Abstract
Development of the central nervous system requires proliferation of neuronal and glial cell precursors followed by their subsequent differentiation in a highly coordinated manner. The timing of neuronal cell cycle exit and differentiation is likely to be regulated in part by inhibitors of cyclin-dependent kinases. Overlapping and sustained patterns of expression of two cyclin-dependent kinases, p19Ink4d and p27Kip1, in postmitotic brain cells suggested that these proteins may be important in actively repressing neuronal proliferation. Animals derived from crosses of Ink4d- null with Kip1-null mice exhibited bradykinesia, proprioceptive abnormalities, and seizures, and died at about 18 days after birth. Metabolic labeling of live animals with bromodeoxyuridine at postnatal days 14 and 18, combined with immunolabeling of neuronal markers, showed that subpopulations of central nervous system neurons were proliferating in all parts of the brain, including normally dormant cells of the hippocampus, cortex, hypothalamus, pons, and brainstem. These cells also expressed phosphorylated histone H3, a marker for late G2 and M-phase progression, indicating that neurons were dividing after they had migrated to their final positions in the brain. Increased proliferation was balanced by cell death, resulting in no gross changes in the cytoarchitecture of the brains of these mice. Therefore, p19Ink4d and p27Kip1 cooperate to maintain differentiated neurons in a quiescent state that is potentially reversible.
Keywords: Ink4 proteins, Cip, Kip proteins, p19Ink4d, p27Kip1, neuronal development
Exit from the cell division cycle during terminal differentiation requires the inactivation of cyclin-dependent kinases (CDKs), a process that depends in part on polypeptide CDK inhibitors (CKIs). Vivid testament to the roles of CKIs during differentiation comes from studies of mice in which genes encoding different CKIs have been disrupted. In certain cases, the loss of only one such gene is sufficient to perturb mouse development (1–6), whereas in others different CKIs have been revealed to play redundant roles in regulating the differentiation of particular cell lineages (6–8).
There are two distinct families of mammalian CKIs. The first class, so-called inhibitors of CDK4 (Ink4 proteins), is composed of four distinct members (p16Ink4a, p15Ink4b, p18Ink4c, and p19Ink4d), each capable of specifically inactivating the mitogen-dependent cyclin D-dependent kinases CDK4 and CDK6 (reviewed in ref. 9). Ink4 proteins interact only with the CDK subunit of cyclin D-dependent kinases, ultimately displacing D-type cyclins from their CDK partners and leading to the formation of inactive CDK-Ink4 dimers. Members of the Ink4 family do not associate with other CDKs, and hence do not directly inhibit cyclin E- or A-dependent CDK2 or cyclin B-dependent CDK1 (Cdc2). By contrast, the Cip/Kip family of CKIs is composed of three members (p21Cip1, p27Kip1, and p57Kip2), each of which contains independent binding sites for both cyclin and CDK subunits (reviewed in ref. 10). As such, the Cip/Kip proteins can form higher-order complexes with particular cyclin–CDK holoenzymes, thereby preventing both substrate and ATP binding and canceling their enzymatic activity. Although the Cip/Kip proteins are potent inhibitors of cyclin E- and A-dependent CDK2, perhaps paradoxically, their binding to cyclin D-CDK complexes does not inhibit their activity (11, 12), but instead enhances their assembly, stability, and nuclear localization (13, 14). Hence, Cip/Kip proteins remain stably associated with enzymatically active cyclin D-CDK holoenzymes in proliferating cells. The induction of Ink4 proteins disrupts cyclin D-dependent kinases, releasing the bound Cip/Kip proteins and enabling them to inhibit CDK2. Inhibition of all G1 CDKs results in rapid G1-phase arrest, usually within a single cell cycle (reviewed in ref. 10).
Among the Ink4 family members, only p18Ink4c and p19Ink4d are expressed in fetal development (15), and stereotypic patterns of expression have been observed within the central nervous system (CNS) from embryonic day (E) 11.5 onward (16). During development of the neocortex, for example, asymmetric divisions give rise to differentiated neurons that exit the cell cycle and migrate to their final positions in the brain (reviewed in ref. 17). Cells engaged in such divisions express p18Ink4c, which is turned off as cells withdraw from the cell cycle and is replaced by p19Ink4d, whose expression continues in postmitotic neurons and is maintained into adulthood (16). Similar patterns of Ink4d expression can be observed in other areas of the adult brain, including neurons of the dentate gyrus, the pyramidal layer of the hippocampus, and regions of the cerebellum, thalamus, and brainstem. Although CDKs are down-regulated in adult neurons in the CNS, cyclin D1 levels remain elevated (18, 19). The persistence of p19Ink4d may therefore guard against CDK activation, providing an active brake that keeps cells from cycling. Reduced expression of p19Ink4d in the dentate gyrus after kainate-induced seizures also previously demonstrated that excitatory signals could modify Ink4d expression in nondividing cells, an event associated with increased apoptosis in this region (16). Although one might have assumed that disruption of Ink4d might therefore lead to abnormalities during neurogenesis, CNS development in mice lacking this gene was overtly normal (20).
Another CKI, p27Kip1, is temporally expressed in the developing brain at sites similar to those that express p19Ink4d, and its expression is also maintained in postmitotic neurons into adulthood (21). Like Ink4d-deficient mice, animals lacking Kip1, independently derived in three separate laboratories, do not exhibit neurological deficits and show no apparent CNS defects (1–3). This observation left open the possibility that Ink4d and Kip1 may play partially redundant roles in the differentiation of neurons within the CNS. Here, we show that animals lacking both p19Ink4d and p27Kip1 exhibit ectopic neuronal cell divisions and apoptosis in many parts of the brain that are normally quiescent. This result suggests that CKIs actively prevent cell division within the brain and that withdrawal of neurons from the cell division cycle is potentially reversible.
Materials and Methods
Derivation of Mouse Strains.
Mouse strains (C57BL/6 × 129Svj) deficient in Ink4d (20) and Kip1 (1) were described previously. Ink4d-deficient females were bred to Kip1-null males to yield compound heterozygotes. Interbreeding generated wild-type single- and double-null animals at the expected Mendelian frequency. Different crosses were used to generate littermates of the following genotypes and frequencies. (i) An intercross of Ink4d−/−, Kip1+/− mice yielded 25% double-null animals and 25% Ink4d−/−, Kip1+/+ mice; (ii) Ink4d+/−, Kip1−/− (males) crossed with Ink4d+/−, Kip1+/− (females) generated 12.5% double-null and 12.5% Ink4d+/+, Kip1−/− offspring; (iii) Ink4d+/−, Kip1+/+ bred to Ink4d+/−, Kip1+/+ generated 25% wild-type and 25% Ink4d−/−, Kip1+/+ animals; (iv) Ink4d+/+, Kip1−/− (males) crossed to Ink4d+/+, Kip1+/− (females) yielded 50% Ink4d+/+, Kip1−/− mice. Mouse-tail DNA was analyzed by PCR to distinguish Kip1 wild-type and mutant alleles (1), whereas Southern blotting was used to determine Ink4d status (20).
Bromodeoxyuridine (BrdUrd) Labeling and Tissue Preparation.
Mice of all possible genotypes at postnatal day (P)14 and P18 were injected intraperitoneally five times at 2-hr intervals with 50 μg/g body weight of a 5-mg/ml solution of BrdUrd (Sigma) in 7 mM NaOH. Two hours after the final injection, animals were anesthetized with 250 mg/kg of tribromoethanol (Sigma) in normal saline (0.9% NaCl in phosphate buffer) at a concentration of 25 mg/ml and transcardially perfused with normal saline followed by either 95% ethanol/acetic acid (3:1) or 4% paraformaldehyde (Sigma). Brains and eyes were removed from the calvaria and postfixed overnight, dehydrated, defatted, and embedded in paraffin. Serial sections (5 μm) were collected on separate Vectabond coated slides (Vector Laboratories).
Protein Detection, Immunohistochemistry, and Apoptosis Assay.
Sequential immunoprecipitation and immunoblotting were performed as previously described (16). Serial sections from mouse tissues fixed with 95% ethanol/acetic acid were processed for BrdUrd staining [with monoclonal anti-BrdUrd (1:100), Dako] for BrdUrd plus neurofilaments protein (NFP) staining [monoclonal anti-NFP (1:300), Zymed], or for histone H-3 staining [by use of polyclonal antibody to phosphorylated histone H3 (HH3) (1:100), Upstate Biotechnology, Lake Placid, NY]. Material from animals perfused with 4% paraformaldehyde was stained with monoclonal (1:500 dilution) Neuron nuclei (NeuN, Chemicon). Single labeling with anti-BrdUrd, anti-NeuN, or anti-HH3 was detected by using either a mouse or rabbit immunoperoxidase ABC kit (Vector) according to the manufacturer. Double labeling with either NFP or glial fibrillary acidic protein (GFAP) [polyclonal anti-GFAP (1:400), Dako] and BrdUrd was performed by using the Dako EnVision Doublestain System. One series of slides was processed for assay of apoptotic cells by using the Dead End Colorimetric Detection System (Promega). Estimates of total neuronal number (22) were made on every 20th serial section of hippocampus immunolabeled with NeuN. Images were captured by using a Kontron Digital Camera Model 3012 and imported into adobe photoshop (Ver. 3.0, Adobe Systems, Mountain View, CA). Any changes to contrast were applied equally to all images. Photographs were printed on a Fuji Pictography 300 printer.
Results
Derivation of Mice Deficient for Ink4d and Kip1.
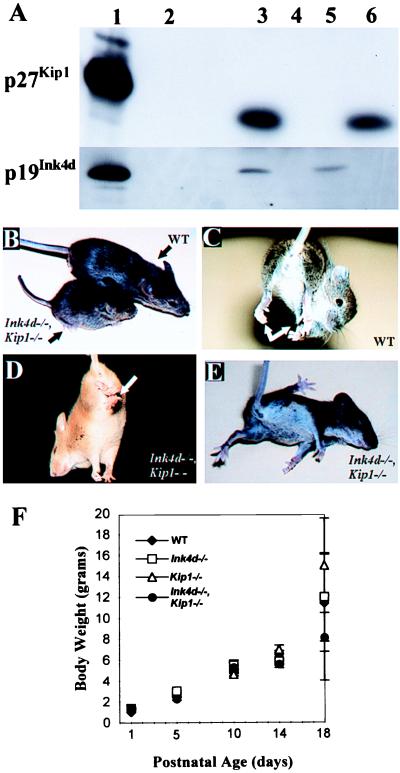
Because p19Ink4d and p27Kip1 are coexpressed in postmitotic neurons in the CNS, we reasoned that these CDK inhibitors might cooperate in maintaining cells in a quiescent state. We bred Ink4d-deficient animals (20) with Kip1-deficient mice (1) and then interbred compound heterozygotes. These crosses yielded the expected Mendelian distribution of wild-type singly- and doubly deficient offspring, indicating that codeletion of both genes permitted viable fetal development. Western blotting analysis of brain homogenates confirmed the expected lack of p19Ink4d and/or p27Kip1 expression in littermates of different genotypes (Fig. 1A). No gross anomalies were observed in young mice that retained either one wild-type Ink4d or Kip1 allele. From birth until P14, all doubly deficient animals also developed normally, but after P14 they failed to thrive and by P18, they were markedly smaller than their Ink4d−/−, Kip1+/+ littermates (Fig. 1 B and F). As early as P10, these mice began to develop progressive neurological abnormalities, eventually leading to paresis, bradykinesia, tremors, hypertonia, abnormal leg clasping reflexes (Fig. 1 C and D), light sensitivity, and seizures (Fig. 1E). By P18 or soon thereafter, all such mice exhibited labored (Cheyne-Stokes) respiration and died. These neurologic signs were observed in all double-null mice arising from two independently derived Ink4d-null embryonic stem cell clones. In turn, Kip1-null mice independently derived in three laboratories have identical phenotypes (1–3), so we have no reason to suspect that the observed outcome in double-null animals depends on the specific embryonic stem cell clone used to derive the mice.
Figure 1.
Postnatal development in Ink4d, Kip1-deficient mice. (A) Immunoblotting of brain extracts from mice of the different genotypes. Recombinant proteins produced in insect Sf9 cells infected with the indicated baculovirus vectors (lanes 1) or wild-type baculovirus (lane 2) were used as controls. Genotypes: wild type (lane 3), double-null (lane 4), Kip1-null (lane 5), and Ink4d-null (lane 6). (B) At P18, Ink4d, Kip1 double-null mice appear consistently smaller than their wild-type littermates. (C) When lifted by their tails, wild-type mice twist, extend their legs, and attempt to find a solid object to grasp, including climbing back up their own tails, whereas (D) double-null mice hold their limbs by their trunk. (E) By P18, Ink4d, Kip1 double-null mice exhibit seizures and have difficulty righting themselves. (F) Body weights of postnatal mice at different ages [± SEM] for wild type (♦), Ink4d-null (□), Kip1-null (Δ), or Ink4d, Kip1 double-null (●) mice.
Proliferation of Cells in the Brains of Postnatal Ink4d−/−/Kip1−/− Mice.
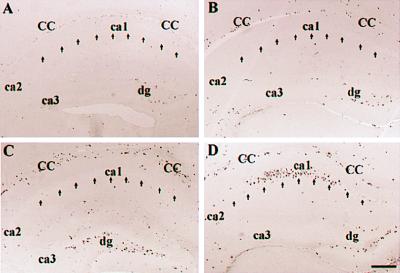
In principle, codeletion of Ink4d and Kip1 might prevent cell cycle exit in populations that are normally quiescent, so we pulse labeled P18 mice of all genotypes with BrdUrd, which allows detection of cells in S phase. Because seizure foci frequently arise in the hippocampus, this region of the brain was analyzed first. In wild-type animals, BrdUrd labeling was restricted, as expected, to the dentate gyrus, which still contains cycling cells at this time, but was undetectable in the pyramidal cell layer (Fig. 2A). Similar results were observed in hippocampal sections taken from Ink4d-deficient animals (Fig. 2B). Brain sections from Kip1-deficient mice showed BrdUrd labeling in the corpus callosum (Fig. 2C), probably resulting from enhanced oligodendrocyte proliferation, as previously reported (23). In contrast, in Ink4d−/−, Kip1−/− animals, we observed prominent BrdUrd labeling not only in the corpus callosum but also in the hippocampal pyramidal cell layer, a region of the brain that is normally dormant by P18 (Fig. 2D).
Figure 2.
BrdUrd labeling of hippocampus at P18 from (A) wild-type, (B) Ink4d–null, (C) Kip1-null, and (D) Ink4d, Kip1 double-null mice. Note prominent labeling in D ca1. CC, corpus callosum; DG, dentate gyrus; ca1 through ca3, corpora Ammon’s sectors 1–3. Bar for all figures (shown in D) = 200 μm.
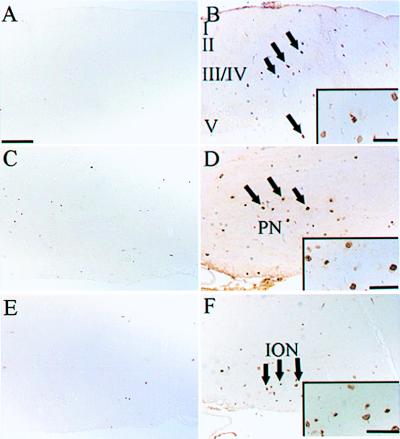
When other regions of the brain were examined, BrdUrd labeling was also detected in layers III and V of the cerebral cortex (Fig. 3B) and in pontine (Fig. 3D) and brainstem nuclei (Fig. 3F). At this age of development, these regions should not contain replicating cells (24, 25), consistent with results obtained with brain sections from wild-type mice (Fig. 3 A, C, and E). Analysis of the hypothalamus, substantia nigra, and retina from Ink4d, Kip1 doubly deficient mice also revealed ectopic cell proliferation (data not shown). Despite widespread BrdUrd labeling in the brains of these animals, the number of positive cells at any one time did not exceed 1% to 5% of the total, except for the pyramidal layer of the hippocampus, where a higher proportion of cells were labeled at P18 (Fig. 2D).
Figure 3.
Brain sections of the (A and B) cortex, (C and D) pons, and (E and F) brainstem in (A, C and E) wild-type and (B, D and F) Ink4d, Kip1 double-null mice. BrdUrd labeling is absent at P18 in the wild-type mouse brains. Ink4d, Kip1 double-null mice show prominent BrdUrd labeling in layers III and V of the (B, arrows) cortex, (D, arrows) pontine nuclei, and (F, arrows) brainstem nuclei including the inferior olive. (Insets) Enlarged fields of labeled cells. I, II, III/IV, and V, cerebral cortical layers; PN, pontine nuclei; ION, inferior olive nucleus. Bars = A–F, 200 μm; B Inset, 50 μm; D Inset, 95 μm; F Inset, 100 μm.
Proliferating Brain Cells Are Differentiated Neurons.
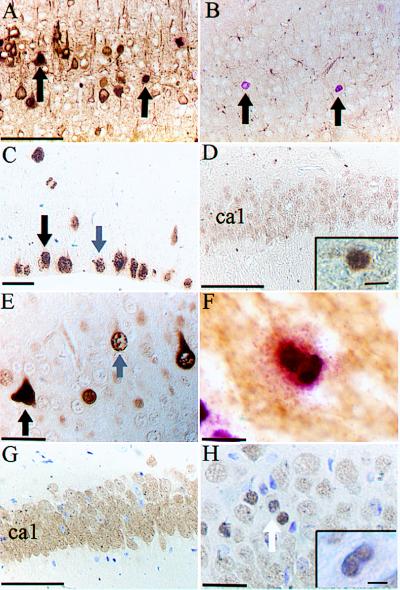
To determine whether BrdUrd-positive cells were neurons or glia, we combined BrdUrd labeling with staining for either a neuron-specific marker, NFP (26), or a glial marker, GFAP (27). The majority of cells that incorporated BrdUrd also expressed NFP (Fig. 4A) but not GFAP (Fig. 4B), indicating the BrdUrd-positive cells in the CNS of mice lacking both Ink4d and Kip1 were differentiated neurons.
Figure 4.
(A) Section of neocortex from an Ink4d, Kip1 double-null mouse shows that BrdUrd-positive cells entering S phase (pink) are also positive for NFP (brown). Arrows point to double-labeled cells. (B) The same cells do not stain for GFAP (arrows indicate BrdUrd-labeled cells). (C) Control brain section from the neocortex of a wild-type E11.5 mouse labeled with HH3 showing cells in G2 (grey arrow) and M phase (black arrow). (D) Hippocampal section from P18 wild-type mouse labeled with HH3 lacking positively stained cells in ca1, but not in the dentate gyrus (Inset). (E) Hippocampal section from P18 Ink4d, Kip1 double-null mouse labeled with HH3 showing cells in G2 phase (gray arrow) and M phase (black arrow). (F) Cortical section from Ink4d, Kip1 double-null mouse showing a doublet of BrdUrd-positive cells unlabeled for NFP. (G) Hippocampal section from P18 wild-type mouse labeled with NeuN showing many positive neuronal cells in ca1. (H) Hippocampal section from P18 Ink4d, Kip1 double-null mouse labeled with NeuN showing a neuronal doublet (arrow) and a NeuN-positive cell undergoing mitosis (Inset). Bars: A and B = 200 μm; C = 20 μm; D = 200 μm; E = 16 μm; F = 12 μm; G = 200 μm; H = 20 μm; Insets: D = 9 μm, H = 5 μm.
Incorporation of BrdUrd alone identifies cells undergoing DNA replication but does not formally provide evidence that these cells are actually capable of division. We therefore examined phosphorylated HH3 expression, which is confined to cells in the G2 and M phases of the cell cycle. In general, cells in G2 phase exhibit punctate HH3 nuclear staining that changes to a more condensed pattern once they enter M phase (28). To visualize these effects in normal proliferating neurons, we studied the subventricular zone (SVZ) of the neocortex at E11.5, which exhibited the expected staining pattern (Fig. 4C). As also expected, HH3 staining was observed in the SVZ, in some cells in the rostromedial migratory stream, and in the dentate gyrus of wild-type animals at P18, regions where neuronal proliferation normally occurs (Fig. 4D Inset), but not in the pyramidal cells of the hippocampus (Fig. 4D). In brain sections from animals lacking both Ink4d and Kip1, HH3 staining was also seen in those regions that were labeled with BrdUrd, particularly within the pyramidal layer of the hippocampus (Fig. 4E), suggesting that the neuronal cells progressed through S phase into G2/M phase.
BrdUrd-positive mitotic figures and labeled cell doublets were observed in brains of Ink4d, Kip1-deficient mice (Fig. 4F). Because mitotic cells do not express NFP, we could not confirm their origin by costaining for this indicator. Therefore, we used an alternative neuron-specific marker, NeuN (29), which can detect neurons undergoing mitosis. As expected, NeuN-positive mitotic cells were not observed in the pyramidal cell layer of the hippocampus of wild-type animals (Fig. 4G), whereas sections from Ink4d−/−, Kip1−/− animals showed NeuN-positive mitotic figures and cell doublets (Fig. 4H). Therefore, we could document that, at P18, neurons in Ink4d, Kip1 doubly deficient mice completed S phase, progressed through G2 and M phase, and underwent cytokinesis.
Proliferation in the Brain of Ink4d Kip1 Double-Null Mice Is Balanced by Apoptosis.
Although many organs, including the brains, of Kip1-null mice are enlarged 4–6 wk after birth (1–3), comparison of brain weights and histology did not reveal evidence of organomegaly in younger Kip1−/− mice or in those lacking both Ink4d and Kip1. Moreover, no gross morphological anomalies were noted in the cytoarchitecture of the CNS of either strain. However, because neurons in many regions of the brain of Ink4d, Kip1 double-null mice continued to proliferate, we first performed cell counts in the hippocampus of the doubly deficient animals, comparing them to their wild-type counterparts. We saw no significant increase in the number of cells in any of the regions of the hippocampus (ca1, ca2/3) of the Ink4d−/−, Kip1−/− mice (data not shown).
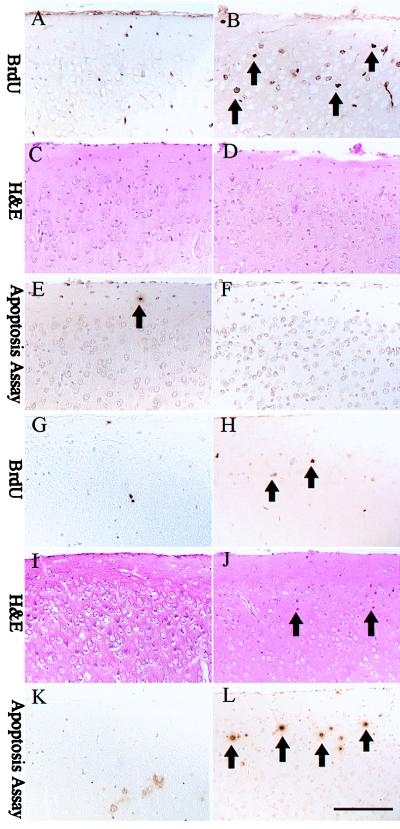
We next investigated whether cell division was offset by cell death at a time when neurological signs were evident (P18). Serial sections of brain from mice metabolically labeled with BrdUrd were stained with hematoxylin and eosin (H&E), which detects pyknotic nuclei, and were scored for apoptotic cells by using a colorimetric assay. At P14, a time when neurological signs were not fully advanced, layer II of the cerebral cortex of doubly deficient animals contained numerous BrdUrd-positive cells (Fig. 5B). H&E staining (Fig. 5D) or apoptotic assays (Fig. 5F) detected no significant cell death. The same was true in the hippocampus at P14 (data not shown). The cortical region of wild-type mice was normal (Fig. 5 A, C, and E). However, at P18, neurons within layer II of the cortex of the double-null mice were undergoing cell death, as determined by the presence of pyknotic nuclei (Fig. 5J) and positive apoptotic staining (Fig. 5L). Pyknotic cells were also present in the outer margins of the pyramidal layer of the hippocampus (data not shown). These data suggest that the neuronal proliferation observed at P14 is later balanced by cell death.
Figure 5.
Cortical sections of (Left) wild-type and (Right) Ink4d, Kip1 double-null mice taken at (A–F) P14 and (G–L) P18. At P14, Ink4d, Kip1 double-null mice show prominent BrdUrd labeling in layer II of the cortex (B) without apoptosis (D, F), whereas at P18, apoptosis is observed in the cortical layer II (J, L) with less BrdUrd labeling (H). Bars = 200 μm.
Discussion
The patterning and development of the nervous system depends on the ability of neuronal progenitors to divide and give rise to cells that exit the cycle, differentiate, and migrate to their proper positions in the brain. However, observations here that postmitotic neurons retain the capability to proliferate is counter to classical neuroscience dogma that dates back to Santiago Ramon y Cajal (30), who stated in 1913 that “in adult centers, the nerve paths are something fixed, ended, immutable. Everything may die, nothing may be regenerated.” By disabling genes encoding negative regulators of the cell division cycle, we found that codeletion of Ink4d and Kip1 can allow neurons to divide at a time when they would have normally exited the cell cycle. That CKIs actively function to maintain neurons in their quiescent state raises the possibility that terminal neuronal differentiation is a more plastic process than previously supposed and is, at least potentially, reversible.
Mice lacking both Kip1 and Ink4d were born with no gross abnormalities but developed bradykinesia, proprioceptive anomalies, and seizures through the early postnatal period. Before P14, the double-null mice were morphologically indistinguishable from their wild-type littermates. However, soon thereafter the animals became neurologically compromised and failed to thrive, and by P18, they exhibited seizure activity and labored respiration and ultimately died. On the basis of prospective examinations performed over the entire time course of their illness, we could not attribute the death of these mice to any specific anatomic anomaly. Bradykinesia, muscle weakness, and seizure activity may have contributed to their inability to secure nourishment, consistent with observations that, near the time of death, the animals appeared cachectic and dehydrated. Apart from a neurologic origin, wasting and dehydration can also result in cardiorespiratory distress (31) and contribute to the development of Cheynes-Stokes respiration (32, 33). Hence, death could have resulted from developing neurologic deficits, from indirect metabolic consequences, or both.
Apart from mild defects in male germ cell development, mice lacking Ink4d alone are fertile, show no other developmental anomalies, and enjoy a normal and uncomplicated life span (20). Although they also develop normally in utero, mice lacking Kip1 have an increased body size, which becomes clearly evident by 4–6 wk after birth (1–3). This is considerably later than the time at which mice deficient in both Ink4d and Kip1 expired. Organomegaly in Kip1-null animals appears to reflect a cell autonomous defect of p27Kip1 function itself rather than hormonal dyscrasia involving insulin-like growth factor or growth hormone. Increased organ size has therefore been attributed to a failure of many cell types to efficiently withdraw from the cell cycle. Females are sterile because of maturation arrest of ovarian follicles and failure to maintain a corpus luteum. The brains, like other organs of Kip1-null mice, contain increased cell numbers with no apparent distortion in cytoarchitecture. Although most organs appear otherwise normal, more subtle defects have been noted. The retinas of Kip1-null mice are hyperplastic and folded (3), whereas hair cells and supporting cells in the inner ear continue to be mitotically active as late as P6 (34, 35). Like mice heterozygous for the retinoblastoma (Rb) locus who lose the normal Rb allele in tumor tissues (36–38), Kip1-deficient animals also spontaneously develop middle-lobe pituitary adenomas, grossly evident by 2–3 months of age and progressing throughout their first year of life. Animals lacking Ink4c also exhibit organomegaly and pituitary tumors later in life in much the same way as Kip1-null mice, but those lacking both these genes develop pituitary tumors with a much earlier onset, indicating that Ink4c and Kip1 play overlapping roles in this organ (6). Thus, Kip1 may collaborate with different Ink4 family members in different cell lineages.
Previous analyses showed that Ink4d and Kip1 are coexpressed in similar regions of the developing brain, particularly in the cerebral cortex (16, 21). Mice deficient for both genes displayed abnormal proliferation of discrete populations of differentiated neurons. Pulse labeling with BrdUrd at P18 revealed widespread proliferation in several regions of the brain, including hippocampal pyramidal cells, the cerebral cortex, hypothalamus, pons, and brainstem. Proliferation in these regions is normally completed by E17.5 (24, 25). Because we pulsed with BrdUrd for 8 hr at P18 and sacrificed the animals immediately thereafter, it is unlikely that labeled progenitor cells could have divided and then migrated from germinal zones to their final positions in the brain during this interval. Hence, the onset of S phase likely occurred in postmigratory differentiated neurons. We confirmed that BrdUrd-positive cells were neurons by double labeling with NFP. On the basis of HH3 and NeuN staining, these cells progressed through G2 and M phase and underwent cytokinesis.
The cytoarchitecture and histology of the brains of the doubly deficient animals appeared unremarkable, suggesting that neuronal proliferation might be compensated by increased cell death. Compensation by apoptosis appeared to be true for the cerebral cortex and hippocampus, in the sense that regions that had manifested ectopic mitoses showed an increased apoptotic index at a later time. The onset of frank neurological deficits by P14 corresponded to the appearance of proliferating cells, whereas the timing of neuronal death appeared to be more closely correlated with mouse lethality. In cultured neurons, cell death induced by cyclin D1 overexpression can be inhibited by p16Ink4a (19), consistent with our results in living animals. On the other hand, inactivation of cyclin D2 led to cerebellar hypoplasia in specific lineages, because of defects in primary neurogenesis and terminal differentiation with increased apoptosis (39), implying that different D-type cyclins exert different effects in CNS development.
On the basis of the critical role that Ink4 and Cip/Kip inhibitors are thought to play in preventing Rb phosphorylation, a naive prediction might have been that the phenotype of Ink4d, Kip1 double-null animals would resemble that of Rb-deficient mice. Indeed, Rb-null fetuses exhibit enhanced proliferation associated with massive apoptosis of neurons, but they die because of defects in differentiation of the erythroid lineage by E14.5 of embryogenesis (36–38). In contrast, Ink4d, Kip1 double-null mice survived well beyond birth, while exhibiting neuronal apoptosis at a much later age. Because analysis of brain sections at P14 showed that neurons were not actively dying, the mechanisms governing neuronal apoptosis in Rb-null and Ink4d, Kip1 doubly null animals are likely to be different.
The phenotype of Ink4d, Kip1 double-null mice is unique and does not approximate that observed in mice lacking other CKIs, either alone or in combination (1–8). We therefore conclude that both these CKIs are rate limiting for neuronal terminal differentiation, and that other CKIs cannot substitute for the absence of these family members. We suggest that Ink4d and Kip1 are integral components of the molecular machinery that keeps postmitotic neurons from reentering the cell cycle after they migrate to their final position. Inhibition of the function of these CKIs in the adult brain may eventually provide an avenue for stimulating the growth of neuronal populations that are lost in degenerative diseases or through traumatic injury.
Acknowledgments
We thank Matthew Fero and Jim Roberts (Fred Hutchinson Cancer Research Center, Seattle) for the generous gift of the p27Kip1-null mice; Tom Curran, Jim DeCaprio (Dana Farber Cancer Center, Boston), and Jack Pettigrew (Vision, Touch and Hearing Research Center, Brisbane) for advice and helpful criticism; Zhen Lu and Willem den Besten for animal genotyping; and David Carey for handling of the animals. This work was supported in part by National Institutes of Health grants CA-71907 (M.F.R.), Cancer Center Core Grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital. C.J.S. is an Investigator of Howard Hughes Medical Institute.
Abbreviations
- CDK
cyclin-dependent kinase, CKI, CDK inhibitor, Rb, retinoblastoma protein
- Pn
postnatal day n, En, embryonic day n
- CNS
central nervous system
- NFP
neurofilaments protein
- HH3
histone H3
- NeuN
Neuron nuclei
- GFAP
glial fibrillary acidic protein
References
- 1.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L-H, Broudy V, Perlmutter R M, et al. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 2.Kiyokawa H, Kineman R D, Manova-Todorova K O, Soares V C, Hoffman E S, Ono M, Khanam D, Hayday A C, Frohman L A, Koff A. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh D Y. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 4.Yan Y, Frisen J, Lee M-H, Massagué J, Barbacid M. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 5.Zhang P, Liegeois N J, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, DePinho R A, Elledge S J. Nature (London) 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 6.Franklin D S, Godfrey V L, Lee H, Kovalev G I, Schoonhoven R, Chen-Kiang S, Su L, Xiong Y. Genes Dev. 1998;12:2899–2911. doi: 10.1101/gad.12.18.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P, Wong C, DePinho R, Harper J W, Elledge S J. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Wong C, Liu D, Finegold M, Harper J W, Elledge S J. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruas M, Peters G. Biochim Biophys Acta Reviews in Cancer. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 10.Sherr C, Roberts J M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 11.Soos T J, Kiyokawa H, Yan J S, Rubin M S, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A. Cell Growth Differ. 1996;7:135–146. [PubMed] [Google Scholar]
- 12.Blain S W, Montalvo E, Massagué J. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 13.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 14.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zindy F, Quelle D E, Roussel M F, Sherr C J. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 16.Zindy F, Soares H, Herzog K-H, Morgan J, Sherr C J, Roussel M F. Cell Growth Differ. 1997;8:1139–1150. [PubMed] [Google Scholar]
- 17.Caviness V S, Jr, Takahashi T, Nowakowski R S. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- 18.Freeman R S, Johnson E M., Jr Neuron. 1994;12:343–355. doi: 10.1016/0896-6273(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 19.Kranenburg O, van der Eb A J, Zantema A. EMBO J. 1996;15:46–54. [PMC free article] [PubMed] [Google Scholar]
- 20.Zindy, F., van Deursen, J., Grosveld, G., Sherr, C. J. & Roussel, M. F. (1999) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 21.Lee M-H, Nikolic M, Bapista C A, Lai E, Tsai L-H, Massagué J. Proc Natl Acad Sci USA. 1996;93:3259–3263. doi: 10.1073/pnas.93.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smeyne R J, Goldowitz D. J Neurosci. 1989;9:1608–1620. doi: 10.1523/JNEUROSCI.09-05-01608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Jr, Chao M V, Koff A. Genes Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman J, Bayer S A. J Comp Neurol. 1978;179:49–75. doi: 10.1002/cne.901790105. [DOI] [PubMed] [Google Scholar]
- 25.Stanfield B B, Cowan W M. In: The Development of the Hippocampal Region. Peters A, Jones E G, editors. Vol. 7. New York: Plenum; 1988. pp. 99–131. [Google Scholar]
- 26.Lee V M, Carden M J, Trojanowski J Q. J Neurosci. 1986;6:850–858. doi: 10.1523/JNEUROSCI.06-03-00850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bignami A, Dahl D. J Histochem Cytochem. 1977;25:466–469. doi: 10.1177/25.6.69656. [DOI] [PubMed] [Google Scholar]
- 28.Hendzel M J, Wei Y, Mancini M A, Hooser A V, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 29.Mullen R J, Buck C R, Smith A N. Development (Cambridge, UK) 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 30.Ramon y Cajal S, May R T. Degeneration and Regeneration of the Nervous System. II. New York: Hafner; 1959. p. 750. [Google Scholar]
- 31.Webb J G, Kless M C, Chan-Yan C C. Can Med Assoc J. 1986;135:753–758. [PMC free article] [PubMed] [Google Scholar]
- 32.Hanly P J, Zuberi-Kohokhar N S. Am J Respir Crit Care Med. 1996;153:272–276. doi: 10.1164/ajrccm.153.1.8542128. [DOI] [PubMed] [Google Scholar]
- 33.Lieber C, Mohsenin V. Yale J Biol Med. 1992;65:39–50. [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P, Segil N. Development (Cambridge, UK) 1999;126:1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- 35.Lowenheim H, Furness D N, Kil J, Zinn C, Gultig K, Fero M L, Frost D, Gummer A W, Roberts J M, Rubel E W, et al. Neurobiology. 1999;96:4084–4088. doi: 10.1073/pnas.96.7.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke A R, Maandag E R, van Roon M, van der Lugt N M T, van der Valk M, Hooper M L, Berns A, te Riele H. Nature (London) 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 37.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 38.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C J, Lai C-C, Herrup K, Lee W-H, Bradley A. Nature (London) 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 39.Huard J M, Forster C C, Carter M L, Sicinski P, Ross M E. Development (Cambridge, UK) 1999;126:1927–1935. doi: 10.1242/dev.126.9.1927. [DOI] [PubMed] [Google Scholar]