Abstract
Eukaryotic mRNAs are in a dynamic equilibrium between different functional and subcellular locations. Translating mRNAs can be found in polysomes, while mRNAs stalled in translation initiation accumulate in stress granules, and mRNAs targeted for degradation or translation repression can accumulate in P-bodies. Partitioning of mRNAs between polysomes, stress granules, and P-bodies can affect rates of translation and mRNA degradation. Genetic evidence now indicates that critical proteins within P-bodies or stress granules can enhance or limit viral infection. Moreover, some viral RNAs and proteins, as well as host antiviral defense proteins, accumulate in P-bodies and/or stress granules. These results suggest an important interplay between P-bodies, stress granules and viral life cycles that is just beginning to emerge.
INTRODUCTION
Viral life cycle completion requires a series of events that are often separated in time and occur in specific subcellular compartments of the host. One important aspect of viral-host interactions is how the viral transcripts interact with host machinery for translation, localization, and degradation of mRNAs. Moreover, since many RNA viruses use viral transcripts for both mRNA and genomic RNA (gRNA), mechanisms are required to segregate replicative and packaging events away from translation of the RNAs, thereby avoiding competition between elongating ribosomes and the packaging or replicative machineries. Although of significant importance to the viral replicative process, the mechanisms by which viruses segregate translation from replication and assembly, and the host factors involved, are not well understood. However, recent insights into host mechanisms for the control of mRNA degradation and translation suggest possible means by which the activities or functions of viral RNAs may interact with normal host mRNA biology.
In eukaryotes, two general mRNA decay pathways have been identified (reviewed in Meyer et al., 2004; Parker and Song, 2004), both of which begin with shortening of the 3′ poly (A) tail in a process referred to as deadenylation. The Ccr4p/Pop2p deadenylase complex primarily carries out deadenylation, although additional deadenylases can contribute. Following deadenylation, mRNAs can be degraded in a 3′–5′ direction by an exonuclease complex termed the exosome. Alternatively, after deadenylation the mRNA can be decapped by the Dcp1p/Dcp2p decapping enzyme, leading to 5′ to 3′ degradation by the exonuclease Xrn1p. An important point is that targeting an mRNA for decapping involves the formation of a translationally repressed mRNP (mRNA and protein complex), which can also aggregate into specific cytoplasmic foci referred to as Processing bodies or P-bodies (reviewed in Parker and Sheth, 2007; Eulalio et al., 2007; Anderson and Kedersha, 2006). Transcripts that assemble into an mRNP targeted to P-bodies can either be degraded, or stored for later return to translation.
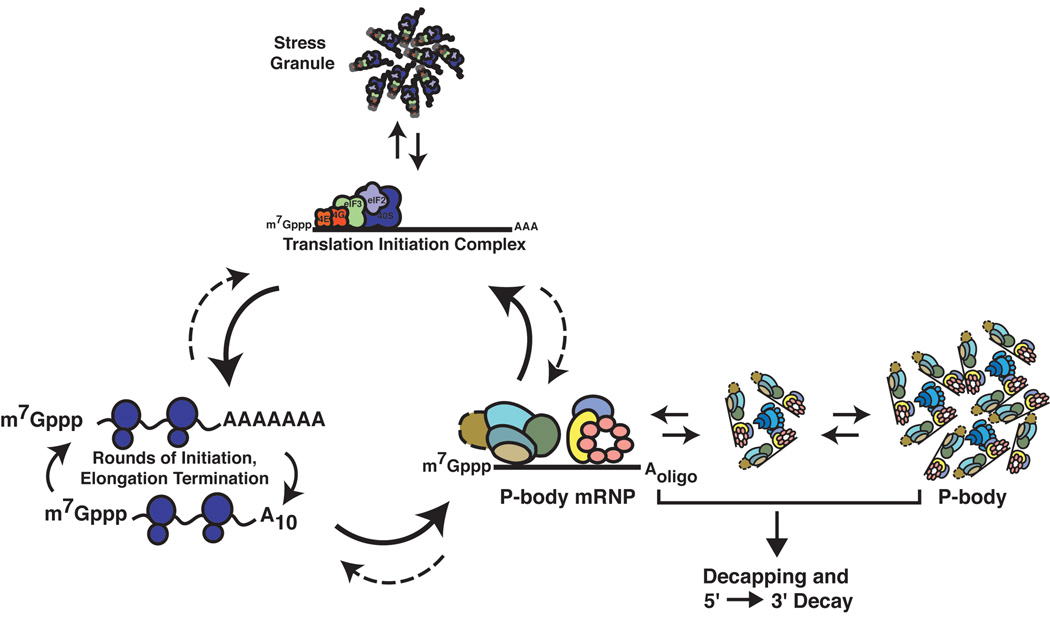
The analysis of P-bodies and the fate of transcripts associated with P-bodies has suggested a model for how host mRNAs enter and exit translation with the following key points (Figure 1) (reviewed in Parker and Sheth, 2007; Eulalio et al., 2007; Anderson and Kedersha, 2006). First, host mRNAs are in dynamic exchange between translating and non-translating pools. Second, many non-translating mRNAs are packaged into a repressed mRNP in conjunction with a set of conserved P-body proteins that mediate translation repression and mRNA degradation, as well as aggregate into P-bodies. Third, once associated with the P-body proteins, an mRNA can either be decapped and degraded, or be stored for later translation. Thus, the transcripts in a P-body are a biochemically distinct, and related, pool of mRNPs that are not engaged in translation and can be subjected to different fates. Finally, mRNAs that are stalled in translation initiation can aggregate into stress granules, which are often seen associated with P-bodies, and represent an intermediate in the transition of mRNAs between P-bodies and polysomes.
Figure 1. Cytoplasmic cycling of mRNA.
mRNA in the cytoplasm of eukaryotic cells may either engage in translation or be sequestered into translationally repressed mRNPs. Stress granules in mammals are formed during various cellular stresses and include mRNA, translation initiation factors and TIA-R proteins. Alternatively, mRNA can be sequestered into P-bodies where the mRNA is stored for future use in translation, or decapped and degraded.
A set of conserved "core" P-body components has been identified as being critical for the translational repression, accumulation and subsequent degradation or storage of mRNAs in P-bodies. These include the decapping enzyme (Dcp1p & Dcp2p) and the 5′ to 3′ exonuclease Xrn1p (Sheth and Parker, 2003; Ingelfinger et al., 2002; Lykke-Anderson, 2002 and van Dijk et al., 2002). P-bodies also contain a set of decapping activators including the heteroheptameric Lsm1p-7p complex, Dhh1p/Rck, and Pat1p (Sheth and Parker, 2003 and Ingelfinger et al., 2002). Dhh1p/Rck and Pat1p have also been shown to be involved in translational repression of mRNA and thereby facilitate the recruitment of repressed mRNA to P-bodies (Coller and Parker, 2005). Other core components of P-bodies include Scd6p/RAP55/CAR-1, Edc3p, and the deadenylase complex Ccr4p/Pop2p/Not1p-5p (Parker and Sheth, 2007; Eulalio et al., 2007). More recently, the yeast Ded1p protein, whose mammalian ortholog is Ddx3, has been identified as being a component of P-bodies and of influencing the movement of mRNAs between P-bodies and polysomes (Beckham et al., 2008). P-bodies in metazoans also accumulate proteins involved in miRNA and siRNA function including Argonaute proteins and GW182 and their orthologs (Parker and Sheth, 2007; Eulalio et al., 2007; Anderson and Kedersha, 2006), which are known to contribute to host antiviral defenses (Dykxhoorn, 2007).
Stress granules are dynamic aggregates of untranslating mRNAs in conjunction with a subset of translation initiation factors (eIF4E, eIF4G, eIF4A, eIF3, eIF2), the 40S ribosomal subunit, and several RNA binding proteins including the poly(A) binding protein (Anderson and Kedersha, 2006). Notable RNA binding proteins in stress granules include TIA-1, TIA-R and G3BP, all of which have self-interaction domains that can contribute to stress granule formation (Gilks et al., 2004; Tourriere et al., 2003). Stress granules form in response to defects in translation initiation including decreased function of eIF2, eIF4A, or eIF4G (Anderson and Kedersha, 2002; Mazouri et al., 2006; Dang et al., 2006), and are therefore thought to be aggregates of mRNA that are stalled in the process of translation initiation, that are then aggregated by the self-interaction domains of TIA-1, TIA-R, G3BP, and possibly other proteins. Because stress responses often involve a transient inhibition of translation initiation, stress granules accumulate during a wide range of stress responses. Stress granules are often docked to P-bodies suggesting mRNAs may move between these two compartments. Whether mRNPs are moving from P-bodies to stress granules, from stress granules to P-bodies, or both, remains to be determined.
The fundamental role of stress granules and P-bodies in the translation repression and degradation of host mRNAs suggests that these complexes, and the proteins within them, will affect the metabolism of viral mRNAs. Indeed, in some cases key components of stress granules and/or P-bodies are involved in limiting viral infection (see below). More surprisingly, genetic evidence demonstrates that proteins within P-bodies are required for the successful completion of the life cycles of some retroviral, retrotransposon, or +strand RNA viruses (summarized in Table 1 and discussed further below). This suggests that P-bodies, at least, will also be important for some viral life cycles, which is also supported by the accumulation of some viral RNAs and proteins in P-bodies. In this review, we discuss the emerging evidence suggesting that interactions between viral transcripts, stress granules and P-bodies can be important both for viral replication and for host antiviral defense.
Table 1.
Mutations in Core P-body Components affecting viral life-cycles
| P-body component | Virus (organism) | Phenotype | Reference |
|---|---|---|---|
| lsm1Δ (Defective Lsm1-7p complex) | Ty1 and Ty3 (yeast) | Reduced retrotransposition | Irwin et al., 2005; Aye et al., 2004; Griffith et al., 2003 |
| Brome mosaic virus (yeast) | Reduced translation & recruitment to replication | Noueiry et al., 2003; Kushner et al., 2003; Diez et al., 2000 | |
| pat1Δ | Ty1 and Ty3 (yeast) | Reduced retrotransposition | Irwin et al., 2005; Aye et al., 2004; Griffith et al., 2003 |
| Brome mosaic virus (yeast) | Reduced translation & recruitment to replication | Noueiry et al., 2003; Kushner et al., 2003; Mas et al., 2006 | |
| dhh1Δ | Ty1 and Ty3 (yeast) | Reduced retrotransposition | Irwin et al., 2005; Aye et al., 2004; Griffith et al., 2003 |
| Brome mosaic virus (yeast) | Reduced translation & recruitment to replication | Noueiry et al., 2003; Kushner et al., 2003; Mas et al., 2006 | |
| xrn1Δ | Ty3 (yeast) | Reduced retrotransposition | Irwin et al., 2005; Beliakova-Bethell et al., 2006 |
| pop2Δ | Ty3 (yeast) | Enhanced retrotransposition | Irwin et al., 2005 |
| ded1-2 (hypomorphic allele) | Brome mosaic virus (yeast) | Reduced Translation of RNA2 | Noueiry et al., 2000 |
| Ddx3 (antisense inhibition) | HIV (mammalian cell culture) | Reduced nuclear export and translation of unspliced HIV1 transcripts | Yedavilli et al., 2004 |
| Ddx3 (siRNA knockdown) | HCV mammalian cell culture) | Reduced Replication due to unknown defect | Ariumi et al., 2007 |
RETROVIRUS-LIKE ELEMENTS AND RETROVIRUSES
Retrotransposons and P-bodies
Initial evidence suggesting a connection between P-bodies and retroelements came from the analysis of the yeast retrotransposons Ty1 and Ty3. Ty1 and Ty3 are endogenous Ty retrotransposons in yeast cells and are model systems for the analysis of retroviruses and retroelements (Reviewed in Roth 2000 and Sandmeyer 1998). Ty1 is a member of the copia-like family of retrotransposons, while Ty3 is a member of the gypsy-like family of retrotransposons (Sandmeyer 1998).
Evidence that Ty elements required P-bodies for some aspect of their life cycle came from genetic data regarding host-viral interactions. Using retrotransposition assays, the Lsm1-Lsm7p complex, Pat1p, and Dhh1p were identified as being required for efficient retrotransposition for both Ty1 and Ty3 (Devine, 2003; Aye et al., 2004; Irwin et al., 2005; Beckham and Parker unpublished). In addition, deletions in the deadenylase complex component, Pop2p resulted in enhanced retrotransposition (Irwin et al., 2005 and Beckham and Parker unpublished). Because Lsm1-7p, Pat1p and Dhh1p promote the formation of P-bodies, and Ccr4p/Pop2p normally limits P-body formation (Coller and Parker, 2005, Teixeira and Parker, 2007), these genetic observations suggested that targeting of Ty transcripts to P-bodies might be important for retrotransposition.
One possibility is that P-bodies, or the mRNPs within them, may serve important roles in the assembly or maturation of the Ty virus-like particles. This is supported by the observation that tagged versions of Ty3 viral RNA and proteins accumulated in P-bodies (Beliakova-Bethell et al., 2006). Moreover, mutations in key components of P-bodies that affected retrotransposition also altered the subcellular distribution of viral components. Specifically, in dhh1Δ yeast strains Ty3 proteins were more diffuse and twice as many cells had multiple smaller foci versus the wild type control (Beliakova-Bethell et al., 2006). In an xrn1Δ strain, Ty3 proteins accumulated in larger foci, paralleling the enhanced P-body formation seen in xrn1Δ mutants (Beliakova-Bethell et al., 2006). These data suggest a role for constituents of P-bodies in the accumulation and/or targeting of viral components to P-bodies.
The precise function of the Lsm1-7p complex, Pat1p and Dhh1p in the Ty3 life cycle remains unclear. Mutational analysis of the capsid and NC components of the Ty3 Gag protein is consistent with P-bodies participating in some aspect of Ty3 assembly or maturation (Larsen et al., 2007; Larsen et al., 2008). However, because lsm1Δ, pat1Δ, and dhh1Δ strains produce at least some Ty3 cDNA (Irwin et al., 2005), the role of P-bodies may be in a late step of viral assembly and/or cDNA transport to the nucleus.
It is possible that other retrotransposon elements will interact with P-bodies during their life cycles. For example, similar to Ty3, the Gag proteins of the Drosophila Doc and I factor transposons accumulate in cytoplasmic foci, of unknown nature, that look similar to P-bodies (Rashkova et al., 2002).
Retroviruses and P-bodies in mammals
An obvious implication of the work with Ty elements is that additional retrotransposons or retroviruses might utilize P-bodies, or the components within them, for important steps in their life cycles. One intriguing connection between HIV and P-bodies has come from the observation that the highly conserved RNA helicase DDX3 is required for the export, and translation, of unspliced HIV-1 from the nucleus (Yedavilli et al., 2004). This observation is relevant for two reasons. First, DDX3 is a highly conserved protein and the yeast ortholog of DDX3, Ded1p, can accumulate in P-bodies and affect their formation (Beckham et al., 2008). Second, the unspliced form of HIV1 RNA serves as gRNA. This suggests that DDX3 may recruit HIV1 gRNA to the P-body for subsequent steps in the packaging into virions. Interestingly, some components of P-bodies are nuclear-shuttling proteins and have been proposed to play a role in the export of mRNAs directly into the P-body state (Parker and Sheth, 2007). Moreover, the export of P-body components in yeast is dependent on the Crm1p protein (Pilkington and Parker, unpublished), which is the nuclear export system that Rev targets for export of HIV transcripts from the nucleus (Yedavilli et al., 2004). These data suggest a model wherein Ded1p first engages HIV-1 in the nucleus leading to export in a manner that targets the HIV-1 RNA to a P-body following export, which might be important for subsequent steps in viral function.
Understanding the relationships between P-bodies and mammalian retroviruses will require additional experiments. Many retroviruses are known to undertake key steps in their life cycles in discrete subcellular foci, although the identity of these foci is generally unknown. For example, Mason-Pfizer Monkey virus Gag protein is found in cytoplasmic foci of unknown identity (Weldon et al., 2003), while for Foamy viruses both Env and Gag proteins are found in cytoplasmic foci, which are thought to be associated with the centriole (Yu et al., 2006). An interesting area of future work will be to determine the nature of these cytoplasmic aggregates and any possible relationship to P-bodies.
POSITIVE STRANDED RNA VIRUSES
Another set of viruses that show interactions with P-bodies are positive-strand RNA viruses (+RNA viruses), which encompass over one third of all virus genera and include numerous well-known pathogens such as Hepatitis C (HCV) and West Nile Virus. Despite their differences, all positive-strand RNA viruses share a similar life cycle following infection where the +RNA first serves as an mRNA to produce viral replication factors, and then exits translation and is selectively recruited to a membrane-associated replication complex. An unresolved issue is the mechanism(s) bringing the viral RNAs and proteins together and thus facilitating the assembly of these replication complexes.
Brome Mosaic Virus and P-bodies
One connection between a + strand RNA virus and P-bodies came from genetic studies in yeast using the positive-stranded RNA virus Brome Mosaic Virus (BMV), which can complete all stages of the viral life cycle in yeast (Janda and Ahlquist, 1993). BMV is a member of the alphavirus-like superfamily and has a tri-partite genome consisting of viral transcripts RNA1, RNA2 and RNA3. A subgenomic RNA, termed RNA4, is produced from RNA3 during replication at the endoplasmic reticulum. The transcripts are capped, but lack poly(A) tails (Reviewed in Noueiry and Ahlquist, 2003). Segregation of genes into separate transcripts allows for the independent analysis of function and subcellular localization of each transcript and gene product.
Using genetic screens to find host factors involved in BMV function, two important roles for components of P-bodies were revealed (Kushner et al., 2003). First, it was seen that the P-body components Pat1p, Dhh1p and the Lsm1p-7p complex were required for efficient translation of the RNA1, RNA2 and RNA3 (Noueiry et al., 2003). Similarly, the essential DEAD-box RNA helicase Ded1p, which can also be a component of P-bodies (Beckham et al., 2008), was required for the translation of viral RNA2, which encodes the RNA dependent RNA polymerase (RdRp) protein (Noueiry et al., 2000). Since Pat1p, Dhh1p, and Lsm1-7p are generally translational repressors, it is not clear why they are required for the translational activation of the viral transcripts. One possibility is that these proteins can function as translation activators in some contexts.
A second intriguing connection between the BMV life cycle and P-bodies is that Pat1p, Dhh1p, and the Lsm1-7p complex are all required for entry of the gRNAs into replication, which for BMV involves the formation of a membrane bound replication complex (Noueiry et al., 2003 and Mas et al., 2006). Moreover, three observations suggest these proteins are required for replication because P-bodies may play a role in concentrating the gRNAs, the viral proteins and promoting their interaction with membranes (Beckham et al., 2007). First, RNA2 and RNA3 accumulate in P-bodies. Moreover, the P-body accumulation of RNA3 is dependent on cis-acting signals that contribute to replication efficiency suggesting P-body accumulation is related to replication in some manner. Second, the RNA-dependent RNA polymerase (RdRp) also accumulated in P-bodies and co-immunoprecipitated with Lsm1p. Finally, at least some P-bodies are localized and can be biochemically associated with membranes where BMV and other viruses replicate (Beckham et al., 2007; Wang et al., 2005 and Wilhelm et al., 2005).
Taken together, these results suggest a model for formation of BMV replication complexes wherein the gRNAs and viral replication factors accumulate in P-bodies. Subsequently, interaction of P-bodies containing viral components with membranes could facilitate interactions between the membrane bound replication factor protein 1a, and the components in P-bodies thereby leading to the assembly of a replication complex. Importantly, since all +strand RNA viruses share a similar life cycle, one possibility is that this role of P-bodies in viral replication may be present in other +strand RNA viruses.
Possible connections between HCV and P-bodies
Several observations raise the possibility of a connection between HCV and P-bodies. First, the HCV core protein physically interacts with DDX3 and colocalizes in cytoplasm foci reminiscent of P-bodies (Mamiya and Worman, 1999 and You et al., 1999). Second, knockdowns of DDX3 reduce HCV replication by an undetermined mechanism (Ariumi et al., 2007). Third, the replication of HCV is increased by interaction of the viral RNA with the liver-specific miRNA, miR122, which binds to the 5′NCR of the HCV genome (Jopling et al., 2005). This is striking since one function of miRNAs appears to be the ability to target mRNAs into P-bodies (Liu et al., 2005 and Pillai, 2005). This raises the speculative model that assembly of an RNP containing P-body components and the HCV RNA might be important for efficient replication.
P-BODIES, STRESS GRANULES, AND ANTIVIRAL DEFENSE
In addition to the possible positive roles for P-bodies in viral life cycles, the components of P-bodies and stress granules may also function in host defenses against viruses and transposable elements. For example, siRNAs and miRNAs can contribute to antiviral defense in mammals and plants (Dykxhoorn, 2007). This is relevant since miRNAs, and likely siRNAs, function, at least in part, by recruiting components of P-bodies to target mRNAs thereby leading to both translation repression and mRNA decapping and degradation (reviewed in Valencia-Sanchez et al., 2006). In addition, protein kinase R (PKR), which is activated by dsRNA and contributes to antiviral defense, is observed to accumulate in P-bodies during human papilloma infection, although the significance of this observation is unclear (Hebner et al., 2006).
Another interesting connection between antiviral defense, stress granules and P-bodies is that the antiviral proteins, APOBEC3G and 3F are concentrated in P-bodies and can accumulate in stress granules during stress (Gallois-Montbrun et al., 2007; Wichroski et al., 2006). APOBEC (apolipoprotein B mRNA-editing enzyme catalytic polypeptide like) proteins belong to a family of cytidine deaminases (reviewed in Wedekind et al., 2003). These proteins are thought to play an antiviral role by deaminating cytidines in retroviral or retrotransposon genomes. For example, APOBEC3G can be incorporated into HIV-1 virions and inhibits replication by deaminating cytidines on viral minus strand (first strand) transcripts. Ultimately, these mutations result in guanosine to adenosine transitions in the plus stranded cDNA (Esnault et al., 2005; Griffith et al., 2003 and Yu et al., 2004). Some retroviruses, in turn, make proteins that inhibit the function of APOBECs. For example, the vif protein of HIV-1 binds to APOBEC3G (3G) and triggers its degradation (Yu et al., 2004).
The accumulation of APOBEC proteins in P-bodies could be explained by several possible mechanisms. For example, it could be that P-bodies are sites of viral repression and APOBEC is concentrated there to assist in that process (Wichroski et al., 2006). A simpler possibility is that the concentration of APOBEC3G and 3F in P-bodies reflects their binding to a subset of endogenous RNAs, which are translationally repressed and accumulate in P-bodies. For example, APOBEC3G might interact with transcripts from endogenous retroelements and those transcripts might be expected to be translationally repressed by piRNAs or miRNAs, thereby accumulating in P-bodies along with APOBEC proteins. The ability of APOBEC proteins to interact with a wide variety of RNA binding proteins, to be found on polysomes and in stress granules (Gallois-Montbrun et al., 2007; Marin et al., 2007; Wang et al., 2007; Kozak et al., 2006) is all consistent with the subcellular distribution of APOBEC proteins being determined by the accumulation of specific mRNAs in subcellular sites.
Some viral infections are known to transiently trigger stress granule formation, at least in some cases by the activation of kinases that phosphorylate eIF2 and thereby limit translation initiation (Smith et al., 2006; McInerney et al., 2005; White et al., 2007; Mazouri et al., 2006). Some observations suggest that stress granules, or at least some of the key proteins within them, function to limit a range of viral infections. For example, mouse embryo fibroblasts lacking the TIA-1 protein show increased rates of viral production from a wide range of different viruses including a VSV (a −strand RNA virus), Sindbis virus (a +strand RNA virus), and HSV (a DNA virus replicating in the nucleus (Li et al., 2002). Similarly, polio viral production appears to be limited by the G3BP protein (White et al., 2007). It should be noted that the RNA binding components of stress granules can also have a positive impact in some viral infections. For example, the TIA-R protein binds to a 3′ stem-loop structure in the West Nile Virus -strand and promotes + strand synthesis (Li et al., 2002).
Some viruses also inhibit the host cells ability to form stress granules during the infection. For example, the polio encoded 3C protease cleaves the G3BP protein during infection and thereby prevents stress granule formation at later times of infection (White et al., 2007). The flaviviruses, West Nile and Dengue, also interfere with stress granule formation later in infection, which might be due to reduced levels of host mRNAs at these times (Emara and Brinton, 2007). Finally, the Semliki Forest Virus (SFV) appears to reduce stress granules in the regions of cells that accumulate viral proteins (McInerney et al., 2005). It should be noted that because stress granules and P-bodies require a pool of untranslating mRNA for their assembly, any virus that leads to degradation of host mRNAs may inhibit the assembly of these structures due to reductions in the pool of cytoplasmic mRNAs.
Taken together, the above results suggest that stress granules, and/or the components within them, may primarily function to limit viral infections. One possibility is that the formation of stress granules is part of the host response to limit translation by sequestration of limiting translation factors and thereby reduces viral replication (Schütz and Sarnow, 2007). Alternatively, the TIA-1 and G3BP proteins might limit viral function simply by repressing the function of specific host or viral transcripts required for viral life cycles.
DIVERSITY OF INTERACTIONS BETWEEN STRESS GRANULES, P-BODIES AND VIRUSES
The evidence discussed above suggests that stress granules, P-bodies and the components within them will influence viral life cycles in a variety of manners. First, the stress granule and P-body components may play important roles in host defenses against viruses by repressing the function of viral transcripts. Alternatively, the translational control of viral transcripts might be important for maintaining proper stoichiometry of viral components. Finally, since many P-body components can shuttle between the nucleus and the cytoplasm, P-bodies may function in the nuclear cytoplasmic transport and remodeling of some viral mRNPs.
An intriguing possibility is that the concentration of mRNAs at specific subcelluar sites by P-bodies and/or stress granules might be important in directing the translation of viral mRNAs in specific subcellular regions. For example, during vaccinia virus infection, viral mRNAs are recruited to cytoplasmic "factories" where translation, replication and viral assembly proceeds (Katsafanas and Moss, 2007). Strikingly, these factories include the G3BP protein, which also appears to affect viral transcription in vitro (Katsafanas and Moss, 2004; 2007). Since G3BP is an important component of stress granules, this raises the possibility that the assembly of vaccinia factories and stress granules may share common assembly mechanisms.
P-bodies and the translationally repressed mRNPs that accumulate within them may also provide a pool of translationally repressed viral transcripts for efficient packaging or formation of viral replication complexes. This model builds on the role of P-bodies as important sites for host mRNAs to enter different fates including translation, storage, or degradation. In an extension of this model to viruses, viral transcripts or genomes, similar to host mRNAs, would localize to P-bodies when not engaged in active translation. Following accumulation in P-bodies, interactions between the RNAs and viral proteins, in the absence of competing translation machinery, might allow for efficient packaging or formation of replication complexes. Moreover, given the ability of P-bodies to aggregate a number of mRNAs in a single subcellular location, P-bodies may be particularly important in the coordination of packaging for multi-segmented RNA viruses, such as flu, and may possibly expedite the genetic diversity of multi-segmented viruses by bringing together the diverse assortment of genomic transcript possibilities.
It should be noted that in cases where viral mRNAs are targeted to P-bodies for packaging or replication complex formation, particular mRNP complexes must form that prevent their decapping. For example, adenoviral mRNAs contain a 5′ poly(A) tail that binds to the Lsm1-7p complex and prevents mRNA decapping (Bergman et al., 2007), presumably by limiting interaction of the 5′ end of the mRNA with the RNA binding site on the decapping enzyme (Steiger et al., 2003). Thus, work examining understanding how viral mRNAs interact with, and evade, the host mRNA degradation machinery may shed new light on viral function and their interactions with P-bodies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus replication. J. Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye M, Irwin B, Beliakova-Bethell N, Chen E, Garrus J, Sandmeyer S. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics. 2004;168:1159–1176. doi: 10.1534/genetics.104.028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko AM, Noueiry A, Ramasawami M, Parker R. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. MBC Epub ahead of Print. 2007 doi: 10.1091/mbc.E07-09-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C, Light H, Nissan T, Ahlquist P, Parker R, Noueiry A. Interactions between Brome Mosaic Virus RNAs and Cytoplasmic Processing Bodies. J. Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N, Beckham C, Giddings TH, Winey M, Parker R, Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman N, Moraes K, Anderson J, Zaric B, Kambach C, Schneider R, Wilusz C, Wilusz J. Lsm proteins bind and stabilize RNAs containing 5′ poly(A) tracts. Nat Struct Mol Biol. 2007;14:824–831. doi: 10.1038/nsmb1287. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- Devine SE. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics. 2003;164:867–879. doi: 10.1093/genetics/164.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM. MicroRNAs in viral replication and pathogenesis. DNA Cell Biol. 2007;26:239–249. doi: 10.1089/dna.2006.0559. [DOI] [PubMed] [Google Scholar]
- Emara M, Brinton M. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. PNAS. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Deqannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Gallois-Monbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;12:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Hebner C, Wilson R, Rader J, Bidder M, Laimins L. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J. Gen. Virol. 2006;87:3183–3193. doi: 10.1099/vir.0.82098-0. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–14501. [PMC free article] [PubMed] [Google Scholar]
- Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 2005;15:641–654. doi: 10.1101/gr.3739005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- Jopling C, Yi M, Lancaster A, Lemon S, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPas-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 2004;279:52210–52217. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host and Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- Kushner DB, Lindenbach BD, Grdzelishvilli VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc Natl Acad Sci USA. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen LS, Beliakova-Bethell N, Bilanchone V, Zhang M, Lamsa A, Dasilva R, Hatfield GW, Nagashima K, Sandmeyer S. Ty3 nucleocapsid controls localization of particle assembly. J Virol. Epub ahead of print. 2007 doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen LS, Kuznetsov Y, McPherson A, Hatfield GW, Sandmeyer S. TY3 GAG3 protein forms ordered particles in Escherichia coli. Virology. 2008;370:223–227. doi: 10.1016/j.virol.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. Journal of virology. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez M, Hannon G, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274:1565–1576. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- Marin M, Golem S, Rose KM, Kozak SL, Kabat D. HIV-1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J Virol. Epub ahead of print. 2007 doi: 10.1128/JVI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas A, Alves-Rodriques I, Noueiry A, Ahlquist P, Diez J. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J Virol. 2006;80:246–251. doi: 10.1128/JVI.80.1.246-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazouri R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka T, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;10:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus tranlation regulation. Mol Biol Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Noueiry AO, Chen J, Ahlquist P. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc Natl Acad Sci USA. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry AO, Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- Noueiry AO, Diez J, Falk SP, Chen J, Ahlquist P. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol Cell Biol. 2003;23:4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pillai R. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova S, Karam SE, Pardue ML. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc Natl Sci USA. 2002;99:3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JF. The yeast Ty virus-like particles. Yeast. 2000;16:785–795. doi: 10.1002/1097-0061(20000630)16:9<785::AID-YEA550>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Sandmeyer S. Targeting transposition: at home in the genome. Genome Res. 1998;8:416–418. doi: 10.1101/gr.8.5.416. [DOI] [PubMed] [Google Scholar]
- Schütz S, Sarnow P. How viruses avoid stess. Cell Host Microbe. 2007;2:284–285. doi: 10.1016/j.chom.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Schmechel sC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an intergrated cellular stress response. J. Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol. Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;1600:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;200:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang Q, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lee WM, Watanabe T, Schwart M, Janda M, Ahlquist P. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J Virol. 2005;79:13747–13758. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- Weldon RA, Sarkar P, Brown SM, Weldon SK. Mason-Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc9. Virology. 2003;314:62–73. doi: 10.1016/s0042-6822(03)00348-9. [DOI] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen W, Lloyd RE. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host and Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. RNA 12, 547–554. [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH, Lee YH. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. 1999;73:2841–2852. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. The Journal of biological chemistry. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Yu SF, Eastman SW, Linial ML. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic (Copenhagen, Denmark) 2006;7:966–977. doi: 10.1111/j.1600-0854.2006.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]