Abstract
The S phase-specific adaptor protein Claspin mediates the checkpoint response to replication stress by facilitating phosphorylation of Chk1 by ataxia-telangiectasia and Rad3-related (ATR). Evidence suggests that these components of the ATR pathway also play a critical role during physiological S phase. Chk1 is required for high rates of global replication fork progression, and Claspin interacts with the replication machinery and might therefore monitor normal DNA replication. Here, we have used DNA fiber labeling to investigate, for the first time, whether human Claspin is required for high rates of replication fork progression during normal S phase. We report that Claspin-depleted HeLa and HCT116 cells display levels of replication fork slowing similar to those observed in Chk1-depleted cells. This was also true in primary human 1BR3 fibroblasts, albeit to a lesser extent, suggesting that Claspin is a universal requirement for high replication fork rates in human cells. Interestingly, Claspin-depleted cells retained significant levels of Chk1 phosphorylation at both Ser317 and Ser345, raising the possibility that Claspin function during normal fork progression may extend beyond facilitating phosphorylation of either individual residue. Consistent with this possibility, depletion of Chk1 and Claspin together doubled the percentage of very slow forks, compared with depletion of either protein alone.
INTRODUCTION
Dividing cells are constantly challenged with DNA damage and other difficult conditions for DNA replication and chromosome segregation. DNA damage response pathways maintain genomic integrity by coordinating the regulation of cell cycle progression with DNA repair. During S phase, replication fork progression can be impaired by insufficient nucleotide supply or lesions and obstacles on the DNA template. This activates the DNA damage sensor kinase ataxia-telangiectasia and Rad3-related (ATR), which activates its major downstream effector kinase Chk1 by phosphorylation on the serine residues Ser317 and Ser345 (Liu et al., 2000; Zhao and Piwnica-Worms, 2001). Together, ATR and Chk1 suppress further origin firing and stabilize stalled replication forks. The ATR-Chk1 pathway is vital even in the absence of exogenous stresses, during an “unperturbed” cell cycle (Petermann and Caldecott, 2006). Our own findings demonstrate that Chk1 is required for high rates of replication fork progression during an unperturbed S phase (Petermann et al., 2006), and we and others have shown that inhibition or depletion of Chk1 in human cells leads to increased origin firing, accumulation of single-stranded DNA (Syljuasen et al., 2005; Maya-Mendoza et al., 2007), activation of the ATR pathway and the generation of DNA breaks (Syljuasen et al., 2005).
ATR depends on the S/G2 phase-specific adaptor protein Claspin to efficiently phosphorylate Chk1 in Xenopus and several human cell lines (Kumagai and Dunphy, 2000; Chini and Chen, 2003; Lin et al., 2004; Chini et al., 2006; Liu et al., 2006). Human Claspin is constitutively associated with ATR, and phosphorylation of Claspin, presumably by ATR, facilitates its interaction with Chk1 (Chini and Chen, 2003). This interaction with Chk1, which occurs via the active site of Chk1, is required for the promotion of Chk1 phosphorylation (Lee et al., 2005). Chk1, in turn, stabilizes Claspin in HeLa cells (Chini et al., 2006). There is evidence for a role of Claspin during normal DNA replication. Claspin is a ring-shaped protein that binds to replication fork structures (Sar et al., 2004). Xenopus Claspin binds to chromatin in a pre-replicative complex (RC) and Cdc45-dependent manner, suggesting binding to origins during unwinding, and it interacts with several replication proteins, including RPA, RFC, and Cdc45 (Lee et al., 2003, 2005). Depletion of Claspin from Xenopus egg extracts leads to modestly reduced DNA synthesis rates in vitro (Lee et al., 2003). In human cells, overexpression or small interfering RNA (siRNA)-mediated depletion of Claspin increases or reduces, respectively, cell proliferation and the percentage of 5-bromo-2′-deoxyuridine (BrdU)-positive cells (Lin et al., 2004). Similar to depletion of Chk1, depletion of human Claspin induces DNA damage (Liu et al., 2006). Finally, Mrc1, the functional homologue of Claspin in Saccharomyces cerevisiae, travels with the replication fork (Osborn and Elledge, 2003), and it is required for normal fork progression rates during unperturbed S phase. Replication forks in Mrc1Δ cells progress at ∼50–70% of the normal rate (Szyjka et al., 2005; Tourriere et al., 2005; Hodgson et al., 2007).
Here, we have used DNA fiber labeling to examine, for the first time, whether Claspin is required for normal replication fork progression rates in human cells. We conclude that Claspin is required for high rates of fork progression during normal human S phase, and we suggest that this requirement may extend beyond the reported role of Claspin in mediating the phosphorylation of Chk1 at Ser317 and Ser345.
MATERIALS AND METHODS
Cell Lines
Human HeLa adenocarcinoma cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), human HCT116 colon carcinoma cells were grown in DMEM supplemented with 10% FCS, and 1BR3 primary human fibroblasts were grown in minimal essential medium supplemented with 15% FCS. All cell lines were grown in a humidified CO2 atmosphere at 37°C.
DNA Fiber Experiments
To knock down human Claspin, we used an siRNA duplex oligonucleotide (QIAGEN, Hilden, Germany) directed against the Claspin target sequence (sense): CCUUGCUUAGAGCUGAGUC (Chini et al., 2006). To knock down human Chk1, we used siRNA duplex oligonucleotides (Invitrogen, Paisley, United Kingdom) directed against the Chk1 target sequence (sense) UCGUGAGCGUUUGUUGAAC (Zhu et al., 2004). siRNA duplex oligonucleotides directed against the firefly luciferase target sequence CUUACGCUGAGUACUUCGA (Hoek and Stillman, 2003) were used as a control. siRNA duplexes (10 nM) were transfected using siPORT NeoFX reverse transfection reagent (Ambion, Austin, TX) (0.25 μl reagent/104 cells, at a cell concentration of 1 × 105/ml). 1BR3 cells were transfected with 50 nM siRNA duplex and 1.2 μl of reagent/104 cells, at a cell concentration of 2.5 × 104/ml.
Twenty-four hours after siRNA transfection (or 48 h after transfection for 1BR3 cells), exponentially growing cells were pulse labeled with 25 μM BrdU for 15 min followed by 250 μM iododeoxyuridine (IdU) for 20 min, as indicated. Labeled cells were harvested, and DNA fiber spreads prepared as described previously (Petermann et al., 2006). For immunodetection of BrdU-labeled tracts, acid treated fiber spreads were incubated with rat anti-BrdU monoclonal antibody (mAb) (Oxford Biotechnology, Oxon, United Kingdom) at 1:600 dilution for 1 h at room temperature. Slides were then fixed with 4% paraformaldehyde and incubated with an AlexaFluor 488-conjugated donkey anti-rat immunoglobulin G (IgG) (Invitrogen) at 4 μg/ml for 1.5 h at room temperature. To detect both BrdU- and IdU-labeled patches, a mouse mAb that recognizes both IdU and BrdU (BD Biosciences, Franklin Lakes, NJ) was used at 17 ng/ml over night at 4°C, followed by an AlexaFluor 555-conjugated goat anti-mouse IgG (Invitrogen) at 4 μg/ml for 1.5 h at room temperature. Fibers were examined with an LSM 510 confocal microscope (Carl Zeiss, Jena, Germany) using a 100× (1.4 numerical aperture) lens. The lengths of BrdU- (Alexa Fluor [AF] 488, green) and IdU (AF 555, red)-labeled patches were measured using the LSM software (Carl Zeiss), and micrometer values were converted into kilobase using the conversion factor 1 μm = 2.59 kb (Henry-Mowatt et al., 2003). Measurements were recorded from fibers in well spread (untangled) areas of the slides to prevent the possibility of recording labeled patches from bundles of fibers.
Immunoblotting
Cells were transfected with 20 nM Claspin siRNA or ON-TARGETplus siCONTROL Nontargeting Pool (Dharmacon RNA Technologies, Lafayette, CO) as a control using Dharmafect 1 transfection reagent (Dharamcon RNA Technologies) (0.6 μl reagent/104 cells, at a cell concentration of 5 × 104/ml). Forty hours after transfection, cells were treated with 10 mM hydroxyurea (HU) for the times indicated. Afterward, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 5% Na-deoxycholate, and 0.1% SDS) containing 1× protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland) and 1× phosphatase inhibitor cocktail 2 (Sigma-Aldrich, Poole, Dorset, United Kingdom). Lysates from 5.6 × 104 cells per lane were resolved by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Total Claspin was detected using a rabbit polyclonal anti-Claspin antibody, a kind gift from Dr. Junjie Chen (Yale School of Medicine, New Haven, CT), at a 1:1000 dilution. Total Chk1 was detected using rabbit polyclonal anti-Chk1 antibody (Cell Signaling Technology, Danvers, MA) at a dilution of 1:300. Chk1 phosphorylated at Ser345 or Ser317 was detected using rabbit polyclonal anti-phospho-Ser345 or rabbit polyclonal anti-phospho-Ser317 antibody at a dilution of 1:300 (Cell Signaling Technology). All incubations with primary antibodies were performed at 4°C overnight. Band intensities were quantified by densitometry by using the ImageJ software (http://rsb.info.nih.gov/ij/).
RESULTS
HeLa cells were transfected with siRNA duplexes directed against human Claspin or human Chk1, or against luciferase as a control (Figure 1A). Twenty-four hours later, the cells were pulse labeled for 15 min with 25 μM BrdU followed by 250 μM IdU for 20 min, and the length of the labeled DNA replication tracts in DNA fiber spreads was then quantified by immunofluorescence. A visual comparison of DNA fibers from control- and Claspin siRNA-transfected cells revealed a striking difference in the overall length of their replication tracts (Figure 1B). When the distribution of fork rates within populations of forks was quantified and plotted, the entire distribution of fork rates in Claspin-depleted cells shifted leftward, to slower fork rates, during both pulse labels (Figure 1C). The extent of this leftward shift was the same in Claspin- and Chk1-depleted cells (Figure 1D). Mean fork rates obtained from these experiments are listed in Table 1. These data suggest that in Claspin-depleted cells, as in Chk1-depleted cells, replication forks progress at a slower rate than in wild-type cells.
Figure 1.
Comparison of replication fibers and fork rates in HeLa cells depleted of Claspin and Chk1. (A) Claspin, Chk1, and XRCC1 (control) levels in total cell extract from HeLa cells treated with luciferase (control), Claspin, or Chk1 siRNA for 24 h. (B) Representative images of replication tracts from control (luciferase)-, Claspin-, or Chk1-depleted HeLa cells pulse labeled with 25 μM BrdU for 15 min (green track) followed by 250 μM IdU for 20 min (red track), and then processed for DNA fiber spreads as described in Materials and Methods. Fork direction is indicated by a black arrow and the junction between pulse labels is indicated by white arrows. (C) Distribution of replication fork rates in control- or Claspin-depleted HeLa cells pulse labeled and processed as described above. (D) Distribution of replication fork rates in Chk1- or Claspin-depleted HeLa cells. For each panel, the distribution of fork rates during the entire labeling period is shown. Data bars are the mean of three independent experiments, with similar results observed in each, and error bars represent 1 SD. The total number of forks scored for each is indicated in parentheses.
Table 1.
Comparison of mean fork rates in control-, Claspin-, or Chk1-siRNA–treated human cell lines
| siRNA | Mean fork rates in kb/min (±1 SD) |
||
|---|---|---|---|
| HeLa | HCT116 | 1BR3 | |
| Control | 0.81 (0.09) | 0.98 (0.03) | 0.73 (0.02) |
| Claspin | 0.60 (0.08) | 0.66 (0.02) | 0.63 (0.08) |
| Chk1 | 0.63 (0.04) | 0.78 (0.08) | 0.57 (0.02) |
| Claspin + Chk1 | 0.49 (0.07) | N.D.a | N.D. |
Claspin- or Chk1-depleted cells were pulse labeled with 25 μM BrdU for 15 min followed by 250 μ M IdU for 20 min, and then they were processed for DNA fiber spreads. Replication fork rates were measured and the mean fork rates were calculated. Data are combined from two independent experiments or three independent experiments for HeLa cells.
a N.D., not determined.
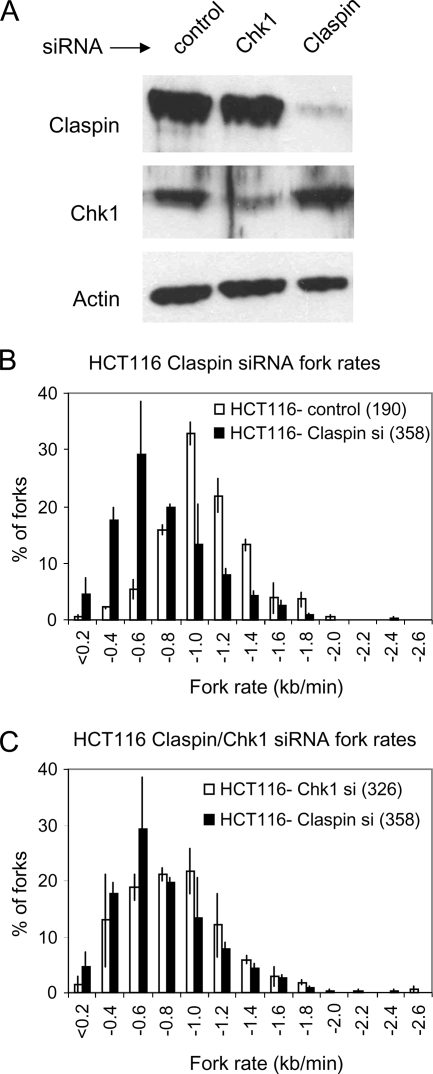
We next examined whether Claspin and Chk1 were also required for high rates of replication fork progression in a different human cancer cell line, HCT116 colon carcinoma cells. HCT116 cells were mock transfected or transfected with Claspin or Chk1 siRNA (Figure 2A). Twenty-four hours later, cells were pulse labeled as described above, and fork rate distributions were quantified and plotted. Claspin depletion strongly reduced overall fork progression rates in HCT116 cells (Figure 2B). The reduction in fork rates was slightly more pronounced in Claspin- compared with Chk1-depleted HCT116 cells (Figure 2C and Table 1), which might be due to the very efficient depletion of Claspin in this cell line (Figure 2A).
Figure 2.
Replication fork rates in HCT116 cells depleted of Claspin and Chk1. (A) Claspin, Chk1, and actin levels in total cell extract from HCT116 (p53+/+) cells mock treated or treated with Chk1 or Claspin siRNA for 24 h. (B) Distribution of replication fork rates in control- or Claspin-depleted HCT116 cells pulse labeled and processed as described above. (C) Distribution of replication fork rates in Chk1- or Claspin-depleted HCT116 cells. For each panel, the distribution of fork rates during the entire labeling period is shown. Data are combined from two independent experiments. In one experiment, mock transfection was used as a control, and in the other experiment, cells were treated with luciferase siRNA as control. For each data set, the similar results were observed in each experimental repeat. The total number of forks scored for each cell line is indicated in parentheses.
Both HeLa and HCT116 cells are cancer cell lines; therefore, they might have altered checkpoint pathways and replication machineries as a part or result of tumorigenesis. We therefore analyzed whether Claspin and Chk1 are also required for the maintenance of high rates of replication fork progression in primary human cells. 1BR3 primary human fibroblasts were control transfected or transfected with Claspin or Chk1 siRNA duplexes (Figure 3A). Forty-eight hours later, cells were pulse labeled as described above, and fork rate distributions were quantified and plotted. Claspin depletion had a very mild effect on overall fork progression rates in 1BR3 cells (Figure 3B). This reduction in fork rates was slightly more pronounced in Chk1-depleted cells than in Claspin-depleted 1BR3 cells (Figure 3C and Table 1). These data suggest that both Claspin and Chk1 are required for high rates of replication fork progression in many human cell lines, including primary cells, although the requirement is less pronounced in primary fibroblasts. The requirement for Claspin and Chk1 is regardless of p53 status, because both HCT116 and 1BR3 cells are proficient in p53, whereas HeLa cells are p53 deficient.
Figure 3.
Replication fork rates in primary human skin fibroblasts (1BR3) depleted of Claspin and Chk1. (A) Claspin, Chk1, and XRCC1 (control) levels in total cell extract from 1BR3 cells treated with luciferase (control), Chk1, or Claspin siRNA for 48 h. (B) Distribution of replication fork rates in control- or Claspin-depleted 1BR3 cells pulse labeled and processed as described above. (C) Distribution of replication fork rates in Chk1- or Claspin-depleted 1BR3 cells. For each panel, the distribution of fork rates during the entire labeling period is shown. Data are combined from two independent experiments. For each data set, the similar results were observed in each experimental repeat. The total number of forks scored for each cell line is indicated in parentheses.
Claspin was first described as a mediator of Chk1 phosphorylation by ATR, and the similar phenotypes of Claspin- and Chk1-depleted cells might arise because Claspin functions upstream of Chk1, facilitating its phosphorylation by ATR. We therefore analyzed whether replication stress-induced phosphorylation of Chk1 was impaired in Claspin-depleted cells. HeLa or HCT116 cells were transfected with Claspin or control siRNA and 40 h after transfection, cells were treated with HU for 1–6 h (Figure 4). Phosphorylation of Chk1 was analyzed by immunoblotting using specific antibodies directed against Chk1 phosphorylated on Ser317 or Ser345 (Figure 4). Claspin depletion decreased phosphorylation of Chk1 at Ser317 and Ser345 in both HeLa (Figure 4A) and HCT116 cells (Figure 4B). Phosphorylation at Ser317 was decreased by <50% and phosphorylation at Ser345 was decreased by up to 70% in Claspin-depleted cells. In agreement with observations by Chini et al. (2006), we observed decreased hyperphosphorylation of Chk1 in Claspin-depleted cells (Figure 4, A and B, supershifted bands indicated by arrows with asterisks). Thus, Claspin depletion has a greater impact on phosphorylation of Chk1 at Ser345 than at Ser317, which is in good agreement with observations in U2OS cells (Liu et al., 2006), but significant phosphorylation at both sites remains in Claspin-depleted cells.
Figure 4.
Effects of Claspin depletion on Chk1 phosphorylation in HeLa and HCT116 cells treated with the replication inhibitor hydroxyurea. HeLa cells (A) and HCT116 cells (B) were treated with nontargeting siRNA pool or Claspin siRNA for 40 h, and then they were treated or not with 10 mM HU for 1, 2, 4, and 6 h as indicated. Claspin and Chk1 levels and phosphorylation of Chk1 on Ser317 and Ser345 were visualized using specific antibodies. Phosphorylated bands of Chk1 are denoted by arrows, and hyperphosphorylated bands are denoted by arrows with asterisks. Relative phosphorylated Chk1 levels were determined by densitometry and normalized to total Chk1 levels. Subsequently, phosphorylated Chk1 levels in Claspin-depleted cells were normalized to phosphorylated Chk1 levels in control cells.
To analyze further the relationship between Claspin and Chk1 during normal replication fork progression, we cotransfected HeLa cells with Claspin and Chk1 siRNA to knock down both proteins simultaneously (Figure 5A). Comparison of the fork rate distributions between Claspin only- and Claspin- and Chk1-depleted cells revealed that simultaneous knockdown of both proteins increased the percentage of very slow-progressing forks (<400 base pairs/min) about twofold (Figure 5B). A comparison of fork rate distributions between Chk1- and double-depleted cells yielded the same result (Figure 5C). Quantification of depletion efficiencies for Claspin and Chk1 confirmed that this increase in very slow forks was not due to more efficient depletion of either protein in double-depleted samples (Figure 5A). These data are consistent with Claspin and Chk1 fulfilling distinct, but partially overlapping, roles during the maintenance of normal replication fork rates.
Figure 5.
Comparison of replication fork rates in HeLa cells depleted of both Claspin and Chk1 and depleted of Claspin or Chk1 alone. (A) Claspin, Chk1 and XRCC1 (control) levels in total cell extract from HeLa cells treated with luciferase siRNA (control), Chk1 siRNA, Claspin siRNA, or both Claspin and Chk1 siRNA for 24 h. Relative Claspin and Chk1 levels were determined by densitometry and normalized to levels in control cells. Depletion efficiencies are given for the blot shown and as averages of all three independent experiments. (B) Distribution of replication fork rates in Claspin and Chk1- or Claspin alone-depleted HeLa cells processed as in Figure 1. (C) Distribution of replication fork rates in Claspin and Chk1- or Chk1 alone-depleted HeLa cells. Data bars are the mean of three independent experiments, with similar results observed in each, and error bars represent 1 SD.
DISCUSSION
Here, we report that Claspin is required for normal rates of replication fork progression in human cells. Claspin has repeatedly been proposed to play a role in monitoring DNA replication for many reasons. Claspin binding to chromatin is dependent on the pre-RC and Cdc45, which is similar to the chromatin binding behavior of DNA polymerase ε and which suggests binding during the initial unwinding step (Lee et al., 2003). Depletion of Claspin from Xenopus egg extracts decreases in vitro replication rates (Lee et al., 2003), and Mrc1, the functional Claspin homologue in S. cerevisiae, is required for normal fork progression rates (Szyjka et al., 2005; Tourriere et al., 2005). Here we show that Claspin is indeed critical for normal replication fork progression, suggesting that it acts as a replication factor. This is the first demonstration of a role for Claspin in maintaining normal fork rates in vertebrate cells.
Claspin, as well as Chk1 depletion, affects replication fork rates in a variety of human cell lines, including p53-deficient and -proficient cells and primary fibroblasts (Figures 1–3). The effect on replication fork rates in primary fibroblasts is however small (Figure 3), and we speculate that the requirement for Claspin and Chk1 for normal fork progression is more pronounced in fast-growing cell lines with high global fork progression rates (1BR3 cells display relatively slow mean fork rates; Table 1). Alternatively, this result could reflect differences in depletion efficiency and basal Claspin levels in the cell lines used. Interestingly, lack of Bloom syndrome helicase (BLM) was recently reported to result in slow fork progression. This replication slowing in BLM-deficient cells was similar in primary and transformed fibroblasts, suggesting that BLM might fulfill a function that is more routinely required during normal replication, in primary fibroblasts at least, than that fulfilled by Claspin and Chk1 (Rao et al., 2007).
We demonstrated previously that Chk1 maintains high rates of replication fork progression in HeLa and chicken DT40 cells (Petermann et al., 2006). Subsequently, it was shown that Chk1 is required for Claspin stability in HeLa cells and that Claspin levels decrease after 72 h of Chk1 siRNA treatment (Chini et al., 2006). We therefore investigated the possibility that the reduced fork speeds we observed in Chk1-depleted HeLa cells might be due to lack of Claspin. However, Chk1-depleted HeLa cells, as well as HCT116 and 1BR3 cells, retain normal levels of Claspin under our conditions, which include a maximal 48 h of siRNA treatment (Figures 1–3). Reduced fork speeds in Chk1-depleted cells are therefore not due absence of Claspin.
We also considered the possibility that reduced fork rates in Claspin-depleted cells reflect its role in facilitating ATR-mediated phosphorylation of Chk1 at Ser317 and Ser345 (Kumagai and Dunphy, 2000; Chini and Chen, 2003; Chini et al., 2006). Although we observed decreased Chk1 phosphorylation at Ser317 and Ser345 in Claspin-depleted cells, significant residual phosphorylation did still occur, especially at Ser317 (Figure 4). This might be due to incomplete depletion of Claspin, or to redundancy of Claspin with other mediators of Chk1 phosphorylation such as BRCA1 (Lin et al., 2004; Yoo et al., 2006). The more pronounced effect of Claspin depletion on phosphorylation at Ser345 and hyperphosphorylation suggests that Claspin mainly facilitates efficient Chk1 phosphorylation at sites other than Ser317. Although we cannot rule out that the partial reduction in Chk1 phosphorylation (50–70%) is sufficient to account for the slower fork rates observed in Claspin-depleted cells, we favor the possibility that Claspin fulfills one or more additional roles during the maintenance of normal fork rates. It is also noteworthy in this respect that depletion of Chk1 and Claspin together doubled the percentage of very slow forks compared with depletion of each protein alone, supporting the possibility that Claspin maintains normal fork rates through mechanisms other than, or in addition to, Chk1 function (Figure 5). A similar phenomenon has been observed in budding yeast, where the checkpoint function of Mrc1 is not required for its role in normal replication fork progression (Osborn and Elledge, 2003). Interestingly, high Chk1 activity has been found to reduce rather than promote replication fork progression (Seiler et al., 2007), whereas Chk1 promotes fork rates in unperturbed cells when Chk1 phosphorylation and basal activity are low. These observations further support the idea that Claspin promotes fork progression independently of Chk1 phosphorylation.
What might be the mechanisms by which Claspin and Chk1 facilitate replication fork progression? We previously suggested a role for Chk1 in maintaining fork stability based on the observation that Chk1 deficiency reduces fork rates more strongly during the second pulse label, indicating increased irreversible or prolonged fork stalling (Petermann et al., 2006). Our current experiments did again show a greater impact of Claspin or Chk1 depletion during the second pulse label, although this effect was not equally pronounced in all cell lines analyzed (data not shown). Claspin might thus also facilitate replication fork progression by preventing prolonged fork stalling. Currently, we cannot discriminate between an impact of Claspin on fork stabilization, fork restart, or both. However, in budding yeast, Mrc1 is not required to prevent fork collapse during HU treatment, but for restart of stalled replication forks (Tourriere et al., 2005). This suggests that Mrc1 acts together with the replication machinery to facilitate proper replication fork progression, rather than facilitating the repair of damaged replication forks. Claspin may play a similar role, an idea that might be supported by the observation that Claspin seems to bind chromatin during origin unwinding, suggesting that it travels with all replication forks (Lee et al., 2003, 2005).
In summary, we show here that Claspin is required to maintain high rates of replication fork progression in human cells during normal, unperturbed S phase. In addition, we suggest that this requirement may extend beyond the role of Claspin in mediating the phosphorylation of Chk1 at Ser317 and Ser345. Several human cancer cell lines overexpress Claspin (Lin et al., 2004), and it is possible that the role of this important protein in regulating replication fork rates may impact greatly on genome stability.
ACKNOWLEDGMENTS
We thank Dr. Junjie Chen for the generous gift of Claspin antibodies and Dr. Elizabeth Madgett for optimizing the DNA fiber technique. We thank the Medical Research Council (T.H.) for additional funding. E.P. was primarily supported by Biotechnology and Biological Sciences Research Council grant BBS/B/02967 (to K.W.C.).
Abbreviations used:
- BrdU
5-bromo-2′-deoxyuridine
- IdU
Iododeoxyuridine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1035) on March 19, 2008.
REFERENCES
- Chini C. C., Chen J. Human claspin is required for replication checkpoint control. J. Biol. Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- Chini C. C., Wood J., Chen J. Chk1 is required to maintain claspin stability. Oncogene. 2006;25:4165–4171. doi: 10.1038/sj.onc.1209447. [DOI] [PubMed] [Google Scholar]
- Henry-Mowatt J., Jackson D., Masson J. Y., Johnson P. A., Clements P. M., Benson F. E., Thompson L. H., Takeda S., West S. C., Caldecott K. W. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell. 2003;11:1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- Hodgson B., Calzada A., Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol. Biol. Cell. 2007;18:3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M., Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Lee J., Gold D. A., Shevchenko A., Shevchenko A., Dunphy W. G. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Li K., Stewart G. S., Elledge S. J. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. USA. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A., Petermann E., Gillespie D. A., Caldecott K. W., Jackson D. A. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Caldecott K. W. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle. 2006;5:2203–2209. doi: 10.4161/cc.5.19.3256. [DOI] [PubMed] [Google Scholar]
- Petermann E., Maya-Mendoza A., Zachos G., Gillespie D. A., Jackson D. A., Caldecott K. W. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. A., Conti C., Guirouilh-Barbat J., Nakamura A., Miao Z. H., Davies S. L., Sacca B., Hickson I. D., Bensimon A., Pommier Y. Endogenous {gamma}-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol. Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- Sar F., Lindsey-Boltz L. A., Subramanian D., Croteau D. L., Hutsell S. Q., Griffith J. D., Sancar A. Human claspin is a ring-shaped DNA-binding protein with high affinity to branched DNA structures. J. Biol. Chem. 2004;279:39289–39295. doi: 10.1074/jbc.M405793200. [DOI] [PubMed] [Google Scholar]
- Seiler J. A., Conti C., Syed A., Aladjem M. I., Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell Biol. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syljuasen R. G., Sorensen C. S., Hansen L. T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjka S. J., Viggiani C. J., Aparicio O. M. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol. Cell. 2005;19:691–697. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Tourriere H., Versini G., Cordon-Preciado V., Alabert C., Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Jeong S. Y., Dunphy W. G. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 2006;20:772–783. doi: 10.1101/gad.1398806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Chen Y., Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]