphorylation at S25/29, which is necessary for the correct phosphorylation of both Chk2 and BRCA1 and subsequent checkpoint activation. Therefore, MDC1 might regulate these signaling events partially through its ability to control 53BP1 phosphorylation. Using MEFs from MDC1−/− and RNF8−/− mice, we show that 53BP1 localization to the sites of DNA damage requires RNF8, which, in turn, requires MDC1. These results support several recent studies demonstrating that MDC1 recruits RNF8 to the sites of DNA damage and RNF8, in turn, ubiquitinates H2AX, H2A, and probably other substrates at the sites of DNA breaks (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). Ubiquitinated H2AX, H2A, or other RNF8 substrates would then recruit BRCA1 and 53BP1. BRCA1 is proposed to bind ubiquitinated RNF8 substrates indirectly through a mediator protein, Rap80, which has an ubiquitin-binding domain (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007). However, Rap80 is not required for 53BP1 foci formation, suggesting that an unidentified factor mediates the binding of ubiquitinated protein and 53BP1. Therefore, we propose an H2AX–MDC1–RNF8 pathway, which is distinct from methylated H3/H4 that also regulates 53BP1 foci formation (Fig. 5 E). It is not clear why cells use two different mechanisms to regulate the localization of 53BP1 and whether these two mechanisms relate to each other. The fact that 53BP1 but not MDC1 is essential for CSR and that 53BP1−/− cells respond differently from MDC1−/− cells to MMC suggest that 53BP1 does have functions independent of MDC1. It is possible that the mode of 53BP1 recruitment is specific to the functions of 53BP1. These issues need to be further clarified in the future.
Our studies on organism development and tumorigenesis further highlight the fact that genomic instability is a driving force of tumorigenesis, supporting the data that many DDR factors function as potent tumor suppressors. 53BP1 has been show to function as a tumor suppressor in mice (Ward et al., 2003; Morales et al., 2006), whereas our data are the first to suggest a role for MDC1 in tumor suppression. A recent study showed the loss of MDC1 expression in breast and lung cancer samples, supporting a potential role of MDC1 in tumor suppression in humans (Bartkova et al., 2007). However, MDC1 mutations have not yet been identified in human cancer samples. We did not find that loss of both MDC1 and 53BP1 significantly increased tumor incidence compared with MDC1 KO mice. Interestingly, the loss of MDC1 and 53BP1 is mostly mutually exclusive in human cancer samples (Bartkova et al., 2007). These results indicate that the loss of either MDC1 or 53BP1 is sufficient for tumorigenesis, and the loss of both does not provide an overt additional advantage for tumor formation. Collectively, our studies clearly establish MDC1 as a tumor suppressor and an upstream regulator of 53BP1 in the DDR pathway.
Materials and methods
Generation of 53BP1/MDC1 DKO mice, RNF8-deficient mice, and MEFs
MDC1+/− female mice (Lou et al., 2006) were bred with 53BP1−/− male mice (Ward et al., 2003) to generate 53BP1+/− MDC1+/− offspring. Double heterozygous females were then bred back to 53BP1−/− males to generate 53BP1−/− MDC1+/− offspring, which were used for all subsequent breeding. Mice were observed daily for signs of poor health, and moribund mice were killed and screened for tumors in accordance with Mayo Foundation Institutional Animal Care and Use Committee guidelines.
For the generation of RNF8-deficient mice, two embryonic stem cell lines, RRR260 and PT238, were purchased from Bay Genomics. In the RRR260 embryonic stem cell line, the RNF8 gene was disrupted by a neo gene selection cassette inserted between transcripted exon 4 to exon 5 of RNF8. In the PT238 cell line, a neo cassette was inserted between transcripted exon 5 to exon 6. The exact insertion sites were mapped by genomic PCR and DNA sequencing. Both embryonic stem cell lines were injected into C57BL/6 blastocysts to generate two independent chimeric mouse lines. The chimeric mice were then crossed back with C57BL/6 mice to obtain RNF8+/− mice, which were used for subsequent breeding to generate two independent RNF8−/− mice lines. Both RNF8−/− mice lines show identical phenotypes. All MEFs were generated from embryonic day 13.5 or 14.5 embryos using standard procedures.
Metaphase spreads
Passage 1 MEFs were left unirradiated or were treated with 1 Gy and then incubated with 50 ng ml−1 colcemid for 3–4 h. Cells were collected, washed with PBS, resuspended in 75 mM KCl, and incubated at room temperature for 15 min. Cells were fixed in Carnoy's solution (75% methanol and 25% acetic acid), and 15-μl aliquots were dropped onto slides and stained with 5% Giemsa solution. Metaphase spreads were observed using a microscope (Eclipse 80i; Nikon) with a 40× NA 1.3 oil objective lens at room temperature. Spreads were photographed and analyzed using a camera (Spot 2 Megasample; Diagnostic Instruments, Inc.) and Spot software 4.6 (Diagnostic Instruments, Inc.).
Immunofluorescence
Cells were washed with PBS, incubated in 3% PFA for 12 min, and permeabilized in 0.5% Triton X-100 solution for 5 min at room temperature. Samples were blocked with 5% goat serum and incubated with primary antibody for 60 min. Samples were washed and incubated with secondary antibody (FITC and rhodamine labeled) for 60 min. Cells were then stained with DAPI to visualize nuclear DNA. The coverslips were mounted onto glass slides with antifade solution and visualized using an Eclipse 80i fluorescence microscope with a 60× NA 1.3 oil objective lens at room temperature. Spreads were photographed and analyzed using a Spot 2 Megasample camera and Photoshop software (Adobe).
Western blotting
Cell lysates were prepared and Western blots were performed using standard protocols. Phospho-ATM antibodies were obtained from Rockland, and Chk1 and phospho-Chk1 antibodies were purchased from Cell Signaling Technology. Anti–mouse ATM antibodies were generously provided by Y. Shiloh (Tel Aviv University, Tel Aviv, Israel).
The G2/M checkpoint assay
Cells were irradiated (2 Gy) and were harvested 1 h later. Cells were then fixed and stained with antiphospho-H3 antibodies (Cell Signaling Technology). Mitotic population (mitotic index) was determined by FACS analysis.
Online supplemental material
Fig. S1 shows that DKO MEFs do not have more severe defects in the ATM-dependent DNA damage signaling pathway. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801083/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Susanna Stevens (Division of Biostatistics, Mayo Clinic, Rochester, MN) for her assistance with statistical analyses and Xiaochun Yu for initiating the RNF8 KO project.
This work was supported, in part, by grants from the National Institutes of Health (to J. Chen). Z. Lou was supported by a Susan G. Komen Breast Cancer Foundation research grant. M.S.Y. Huen is supported by an Anna Fuller postdoctoral fellowship. J. Chen is a recipient of an Era of Hope Scholars Award from the Department of Defense and is a member of the Mayo Clinic Breast Specialized Program of Research Excellence program.
Abbreviations used in this paper: ATM, ataxia telangiectasia mutated; BRCT, BRCA1 C terminal; CSR, class-switch recombination; DDR, DNA damage response; DKO, double KO; HR, homologous recombination; IR, ionizing radiation; KO, knockout; MEF, mouse embryonic fibroblast; MMC, mitomycin C; WT, wild type.
References
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177–2196. [DOI] [PubMed] [Google Scholar]
- Anderson, L., C. Henderson, and Y. Adachi. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21:1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova, J., I.Z. Horej Sbreve, M. Sehested, J.M. Nesland, E. Rajpert-De Meyts, N.E. Skakkebaek, M. Stucki, S. Jackson, J. Lukas, and J. Bartek. 2007. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 26:7414–7422. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen, S., C. Lukas, F. Melander, J. Bartek, and J. Lukas. 2005. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170:201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmans, L., R. Kanaar, and J. Essers. 2007. Analysis of DNA double-strand break repair pathways in mice. Mutat. Res. 614:95–108. [DOI] [PubMed] [Google Scholar]
- Essers, J., H. van Steeg, J. de Wit, S.M. Swagemakers, M. Vermeij, J.H. Hoeijmakers, and R. Kanaar. 2000. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 19:1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O., H.T. Chen, A. Celeste, I. Ward, P.J. Romanienko, J.C. Morales, K. Naka, Z. Xia, R.D. Camerini-Otero, N. Motoyama, et al. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993–997. [DOI] [PubMed] [Google Scholar]
- Goldberg, M., M. Stucki, J. Falck, D. D'Amours, D. Rahman, D. Pappin, J. Bartek, and S.P. Jackson. 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 421:952–956. [DOI] [PubMed] [Google Scholar]
- Huen, M.S., R. Grant, I. Manke, K. Minn, X. Yu, M.B. Yaffe, and J. Chen. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen, Y., O. Zgheib, R.A. Ditullio Jr., V.G. Gorgoulis, P. Zacharatos, T.J. Petty, E.A. Sheston, H.S. Mellert, E.S. Stavridi, and T.D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 432:406–411. [DOI] [PubMed] [Google Scholar]
- Kim, H., J. Chen, and X. Yu. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 316:1202–1205. [DOI] [PubMed] [Google Scholar]
- Kolas, N.K., J.R. Chapman, S. Nakada, J. Ylanko, R. Chahwan, F.D. Sweeney, S. Panier, M. Mendez, J. Wildenhain, T.M. Thomson, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, Z., B.P. Chen, A. Asaithamby, K. Minter-Dykhouse, D.J. Chen, and J. Chen. 2004. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 279:46359–46362. [DOI] [PubMed] [Google Scholar]
- Lou, Z., K. Minter-Dykhouse, S. Franco, M. Gostissa, M.A. Rivera, A. Celeste, J.P. Manis, J. van Deursen, A. Nussenzweig, T.T. Paull, et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 21:187–200. [DOI] [PubMed] [Google Scholar]
- Mailand, N., S. Bekker-Jensen, H. Faustrup, F. Melander, J. Bartek, C. Lukas, and J. Lukas. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900. [DOI] [PubMed] [Google Scholar]
- Manis, J.P., J.C. Morales, Z. Xia, J.L. Kutok, F.W. Alt, and P.B. Carpenter. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487. [DOI] [PubMed] [Google Scholar]
- Mochan, T.A., M. Venere, R.A. DiTullio Jr., and T.D. Halazonetis. 2003. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 63:8586–8591. [PubMed] [Google Scholar]
- Morales, J.C., Z. Xia, T. Lu, M.B. Aldrich, B. Wang, C. Rosales, R.E. Kellems, W.N. Hittelman, S.J. Elledge, and P.B. Carpenter. 2003. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 278:14971–14977. [DOI] [PubMed] [Google Scholar]
- Morales, J.C., S. Franco, M.M. Murphy, C.H. Bassing, K.D. Mills, M.M. Adams, N.C. Walsh, J.P. Manis, G.Z. Rassidakis, F.W. Alt, and P.B. Carpenter. 2006. 53BP1 and p53 synergize to suppress genomic instability and lymphomagenesis. Proc. Natl. Acad. Sci. USA. 103:3310–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz, I.M., P.A. Jowsey, R. Toth, and J. Rouse. 2007. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res. 35:5312–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, A., and P.L. Chen. 2003. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J. Biol. Chem. 278:8873–8876. [DOI] [PubMed] [Google Scholar]
- Rappold, I., K. Iwabuchi, T. Date, and J. Chen. 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, L.B., N.H. Chehab, A. Malikzay, and T.D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 3:155–168. [DOI] [PubMed] [Google Scholar]
- Sobhian, B., G. Shao, D.R. Lilli, A.C. Culhane, L.A. Moreau, B. Xia, D.M. Livingston, and R.A. Greenberg. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 316:1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, G.S., B. Wang, C.R. Bignell, A.M. Taylor, and S.J. Elledge. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 421:961–966. [DOI] [PubMed] [Google Scholar]
- Stucki, M., and S.P. Jackson. 2004. MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair (Amst.). 3:953–957. [DOI] [PubMed] [Google Scholar]
- Stucki, M., J.A. Clapperton, D. Mohammad, M.B. Yaffe, S.J. Smerdon, and S.P. Jackson. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 123:1213–1226. [DOI] [PubMed] [Google Scholar]
- Wang, B., and S.J. Elledge. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA. 104:20759–20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., S. Matsuoka, P.B. Carpenter, and S.J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science. 298:1435–1438. [DOI] [PubMed] [Google Scholar]
- Wang, B., S. Matsuoka, B.A. Ballif, D. Zhang, A. Smogorzewska, S.P. Gygi, and S.J. Elledge. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 316:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, I.M., K. Minn, J. van Deursen, and J. Chen. 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, I.M., B. Reina-San-Martin, A. Olaru, K. Minn, K. Tamada, J.S. Lau, M. Cascalho, L. Chen, A. Nussenzweig, F. Livak, et al. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, A., A. Hartlerode, M. Stucki, S. Odate, N. Puget, A. Kwok, G. Nagaraju, C. Yan, F.W. Alt, J. Chen, et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 28:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., and D. Baltimore. 1996. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 10:2401–2410. [DOI] [PubMed] [Google Scholar]
- Xu, Y., T. Ashley, E.E. Brainerd, R.T. Bronson, M.S. Meyn, and D. Baltimore. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411–2422. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Z. Ma, A. Treszezamsky, and S.N. Powell. 2005. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat. Struct. Mol. Biol. 12:902–909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.