Abstract
The Cdc14p-like phosphatase Flp1p (also known as Clp1p) is regulated by cell cycle-dependent changes in its subcellular localization. Flp1p is restricted to the nucleolus and spindle pole body until prophase, when it is dispersed throughout the nucleus, mitotic spindle, and medial ring. Once released, Flp1p antagonizes Cdc2p/cyclin activity by reverting Cdc2p-phosphorylation sites on Cdc25p. On replication stress, ataxia-telangiectasia mutated/ATM/Rad3-related kinase Rad3p activates Cds1p, which phosphorylates key proteins ensuring the stability of stalled DNA replication forks. Here, we show that replication stress induces changes in the subcellular localization of Flp1p in a checkpoint-dependent manner. Active Cds1p checkpoint kinase is required to release Flp1p into the nucleus. Consistently, a Flp1p mutant (flp1-9A) lacking all potential Cds1p phosphorylation sites fails to relocate in response to replication blocks and, similarly to cells lacking flp1 (Δflp1), presents defects in checkpoint response to replication stress. Δflp1 cells accumulate reduced levels of a less active Cds1p kinase in hydroxyurea (HU), indicating that nuclear Flp1p regulates Cds1p full activation. Consistently, Δflp1 and flp1-9A have an increased percentage of Rad22p-recombination foci during HU treatment. Together, our data show that by releasing Flp1p into the nucleus Cds1p checkpoint kinase modulates its own full activation during replication stress.
INTRODUCTION
Adequate genome replication and transfer to daughter cells is compromised in the course of each cell cycle by both intra- and extracellular factors that perturb the stability of the genome. Molecular mechanisms to ensure genome integrity and to cope with DNA harassment were developed in eukaryotes likely at early stages of evolution, and they are consequently conserved. Therefore, a replication checkpoint ensures genomic integrity and cell survival when cells are not able to correctly replicate their genetic material, due to limitation in nucleotide pools or to deleterious alterations in template DNA.
Serine/threonine phosphatase Flp1p (cdc fourteen-like phosphatase, also called Clp1p; hereafter referred as Flp1p) has been proved to control rapid degradation of Cdc25p at the end of mitosis (Esteban et al., 2004; Wolfe and Gould, 2004), which results in enhanced inhibitory Y15 phosphorylation of Cdc2p and in the corresponding loss of kinase activity necessary for mitotic exit. Flp1p is required for the ubiquitination of Cdc25p by the anaphase promoting complex/cyclosome at this cell cycle stage, and cells deleted for flp1+ present higher basal levels of Cdc25p. Interestingly, this role may be conserved in higher eukaryotes as hCdc14Ap is involved in the cell cycle regulation of hCdc25Ap stability through dephosphorylation of Serines 115 and 329 of the mitotic inducer in human cells (Esteban et al., 2006). In fission yeast, Δflp1 cells enter mitosis at a reduced cell size, presenting a wee phenotype (Cueille et al., 2001; Trautmann et al., 2001). Overexpression of Flp1p arrests cells in G2 with dephosphorylated Cdc25p, in a process dependent on active Wee1p (Cueille et al., 2001; Esteban et al., 2004). Beside its role in cell cycle progression, Flp1p is required for an efficient cytokinesis checkpoint (Cueille et al., 2001; Trautmann et al., 2001; Trautmann and McCollum, 2005) and for faithful chromosome segregation (Trautmann et al., 2004).
To carry out its distinct functions, Flp1p must be correctly localized within the cell. In the course of a normal cell cycle, Flp1p localizes in the nucleolus and spindle pole bodies in interphase. In prophase, it is released from the nucleolus by an unknown mechanism to occupy the nucleus, medial ring, and mitotic spindle (Cueille et al., 2001; Trautmann et al., 2001; Chen et al., 2006). Although the release of Flp1p from the nucleolus does not depend on the Septation Initiation Network (SIN), its SIN-dependent maintenance in the cytoplasmatic locations is necessary for the cytokinesis checkpoint (Cueille et al., 2001; Mishra et al., 2005). In fact, it has been shown that Flp1p binds to the 14-3-3 protein Rad24p to be retained in the cytoplasm in response to cytokinesis defects (Mishra et al., 2005; Trautmann and McCollum, 2005). Moreover, localization of Flp1p in the kinetochores and mitotic spindle during mitosis is necessary to prevent defects in chromosome segregation (Trautmann et al., 2004).
In fission yeast, the central sensor of both the replication and the DNA-damage checkpoints is Rad3p, a kinase related to mammalian ataxia-telangiectasia mutated and ATM/Rad3-related checkpoint kinases. Replication stress and DNA-damage inflicted during S phase leads to activation of the checkpoint kinase Cds1p (Murakami and Okayama, 1995; Lindsay et al., 1998; Brondello et al., 1999; Xu et al., 2006), whereas DNA-damage activates the checkpoint kinase Chk1p during G2 phase (Walworth et al., 1993; Brondello et al., 1999). Although checkpoint response is branched in two different pathways, both of them are closely related (Boddy et al., 1998; Rhind and Russell, 1998a). Thus, Chk1p is activated by replication stress in cells deleted for cds1+ (Boddy et al., 1998). Moreover, replication fork arrest is thought to lead to DNA-damage that is repaired in G2, which explains the sensitivity of Δchk1 cells to the ribonucleotide reductase inhibitory drug hydroxyurea (HU).
Activation of Cds1p and/or Chk1p delays entry into mitosis with unreplicated or damaged DNA through the inhibition of Cdc25p and enhancement of Mik1p activity (Furnari et al., 1997; Rhind et al., 1997; Boddy et al., 1998; Furnari et al., 1999; Rhind and Russell, 2001). Apart from interfering with cell cycle progression upon genomic threat, Cds1p has essential functions that Chk1p is not able to perform. Active Cds1p prevents stalled DNA replication forks from collapsing in response to replication stress. Significantly, Swi1p, Swi3p, and Cds1p are required to maintain stalled DNA replication forks in a competent state (Noguchi et al., 2003; Noguchi et al., 2004; Matsumoto et al., 2005). Loss of this specific replication arrest recovery function is patent in Δcds1 cells treated with HU, which experience checkpoint block because of Chk1p activation but rapidly loose viability (Boddy et al., 1998; Lindsay et al., 1998; Sommariva et al., 2005). Importantly, Cds1p interacts with Rad3p-phosphorylated Mrc1p; thus, it is recruited to replication forks (Xu et al., 2006), where (it) phosphorylates key proteins (like Mus81p and Mcm4p) to stabilize stalled forks during replication stress, thus, preserving genome integrity (Kai et al., 2005; Bailis et al., 2008).
Stalled forks represent serious threats to genomic integrity because they are prone to collapse or rearrangement (McGlynn and Lloyd, 2002a,b,c). Besides its role in stabilizing stalled forks, Cds1p kinase activity prevents unwanted recombinational events at stalled replication forks through regulation of Mus81p–Eme1p endonuclease complex, Rqh1p helicase and Rad60p. The extent of Cds1p activation determines the degree in which cells tolerate replication stress (Kai et al., 2005). When Cds1p activation is high, recombination repair processes are avoided, in part by means of Mus81p phosphorylation and its dissociation from chromatin. However, when the activation of the checkpoint kinase is low, Mus81p remains chromatin associated and deletions of genomic sequences occur to tolerate replication stress (Kai et al., 2005).
The present work concentrates in the control mechanisms or checkpoints activated by unreplicated DNA in the fission yeast Schizosaccharomyces pombe and the implication of the phosphatase Flp1p. In this report we propose a role for Cds1p in controlling Flp1p during replication stress. On replication stress, Flp1p changes its subcellular localization by a regulatory mechanism controlled by Cds1p checkpoint kinase. Significantly, Flp1p interacts in vivo with and is an in vitro substrate of Cds1p checkpoint kinase. We also show in vivo evidence indicating that Cds1p protein levels and kinase activity are regulated by the Cdc14p-phosphatase homologue Flp1p. Together, our results provide evidence indicating that Cds1p regulates the release of Flp1p from the nucleolus and that this regulatory step is a key event in the full activation of the checkpoint response to replication stress.
MATERIALS AND METHODS
Yeast Strains, Cell Culture, and Flow Cytometry
All strains used and their procedence are specified in Supplemental Table 1. Standard molecular biology and genetic methods were used for manipulation and construction of new strains (Moreno et al., 1991). Cultures were grown in yeast extract (YE) appropriately supplemented (YES media: yeast extract plus supplements [225 mg/l adenine, histidine, leucine, uracil, and lysine hydrochloride]).
S. pombe cells were incubated at 30°C unless otherwise specified. Strains containing repressible nmt1-plasmids were grown in minimal medium (MM) appropriately supplemented (225 mg/l adenine and uracil) containing thiamine. Induction was performed by washing exponentially growing cultures and resuspending in medium without thiamine as reported previously (Cueille et al., 2001).
cdc10-129 or cdc10-M17 and cdc17-K12 strains were grown asynchronously at the permissive temperature (25°C) and shifted to 36.5°C to arrest cells at G1 and S phase, respectively.
Drug treatment was performed in exponentially cultures growing in YES or MM appropriately supplemented for flow cytometry, ∼107 cells were collected by centrifugation, washed once with water, and fixed in 70% ethanol and processed as described previously (Moreno et al., 1991).
Imaging of Cells
In vivo fluorescence microscopy of green fluorescent protein (GFP)-, yellow fluorescent protein (YFP)-, and red fluorescent protein (RFP)-tagged strains and Hoechst or 4′,6-diamidino-2-phenylindole (DAPI) staining was performed in a Leica DM 6000B microscope (63× objective; 1,32 Oil Plan-APO) equipped with a Hamamatsu ORCA-ER c4742-95 digital camera and Openlab 4.0.3 software (Improvision, Coventry, United Kingdom).
In vivo nuclear staining was performed with Hoechst (bisbenzimide H 33342; Sigma, Madrid, Spain). Cells were collected by centrifugation at 3000 rpm and incubated in Hoechst 1× for 15 min.
To quantify Rad22p-YFP foci appearance at least 400 nuclei from two separate experiments were examined for each strain and each time point. To quantify the percentage of binucleated cells, cells were fixed in 70% ethanol, and then they were washed and resuspended in 1× phosphate-buffered saline (PBS) (8.5 mM Na2HPO4·12H2O, 1.88 mM NaH2PO4·H2O, 130 mM NaCl) previously mixed with DAPI to a 1× concentration of the staining agent. At least 400 cells were classified into uni- or binucleated in each count. Mitotic index in wt and flp1-9A cells was determined by in vivo Flp1p-GFP staining.
Immunoprecipitation, Western Blot Analysis, and Kinase Assays
Whole-cell extracts for Western blotting were obtained as described previously (Cueille et al., 2001). Total protein extract (80 μg) was run on 12% standard SDS-polyacrylamide gel electrophoresis gels, transferred to nitrocellulose, and probed with anti-Cdc2 (1:500) polyclonal antibody, anti-tyr15, or anti-hemagglutinin (Ha) (12CA5; Roche Diagnostics, Mannheim, Germany). Tubulin as loading control was detected with mouse TAT1 anti-tubulin monoclonal antibody (1:500). For all antibodies mentioned, goat anti-rabbit (1:3500) or goat anti-mouse (1:2000) conjugated to horseradish peroxidase (GE Healthcare, Chalfont St. Giles, United Kingdom) were used as secondary antibody. Immunoblots were developed using Western Blotting Luminol Reagent (Santa Cruz Biotechnology, Santa Cruz, CA) or SuperSignal (Pierce Chemical, Rockford, IL).
For immunoprecipitation, protein extracts were prepared using immunoprecipitation lysis buffer (50 mM Tris, pH 7.5, 80 mM β-glycerophosphate, 250 mM NaCl, 15 mM nitrophenylphosphate, 50 mM NaF, 5 mM EDTA, 1 mM dithiothreitol [DTT], and 0.1% NP-40 supplemented with aprotinin and leupeptin both at 10 μg/ml final concentration, and vanadate and phenylmethylsulfonyl fluoride both 1 mM final concentration). Anti-Ha (12CA5; Roche Diagnostics) was used for immunoprecipitation. A mix of antibody and 10 μl of protein A (protein A-Sepharose; GE Healthcare) was incubated in lysis buffer for 1 h at 4°C, previous to incubation with 1 mg of extract in 300 μl of lysis buffer for 2 h at 4°C.
Cds1p activity assay over myelin basic protein (MBP) was performed as reported in Lindsay et al. (1998). Kinase assays were performed over glutathione transferase (GST)-purified Flp1p (obtained from a Δcds1 strain) and controls. GST purification has been described previously (Shiozaki and Russell, 1997). Kinase was immunoprecipitated as described above. Next, 2.5 μg of substrate and protein A-bound kinase were incubated with 10 μl of kinase buffer (10 mM HEPES, pH 7.5, 75 mM MgCls, 0.5 mM EDTA, and 1 mM DTT) containing 2.5 μCi of [γ-32P]ATP and 0.2 mM ATP final concentration, at 30°C for 15 min. Reaction was stopped by the addition of 20 μl of 2× SDS sample buffer.
Protein interaction assays were performed by GST purification from 109 cells after 16 h of induction of the target protein and Western blot of the pull-down for the detection of the other Ha-tagged target as mentioned above. Quantification of results was performed using Molecular Imager FX (Bio-Rad, Hemel Hempstead, United Kingdom).
Kinase-Phosphatase Assays
Cds1p was immunoprecipitated after 2 h of 12 M HU treatment as explained above. Phosphorylation assays over GST-Flp1p were performed as mentioned above using nonhydrolyzable ATP (0.2 mM final concentration in buffer) to avoid possible autodephosphorylation. GST-Flp1p was treated with the immunoprecipitation mix without antibody as a control. Samples were washed three times with phosphatase buffer, pH 6.6 (50 mM imidazol, 1 mM DTT, and 1 mM EDTA). A dephosphorylation assay (30 min at 30°C) of treated and control-treated GST-Flp1p by using 250 μM 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) in 200 μl of phosphatase buffer was performed, using immunoprecipitated kinase as a further control. Samples were analyzed in a Tecan Ultra Evolution instrument (Tecan, Grödig, Austria), by using 360- and 465-nm excitation and emission wavelengths, respectively. Readings were normalized taking intro account the quantity of GST-Flp1p contained in each sample, determined by gel electrophoresis and subsequently staining and quantification of the gel.
Mass Spectrometry and Construction of Flp1p-9A
Characterization of phosphorylation sites on Flp1p was performed essentially as described previously (Esteban et al., 2006), by real-time ionization an precursor ion scanning analysis on a 4000 Q-Trap LC-MS/MS hybrid system (Applied Biosystems/MDS Sciex, Foster City, CA) mass spectrometer.
Mutation of the nine RXXS putative phosphorylation sites on Flp1p was performed sequentially by polymerase chain reaction (PCR) from pREP-KZ-Flp1p, by using complementary primers of 30 bases containing the desired mutation (S160A AGC->GCC, S396A TCT->GCT, S408A TCA->GCA, S467A TCA->GCA, S468A AGT->GCT, S493A AGT->GCT, S499A AGT->GCT, S513A AGC->GCC, and S537A TCT->GCT). Each mutation was verified by DNA sequencing analysis.
The flp1-9A gene was obtained by PCR of pREP-KZ-flp1-9A with flanking BglII–XmaI sites and cloned into a pREP41GFP vector in which the nmt1 promoter had been previously substituted by the 1.2-kb endogenous promoter region of flp1 (Pst1–BglII sites). The resulting construction was then integrated in a flp1::kanMX strain by using the unique restriction site SpeI by standard transformation techniques (Moreno et al., 1991). The correct integration in the genome was corroborated by Southern blot analysis and PCR with flanking and internal oligonucleotides.
RESULTS
In Vivo Nuclear Accumulation of Flp1p-GFP in Response to Hydroxyurea-induced S Phase Arrest
In the course of a normal cell cycle, Flp1p changes its subcellular location to carry out its different functions. Flp1p localizes in the nucleolus and spindle pole bodies in interphase. In prophase, it is released from the nucleolus by an unknown regulatory mechanism to occupy the nucleus, medial ring and mitotic spindle (Cueille et al., 2001; Trautmann et al., 2001). To uncover the possible implication of Flp1p in checkpoint response of fission yeast to unreplicated DNA, we were first interested in the subcellular localization of Flp1p in response to depletion of nucleotides by the ribonucleotide reductase inhibitor HU.
An asynchronous culture of a flp1-GFP strain was treated with 12 mM HU. After 2 h of treatment, the majority of the population was unseptated, uninucleated, and partially elongated as a result of the checkpoint-induced block in S phase. To study in detail Flp1p-GFP localization during replication stress, we used as controls Gar2p-RFP as a nucleolar marker (Gulli et al., 1995; Sicard et al., 1998) and Hoechst staining to visualize the non-nucleolar region of the nucleus. In HU-treated cells, Flp1p-GFP staining was nuclear (Figure 1). This change of localization was not general for nucleolar proteins as proved using the nucleolar marker Gar2p-RFP, and no remodeling of the nucleolus takes place during treatment with the HU concentrations used (Figure 1C and Supplemental Figure 1). Importantly, because Flp1p remains located at the nucleolus and spindle pole body (SPB) during S phase in conditions in which the replication checkpoint is not active (Supplemental Figures 2 and 3), this change of subcellular localization seemed to be specifically triggered by the action of the drug and not to be a result of the cell cycle stage at which cells were blocked. Furthermore, as soon as the stress source was removed, Flp1p-GFP returned to its nucleolar and SPB interphase locations (Supplemental Figure 4).
Figure 1.
Flp1p-GFP accumulates in the nucleus during HU induced replication stress. (A) Asynchronous cultures of exponential growing flp1-GFP gar2-RFP cells were treated for 2 h with the ribonucleotide reductase inhibitory drug HU (12 mM). Localization of nucleolar protein Gar2p-RFP and nuclear Hoechst staining are included as reference controls. (B) Flp1p-GFP locates to the nucleolus and SPB in untreated interphase cells. (C) A 3.5× nuclei magnification of untreated interphase (top) and 12 mM HU-treated cells (bottom). Note that no physical nucleolar reorganization can be observed as a result of HU treatment. Bar, 4 μm.
To show that the observed arrest was specific of S phase checkpoint induction, cdc10-129 flp1-GFP cells were synchronized and released in the presence of 12 mM HU (Supplemental Figure 4). Nuclear localization of Flp1p-GFP was observed as expected. This result further suggests the existence of a molecular mechanism that relates nuclear accumulation of Flp1p-GFP and the activation of S phase checkpoint response.
Flp1p Interacts Functionally with the 14-3-3 Rad24p Protein in the Response to Replication Stress
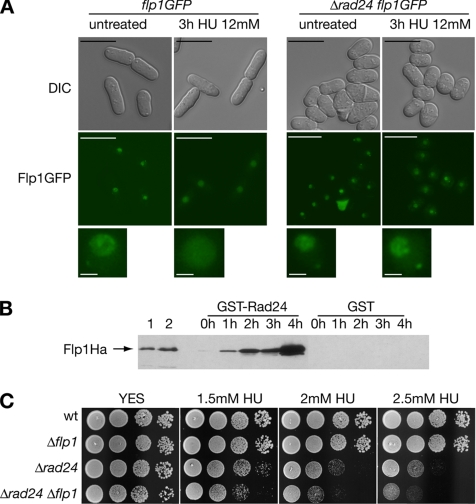
Rad24p and Flp1p have been proved to interact in response to cytokinetic stress (Mishra et al., 2005; Trautmann and McCollum, 2005). Rad24p and Rad25p have been further related to adequate localization and function of proteins implicated in checkpoint response. Association of Chk1p checkpoint kinase with 14-3-3 proteins is stimulated by DNA damage, and such interaction affects localization and checkpoint function of Chk1p (Chen et al., 1999; Dunaway et al., 2005). Furthermore, Rad24p has been reported to mediate nuclear exclusion of Cdc25p in checkpoint response (Lopez-Girona et al., 1999, 2001). We therefore studied whether Rad24p was involved in the nuclear accumulation of Flp1p upon DNA stress. As shown in Figure 2A, nuclear dispersion of Flp1p-GFP relied on a functional rad24+ wild-type allele, suggesting that Rad24p is required for release of the Flp1p phosphatase from the nucleolus in response to replication stress.
Figure 2.
Functional interaction between Rad24p and Flp1p in response to blocks in DNA replication. (A) Flp1p does not change its subcellular localization in response to replicative stress in Δrad24 cells. Live imaging of Flp1p-GFP in a strain deleted for rad24 after 3 h of 12 mM HU treatment. Flp1p remains nucleolar, whereas in control cells the staining is nuclear, as described previously. Bars, 10 μm. A 3× magnification of nuclei is shown in the bottom panel. Bars, 1 μm. (B) Rad24p interacts physically in vivo with Flp1p in checkpoint response to HU. GST-Rad24p was purified from untreated cells and at different times of 12 mM HU treatment. Blots were incubated with α-Ha antibody to detect Flp1pHa. Purified GST was used as a control. In lane 1, 10 μg of whole cell protein extract and in lane 2, 1/200 of the input protein in the pull-down assay shown for reference. No Rad24p–Flp1p interaction is detected in untreated cells, whereas interaction is detected at all treatment points analyzed. (C) Sensitivity assay of Δrad24 and Δrad24 Δflp1 cells to chronic HU exposure at 25°C. Δrad24 is partially sensitive to HU treatment. Δrad24 Δflp1 presents a more sensitive phenotype than each simple mutant.
We were next interested in the possibility of an in vivo interaction of Flp1p with Rad24p upon replication stress. By means of a pull-down assay for GST-tagged Rad24p and Flp1pHa detection, Flp1pHa interaction with the 14-3-3 homologue Rad24p during replication stress was indeed observed (Figure 2B). The interaction between GST-Rad24p and Flp1pHa increased significantly with time during the treatment with HU. In a similar assay Flp1pHa interaction with GST-Rad25p was also observed (Supplemental Figure 5).
As mentioned, 14-3-3 protein homologues Rad24p and Rad25p are required for adequate checkpoint response in fission yeast (Ford et al., 1994). In particular, it has been shown that Δrad24 mutants are defective in the checkpoint response to the DNA replication inhibitor HU at 37°C but proficient at lower temperatures (Forbes et al., 1998). We combined Δrad24 and Δflp1 mutations, and we found that the double mutant was more sensitive to HU than single Δrad24 mutants even at 25°C (Figure 2C). This genetic interaction indicates that deletion of flp1 enhances the replication stress defect of Δrad24 cells and suggests that Flp1 and Rad24p may play certain roles implicated in different branches of the checkpoint response.
Cds1p Triggers Nuclear Accumulation of Flp1p-GFP in Response to Replication Stress
To explain the possible function of Flp1p in checkpoint response to replication stress, we further studied Flp1p-GFP localization in strains deleted for either rad3, cds1, or chk1 (Boddy and Russell, 2001; Nyberg et al., 2002). HU treatment induces an arrest in cell cycle progression in both cds1 or chk1 mutant strains (Murakami and Okayama, 1995; Boddy et al., 1998; Lindsay et al., 1998). However, rad3 mutants are unable to block cell division because they lack the main sensor to genotoxic stress (Boddy et al., 1998; Brondello et al., 1999). In untreated Δcds1 flp1-GFP and Δchk1 flp1-GFP strains Flp1p-GFP localization was indistinguishable from that of wild-type cells (Figure 3, A and B). However, after 2 h of treatment with 12 mM HU the Δcds1 mutant presented nucleolar and SPB staining resembling that of untreated cells and clearly differing from Δchk1 flp1-GFP, which still presented nuclear staining as flp1-GFP cells (Figure 3C). Importantly, in response to HU treatment Flp1p-GFP also remained nucleolar in Δrad3 mutants (Supplemental Figure 6), proving that the release of Flp1p from the nucleolus depends on an active checkpoint. Together, these results indicate that replication stress induced by HU triggers a Cds1p-dependent nuclear accumulation of Flp1p phosphatase.
Figure 3.
Nuclear accumulation of Flp1p depends on checkpoint kinase Cds1p (A) Flp1p-GFP localization and Hoechst costaining in untreated and 12 mM HU treated flp1-GFP cells. (B) Flp1p-GFP localizes normally in both Δcds1 and Δchk1 untreated cells. (C) Flp1p-GFP localization in 12 mM HU-treated cells lacking either Δcds1 or Δchk1. Whereas Δchk1 cells localize Flp1p-GFP to the nucleus in response to replication stress like wild-type cells, the phosphatase remains nucleolar in cells deleted for cds1. Nuclei magnified 3× are shown. Bar, 4 μm.
Flp1p Interacts in Vivo with Cds1p and Is Phosphorylated by the Checkpoint Kinase to Enhance Its Phosphatase Activity In Vitro
Having related Cds1p to Flp1p-GFP localization under HU treatment, an attractive possibility was that Flp1p interacts with Cds1p in vivo. Therefore, we performed a pull-down assay to check for GST-Flp1p/Cds1pHa physical interaction.
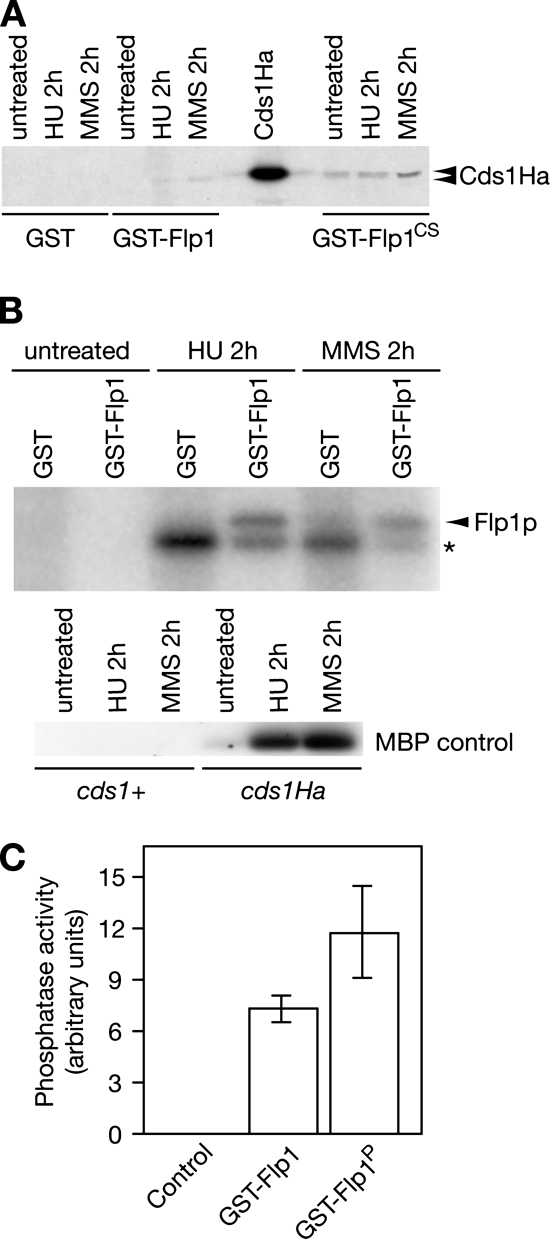
GST-Flp1p and GST-Flp1pCS, an inactive form of Flp1p, were induced and affinity-purified (see Materials and Methods) both from untreated and checkpoint activated cells. Blots were incubated with α-Ha antibody to detect Cds1pHa. We have shown previously that GST-Flp1pCS is more effective in interacting with Cdc25p in vivo than GST-Flp1p (Esteban et al., 2004; Vazquez-Novelle et al., 2005).
Although no interaction was found in untreated cells, our assay showed interaction between Cds1p and Flp1p both in cells treated with 20 mM HU or 0.033% methyl methanesulfonate (MMS). As expected, this result was more evident when expressing the catalytic inactive form of Flp1p, where physical interaction between the studied proteins also can be seen in untreated cells (Figure 4A). These results indicate that Cds1p is able to form in vivo a complex with both the catalytically inactive Flp1pCS mutant and with the wild-type protein in S. pombe cells, suggestive of a substrate/enzyme interaction. The low degree of interaction found with active Flp1p may be explained by the transient nature of phosphatase interactions.
Figure 4.
Cds1p interacts with Flp1p in vivo. (A) Flp1p and Cds1p interact physically in vivo. Pull-down assay showing physical interaction between Cds1p and Flp1p or catalytic inactive Flp1p (Flp1pCS). GST-Flp1p and GST-Flp1pCS were purified both from untreated and checkpoint activated cells, blots were incubated with α-Ha antibody to detect Cds1pHa. Stronger interaction was detected with the catalytic inactive Flp1p. (B) Checkpoint activated Cds1p phosphorylates Flp1p in vitro. Kinase assay of Cds1p by using Flp1p as substrate. Cds1pHa was immunoprecipitated from untreated cells and checkpoint activated cells, respectively, and assayed using purified GST-Flp1p and GSTp (control) as substrates. Activity of checkpoint activated Cds1pHa and of an untagged strain as a control were assayed using MBP as substrate (bottom). Flp1p is phosphorylated by the checkpoint-activated kinase. Phosphorylated Flp1p is marked by an arrow. The band marked by a star corresponds to a protein that coimmunoprecipitates with active Cds1p. (C) Cds1p-mediated Flp1p phosphorylation enhances the phosphatase activity of Flp1p in vitro. Quantification of phosphatase assay using DiFMUP as a substrate. Unphosphorylated and Cds1p phosphorylated GST-Flp1p were assayed in vitro for their ability to dephosphorylate the fluorescent substrate DiFMUP. Active immunoprecipitated Cds1p kinase was used as a negative control.
Next, we were interested in finding out whether Flp1p was a substrate of checkpoint-activated Cds1p. Cds1pHa was immunoprecipitated from checkpoint-activated cultures, and its kinase activity was assayed using MBP as a substrate. Cds1p was activated both as a result of nucleotide depletion (HU) and DNA damage (MMS), reaching a maximum in activity after 2 h of treatment with either drug. Activated Cds1p was assayed using GST-purified GST-Flp1p as a substrate and GST as a control, resulting in specific phosphorylation of Flp1p both by HU- and MMS-activated Cds1p (Figure 4B). Our in vitro observations suggest that Flp1p may be an in vivo substrate for the Cds1p-S phase checkpoint kinase in response to replication arrest or to replication fork-associated DNA damage. We next tested whether the phosphorylation of Flp1p by Cds1p was able to alter its phosphatase activity in vitro. With this purpose, GST-Flp1p was purified to homogeneity from S. pombe Δcds1 cells expressing the GST-fusion protein. In vitro Cds1p-phosphorylated and unphosporylated GST-Flp1p were assayed using DiFMUP as a substrate. As shown if Figure 4C, phosphorylated GST-Flp1p resulted to be more active, proving an enhancement of Flp1p phosphatase activity by the in vitro Cds1p-mediated phosphorylation. In our assays, active Cds1p caused a 37% average increase in the phosphatase activity of Flp1p, which is comparable with the 40% difference observed in Flp1p activity at the onset of mitosis due to inhibitory cyclin-dependent kinase (Cdk1) hyperphosphorylation when compared with interphase cells (Wolfe et al., 2006). However, although Cdk1 phosphorylation was inhibitory Cds1p-mediated phosphorylation of Flp1p activated the phosphatase (Wolfe et al., 2006; and this work).
Mutation of Cds1p Phosphorylation Sites in Flp1p Abolishes the Nuclear Accumulation of the Phosphatase during Replication Stress
Having shown that Flp1p-GFP does not accumulate in the nucleus of cells lacking cds1, also that Flp1p interacts in vivo with the checkpoint kinase and that Flp1p is an in vitro substrate for active Cds1p, we next wished to identify the specific Cds1p-dependent phosphorylated residues of Flp1p. We set up a kinase assay as described in Figure 4, followed by a mass spectrometric analysis to detect phospho-amino acids (see Materials and Methods). We found that serine 468 was phosphorylated by HU-activated Cds1p kinase. Ser468 lies in a RXXS consensus phosphorylation site for Chk2p mammalian kinase (O'Neill et al., 2002). Full-length Flp1p harbors nine different RXXS consensus sites located at serines 160, 396, 408, 467, 468, 493, 499, 513, and 537 (Figure 5A), suggesting that some or all of them may be in vivo targets for the Cds1p kinase. These sites were mutated to nonphosphorylatable alanine residues (9A), and the resulting construction was used as an in vitro substrate for Cds1p phosphorylation. Although able to phosphorylate the GST-Flp1p wild-type control protein, active Cds1p kinase was unable to phosphorylate purified GST-Flp1p-9A, indicating that the mutant protein contained all in vitro Cds1p-phosphorylatable residues (Figure 5B).
Figure 5.
Characterization of the flp1-9A mutant. (A) Schematic representation of Flp1p showing Serine 468 phosphorylation site identified by mass spectrometry (bold) and the RXXS putative phosphorylation sites. (B) Phosphorylation assay of activated Cds1p using GST-Flp1p and RXXS sites mutants as substrates. Mutation of the complete set of sites was performed because phosphorylation of Flp1p is prevented in the flp1-9A mutant. (C) A 10-fold dilution plate assay showing the sensitivity of three different clones of flp1-9A-GFP to HU. It can be observed that the behavior is similar to that of Δflp1, presenting only a decrease on colony size compared with wild-type cells. (D) Mutation of the nine RXXS sites present in Flp1p does not alter the subcellular localization of the protein during the different stages of an unperturbed cell cycle. In vivo Flp1p-9A-GFP staining shows interphase and mitotic cells undergoing normal changes in the subcellular localization of Flp1p-9A as described before for wild-type Flp1p (Cueille et al., 2001). Dot lines represent the cells contour. Bar, 4 μm. (E) Flp1p-9A-GFP is not released to the nucleus in checkpoint response to HU treatment. Bars, 4 μm. Nuclei magnified 3× are shown in inner panels. Note that flp1-9A-GFP cells elongate indicating that they are responding to the checkpoint arrest.
We next used the 9A mutant to replace the endogenous flp1+ locus to generate a flp1-9A-GFP strain with the mutant allele under the control of the native flp1+ promoter (see Materials and Methods). Under physiological conditions flp1-9A-GFP cells were phenotypically identical to wild-type cells (Figure 5, C and D). Accordingly, Flp1p-9A-GFP subcellular localization was identical to that of wild-type Flp1p-GFP during unperturbed cell cycle (Figure 5D).
However, when we assayed the sensitivity of the flp1-9A-GFP strain to HU treatment, we found that the strain behaved like the deletion of flp1 (Figure 5C), basically with a small lag in rate growth compared with wild-type cells when exposed to 5 mM HU. Importantly, mutation of the above-mentioned serines to alanines prevented nuclear accumulation of Flp1p in response to replication stress, because in vivo Flp1p-9A-GFP signal was observed at the nucleolus and SPB in cells treated with HU (Figure 5E and Supplemental Figure 7). Together, these results provide strong evidence that in vivo phosphorylation of Flp1p by Cds1p-checkpoint kinase underlies the nuclear accumulation of the phosphatase in response to HU-induced replication arrest.
Flp1p Does Not Regulate Cdc2p Y15 Phosphorylation in Checkpoint Response
In S. pombe, Flp1p antagonizes mitotic CDK activity by dephosphorylating, thereby down-regulating the Cdc2p Y15 phosphatase Cdc25p (Esteban et al., 2004; Wolfe and Gould, 2004). Cdc25p is also a regulated target of the checkpoint cascade activated by genotoxic stress (Lopez-Girona et al., 1999, 2001). Thus, a role of Flp1p in regulating Cdc25p in response to DNA damage or replication stress seemed reasonable. Having observed that Cds1p promotes Flp1p change of localization as a consequence of replication stress, we were next interested in studying the possible role of Flp1p in degrading Cdc25p and regulating Cdc2p Y15 phosphorylation in a HU-mediated block triggered by DNA replication inhibition and the corresponding replication fork arrest.
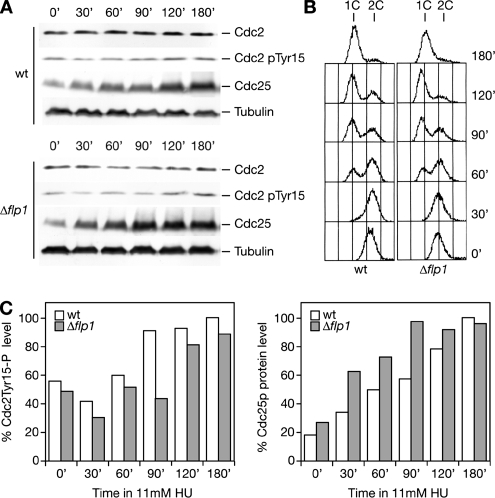
Thus, we analyzed Cdc25p protein levels and Cdc2p Y15 phosphorylation state in a wild-type and a Δflp1 strain during HU treatment by Western blot analysis (Figure 6, A and B). The amount of cellular Cdc25p detected increased as a result of drug treatment both in wild type and Δflp1 (Figure 6A). So did the Cdc2p Y15 inhibitory phosphorylation. Taking into account that Cdc25p basal levels are higher in Δflp1 than in wild type, the kinetics of Cdc25p accumulation seemed to be similar in both strains (Figure 6C). These results allow us to conclude that deletion of flp1 does not significantly affect Cdc25p protein levels or Cdc2p Y15 phosphorylation in response to replication stress caused by nucleotide depletion. Even though in fission yeast the strict requirement of Cdc2 Y15 phosphorylation as the key mechanism for Cdk/cyclin inhibition in response to S phase DNA damage is still in dispute (Rhind et al., 1997; Rhind and Russell, 1998b; Kommajosyula and Rhind, 2006), it is, indeed, a fact that p34cdc2 becomes hyperphosphorylated on tyrosine 15 upon HU, MMS, or UV light treatments (O'Connell et al., 1997; Chu et al., 2007). In this context, it is surprising to see that Flp1p, the phosphatase that reverts Cdk phosphorylation events on Cdc25p down-regulating its activity as cells exit from mitosis, does not seem to exert such control under replicative stress.
Figure 6.
Kinetics of checkpoint-dependent phosphorylation of Cdc2p in Tyrosine 15 and Cdc25p protein accumulation are not significantly altered in Δflp1 cells in response to HU-induced replication arrest. (A) Western blot analysis of Cdc25p, Cdc2p, and Y15 phosphorylation of Cdc2p levels during 12 mM HU treatment in asynchronous cultures of wild-type and Δflp1 strains. (B) FACS analysis showing checkpoint induced S phase arrest of asynchronous wild-type and Δflp1 cells during 12 mM HU treatment. (C) Normalization of quantified Cdc2p Y15P and Cdc25p levels are shown in bar diagrams. Note that Y15P and Cdc25p levels increase during treatment in both strains after similar kinetics.
Cds1p Full Activation Is an Important Event in Replication Stress-induced Flp1p Release from the Nucleolus
Having shown that Flp1p plays no role in regulating Cdc2p tyrosine 15 phosphorylation in response to replication stress, we sought a possible role for Flp1p in the regulation of Cds1p, because we had already found evidence that related the two proteins (Figures 3 and 5) and because of the checkpoint related roles of the kinase independent of cell cycle progression ralentization. Cds1p is activated by phosphorylation during replication stress; therefore, our working hypothesis was that Flp1p, as a phosphatase, could be involved in the modulation of the checkpoint response.
As a first approach, we analyzed whether Cds1p levels were affected by deletion of the flp1 gene. Levels of Cds1pHa (and Chk1pHa) were analyzed by Western blotting in wild-type and Δflp1 cells during a time course treatment with HU. Inspiringly, Δflp1 cells accumulated lower levels of Cds1p than wild-type cells (Figure 7A). In contrast, Chk1pHa levels were not affected by the absence of the flp1 gene (Figure 7A).
Figure 7.
Cds1p protein levels and kinase activity in response to replication stress are altered in Δflp1 cells. (A) Western blot analysis of Cds1pHa and Chk1pHa protein levels in wild-type and Δflp1 cells treated with 20 mM HU. Samples were quantified and normalized to loading controls. Quantification is shown in bar diagrams. Whereas Δflp1 cells accumulate less Cds1p in checkpoint response to HU, Chk1p levels are similar in both strains. (B) Cells deleted for flp1 present decreased in vivo Cds1p activity in checkpoint response. Kinase assay of Cds1p using MBP as substrate. Active Cds1pHa was immunoprecipitated from wild-type and Δflp1 strains, both from HU and MMS treated cells. Activity was normalized to immunoprecipitated Cds1pHa in each sample and represented in a bar diagram. Cds1p-associated kinase activity in wild-type cells, both for HU and MMS treated cells, was arbitrarily normalized to 100%. Error bars are shown for three independent experiments.
We next compared the kinase activity of Cds1pHa in wild-type and Δflp1 cells in an assay using MBP as substrate (see Materials and Methods). Cds1pHa-associated kinase activity was normalized to immunoprecipitated Cds1pHa in every sample. Interestingly, rather than enhancing Cds1pHa activity, absence of Flp1p resulted in reduced kinase activity (Figure 7B). These observations indicate that Δflp1 cells are defective in levels and activity of the Cds1p checkpoint kinase and suggest that Flp1p may act as an activator of Cds1p either directly or indirectly by its action over some modulator of Cds1p activity.
flp1 Mutants Have an Increased Percentage of Rad22p Recombination Foci When Exposed to Replication Stress
S. pombe cells prevent generation of aberrant strand-exchange events during a HU block by activating the replication checkpoint (Meister et al., 2005). Accordingly, S phase checkpoint-deficient Δcds1 mutants harbor multiple recombination-associated foci when exposed to HU (Meister et al., 2005). Concentration of Rad22p, fission yeast Rad52p homologue, into a few bright nuclear spots, or foci, is an indication of the induction of homologous recombination (Lisby et al., 2001; Du et al., 2003; Meister et al., 2003; Noguchi et al., 2003).
We have related full activation of S phase checkpoint kinase Cds1p to the presence of a functional Flp1p phosphatase in response to replication stress. If Flp1p is required to fully activate Cds1p when dealing with genotoxic insults, it would be expected that Δflp1 and flp1-9A mutants would show defects mimicking to some extent those of Δcds1 mutants.
To further characterize flp1 mutant phenotypes, Rad22p-associated recombination foci were visualized in vivo in either untreated or HU-treated wild-type, Δcds1, Δflp1, and flp1-9A cells harboring a single rad22-YFP allele (Figure 8). In untreated cells, the Δflp1 strain presented a single Rad22p-YFP nuclear spot in 19% of the cells, a similar percentage to that observed in Δcds1 cells (25%), but higher than in wild-type and flp1-9A cells (15 and 11%, respectively) (Figure 8). Such single nuclear foci are assumed to be sites of postreplicative DNA repair (Meister et al., 2003; Noguchi et al., 2003). Under these conditions, only Δcds1 cells present a significant percentage of multiple Rad22p (8%). Multiple Rad22p spots are interpreted as recombination foci that occur in G2 or very late in S phase (Meister et al., 2003; Noguchi et al., 2003). Importantly, the percentage of flp1 mutant cells with multiple nuclear Rad22p-YFP spots showed a marked increase when exposed to HU for 2 h (from 1 to 25% in Δflp1, from 1 to 13% in flp1-9A), whereas the number of foci in wild-type cells remained almost constant (from 1 to 2.5%), probably indicating a defect in checkpoint response associated to depletion of Flp1p. It should be noted that the number of recombination foci present in flp1 mutants upon HU treatment is moderate when compared with that of Δcds1 (up to 57%), which again points to a partial loss of checkpoint activity in the Δflp1 strain.
Figure 8.
Hydroxyurea induces Rad22p recombination foci in flp1 mutants. (A) Quantification of nuclei containing single or multiple Rad22p-YFP foci in flp1 mutant strains in asynchronous exponentially growing cells (untreated) and after 2 h of 12 mM HU treatment. Control counts in checkpoint proficient wild-type cells and checkpoint mutant Δcds1 are shown. Results are representative of at least two independent experiments. Standard deviations are as follows: untreated cells, wt % no foci 1.16, % one focus 1.64, % multi foci 0.48, Δflp1 % no foci 1.05, % one focus 0.80, % multi foci 0.24, flp1-9A % no foci 3.02, % one focus 3.40, % multi foci 0.37, Δcds1 % no foci 3.51, % one focus 1.09, % multi foci 2.41; HU treated cells, wt % no foci 1.74, % one focus 1.47, % multi foci 0.27, Δflp1 % no foci 6.32, % one focus 5.16, % multi foci 1.15, flp1-9A % no foci 0.75, % one focus 1.25, % multi foci 0.5, Δcds1 % no foci 0.36, % one focus 1.34, % multi foci 0.97. (B) Live imaging of Rad22p-YFP foci (bright spots) in flp1 mutants and control strains after 2 h of 12 mM HU treatment. Bar, 4 μm. Although recombination foci are rarely seen in a wild-type strain, single and multiple Rad22p-YFP foci are detected in both flp1 mutants nuclei. This effect is however weaker than in checkpoint mutant Δcds1.
Defects on Checkpoint Activation in flp1-9A Cells
We next examined the in vivo localization of Flp1p-GFP in flp1+ wild-type and flp1-9A strains when treated with HU and scored cells with a mitotic Flp1p localization (Supplemental Figure 8, A and B). Although most cells undergoing replication stress located Flp1p-9A-GFP at the nucleolus and SPB as observed previously (Supplemental Figure 8; also see Figure 5), 23% of them presented a mitotic pattern at the 2h time point, suggesting that flp1-9A cells needed more time to mount a robust wild-type-like checkpoint response. The percentage of mitotic cells decreased to wild-type levels after 4 h of treatment most likely as the result of cell division in the absence of DNA replication (Supplemental Figure 8). flp1-9A is a unique mutant harboring two characteristics. First, it behaves like wild-type flp1 in mitotic cycles, and, second, it fails to exit the nucleolus upon replication stress. Accordingly, flp1-9A cells presented a phenotype of recombination foci accumulation comparable with that of Δflp1 (Figure 8). This result further confirms the importance of Cds1p-dependent release of Flp1p from the nucleolus to the nucleus to achieve an adequate checkpoint response to replication stress.
Δflp1 Mutant Cells Are Partially Defective in Checkpoint Response to HU-induced Replication Stress
We reasoned that if Flp1p is required to fully activate Cds1p, mutating flp1+ or preventing the release of Flp1p from the nucleolus might prevent, in turn, robust Cds1p activation and might cause observable defects in the cell cycle arrest induced by HU. Indeed, we have shown here the defective activation of Cds1p and increased percentage in recombination foci in Δflp1 cells. However, we have only found subtle differences on sensitivity in long-term exposure to HU (see plate assay in Figure 5C), suggesting that the control of Flp1p on Cds1p might be only transient, leading to minor defects (in checkpoint response) compared with those observable in fully defective checkpoint mutants. To test this idea, we analyzed the checkpoint arrest to replication stress by testing cell cycle parameters in wild-type, Δflp1, and Δrad3 cells treated with HU.
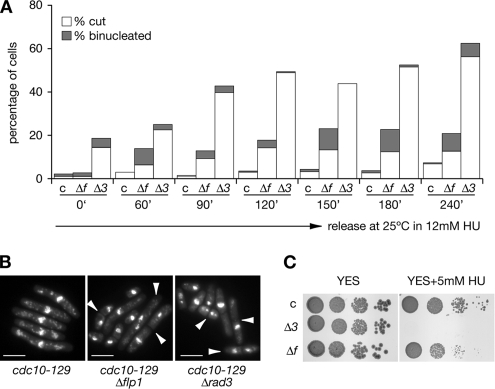
To study whether mutant Δflp1 cells were defective in their response to replication blocks we first checked checkpoint proficiency in cdc10-129 synchronized cells. cdc10-129, cdc10-129 Δflp1, and cdc10-129 Δrad3 strains were synchronized in G1 by incubation at 36.5°C for 4.5 h and then released at the permissive temperature in the presence of 12 mM HU, samples were taken at regular intervals and processed for nuclear staining, DNA content, and cell size analysis (Figure 9). Binucleated (normal and cut) cells were monitored at indicated intervals after release (Figure 9A). Although in synchronized cdc10-129 cells (control) the percentage of binucleated (normal and cut) cells remained low throughout the experiment, cdc10-129 Δflp1 mutants already presented a moderate percentage of cells undergoing mitosis as soon as 60 min after the release in HU (up to 15%) with a steady increase (in percentages) as cells were exposed longer to the replication block (reaching a maximum of 22%). As expected, we found that the checkpoint defects observable in cdc10-129 Δrad3 mutant cells were more severe than those found in cdc10-129 Δflp1 mutant cells, as the percentages of cells undergoing mitosis in the absence of DNA replication ranked from 18%, at the block point, to 62%, at the last time point in Δrad3 mutants. Microscopical examination of cells (Figure 9B) and fluorescence-activated cell sorting (FACS) analysis of DNA content (Supplemental Figure 9) confirmed that cdc10-129 Δflp1 and cdc10-129 Δrad3 were defective in their response to the HU arrest, as cells with less than an haploid content of DNA (1C) were detected. Finally, we also found that the cdc10-129 Δflp1 mutant was slightly more sensitive to HU than control cells (cdc10-129), whereas cdc10-129 Δrad3 mutant cells were hypersensitive (Figure 9C). These results further confirm the postulated partial defect on checkpoint response of flp1 mutants.
Figure 9.
Quantification of checkpoint defects of Δflp1 mutants in synchronized cultures. (A) cdc10-129, cdc10-129 Δflp1, and cdc10-129 Δrad3 strains were synchronized in G1 (4.5 h at 36.5°C), and then they were released from the G1 block (at 25°C) in the presence of 12 mM HU. The percentages of binucleated and cut cells were estimated by counting cell samples stained with DAPI every indicated time point and plotted. At least 200 cells were counted for every strain for each time point and the experiment was repeated twice. Labels: c, cdc10-129 control; Δf, cdc10-129 Δflp1, and Δ3, cdc10-129 Δrad3 mutants. (B) Deletion of flp1+ and rad3+ causes cdc10-129 cells to accumulate binucleated and cut cells upon HU treatment. Nuclear staining (DAPI) of cdc10-129, cdc10-129 Δflp1, and cdc10-129 Δrad3 cells synchronized in G1 and released in the presence of 12 mM HU (120 min of treatment). Note the presence of binucleated and cut cells (indicated by arrows). Bar, 10 μm. (C) A 10-fold dilution plate assay showing the different sensitivity of cdc10-129, cdc10-129 Δflp1, and cdc10-129 Δrad3 strains to chronic exposure to 5 mM HU (at 32°C).
We next analyzed the response to HU in cells synchronized in G1 by using an alternative allele of cdc10, cdc10-M17. Interestingly, we found that Δrad3 and, particularly, Δflp1 mutants were synthetically lethal at intermediate temperatures when combined with cdc10-M17. Several haploids cdc10-M17 Δflp1 and cdc10-M17 Δrad3 strains were selected and they were found partially inviable at 33°C, a temperature at which cdc10-M17 are viable (Supplemental Figure 10A). Remarkably, we also found that rad3+ and, unexpectedly, flp1+ were required to prevent mitosis at the G1 arrest imposed by the cdc10-M17 allele, because cdc10-M17 Δflp1 and cdc10-M17 Δrad3 mutants were found to undergo aberrant mitosis at 36.5°C (Supplemental Figure 10, B and C). Of particular interest is the observation that cdc10-M17 Δflp1 cells are hypersensitive to the chronic presence of HU as cdc10-M17 Δrad3 cells are. All these defects are consistent with a role for Flp1p in checkpoint response. Fully aware that the observed synthetic lethality (and hypersensitivity to HU) could interfere with the interpretation of a block and release experiments, we checked checkpoint proficiency in cdc10-M17 synchronized cells. cdc10-M17, cdc10-M17 Δflp1, and cdc10-M17 Δrad3 strains were synchronized in G1 and then released at the permissive temperature in the presence of 12 mM HU. In keeping with our previous observations with the cdc10-129 allele, we found that cdc10-M17 Δflp1 cells presented a significant increase of cells undergoing aberrant mitosis 90 min after the release in HU. This percentage was moderate compared with data obtained with cdc10-M17 Δrad3 cells (Supplemental Figure 10, C and D). However, even though the results were consistent with those in a cdc10-129 background, as inferred from the severity of the synthetic lethality in cdc10-M17 Δrad3 cells and, to a lesser extent, in cdc10-M17 Δflp1 cells, we observed only a moderate increase in the percentage of aberrant mitosis (cut cells) after the exposure to HU (Supplemental Figure 10 C) compared with cdc10-129 data (Figure 9). Our findings show that in fission yeast G1 presynchronized cells the mutation of flp1+ results in checkpoint defects in the response to replication stress.
DISCUSSION
In the present work, we have explored the consequences of mutating the Flp1p phosphatase in replication checkpoint response. The results of our study indicate that checkpoint-related events control Flp1p localization in response to nucleotide depletion and, importantly, they suggest a close interplay between Flp1p and the main effector of the checkpoint cascade response to replication stress, Cds1p. Interestingly, this interplay results in the full activation of the Cds1p checkpoint kinase.
In cycling cells, Flp1p is located in the nucleolus and SPB during DNA replication at S phase (Cueille et al., 2001; Trautmann et al., 2001). However, upon activation of the checkpoint to replication stress in response to HU, Flp1p is efficiently dispersed throughout the nucleus. This change of location does not occur in a Δrad3 strain, indicating that Flp1p changes its subcellular localization in a checkpoint dependent manner.
Interestingly, whereas the cellular mechanism by which Flp1p is released to the nucleus at mitotic entry during an unperturbed cell cycle remains to be clarified (Chen et al., 2006), evidence presented here shows that under replication stress, Cds1p triggers nuclear accumulation of the fission yeast Cdc14p phosphatase homologue. Furthermore, in HU-treated cells this nuclear accumulation of Flp1p is largely independent of Chk1p kinase. The S phase Cdc2-Tyr15 kinase Mik1p also accumulates in the nucleus of the cell by a mechanism dependent on Rad3p and Cds1p in response to depletion of nucleotides by HU (Boddy et al., 1998; Baber-Furnari et al., 2000; Rhind and Russell, 2001). However, in Saccharomyces cerevisiae Rad53p (a Cds1p homologue) and Chk1p inhibit Cdc14p release from the nucleolus to prevent cell cycle progression in the presence of damaged DNA (Liang and Wang, 2007), indicating that the control mechanism involving Cds1p and Flp1p is not conserved in budding yeast.
Several lines of evidence of our work prove that Flp1p and Cds1p are closely related in the checkpoint response to replication stress. First, reduced Cds1p protein levels as well as down-regulation of Cds1-associated kinase activity in cells lacking flp1 point to an involvement of Flp1p in controlling the checkpoint kinase response to replication perturbation. The relevance of the combined effect of these two facts in Δflp1, which support a role for Flp1p as a net activator of the cascade, is underscored by the finding of multiple Rad22p foci upon HU treatment in this strain, which may stand for enhancement of recombinational repair derived from insufficient checkpoint activation. Second, Cds1p phosphorylates Flp1p in vitro and lack of the kinase or mutation of the corresponding phosphorylation sites in Flp1p prevents the nuclear accumulation of the phosphatase during replication stress. These results strengthen the hypothesis of direct control by the Chk2p homologue. Third, in vivo interaction of Cds1p and Flp1p further account for a direct control mechanism of Cds1p over Flp1p. Whether the control of Flp1p over Cds1p can be explained by a direct interaction is still unclear. A direct control is an interesting possibility because, according to our results, dephosphorylation of the kinase should then act as an activating mechanism. If the phosphatase can enhance the activation of the kinase by playing a role in the dimerization and autophosphorylation steps of Cds1p activation will be addressed in future studies.
Together, our data strongly point to a direct network control involving Cds1p and Flp1p to up-regulate Cds1p kinase in the checkpoint response to replication stress, mechanism which may result in fine-tuning of checkpoint response.
Attenuation of Cds1p/Rad53p levels in S. cerevisiae results in sensitivity to replication stress (Cordon-Preciado et al., 2006). Consistently, fission yeast cells lacking flp1+ fail to mount a robust wild-type-like checkpoint response resulting in a higher number of cells undergoing mitosis in acute HU treatment and increased levels of Rad22p-repair-recombination foci. Cells carrying the flp1-9A mutant allele have similar phenotypes, correlating a proficient checkpoint response to an adequate localization of Flp1p controlled by Cds1p.
Flp1p is not the first cell cycle regulator described to be controlled by checkpoint kinases in response to DNA stress. Cdc2p and Cdc25p are known to be down-regulated by the checkpoint cascade (Walworth et al., 1993; Peng et al., 1997; Rhind et al., 1997; Rhind and Russell, 1998b; Lopez-Girona et al., 1999) and, in contrast, Mik1p/Wee1p kinases are activated to delay cell division (Rhind and Russell, 2001). However, according to our results, Flp1p is unique because it presents the peculiarity of performing a function in response to replication stress that is independent from its regulatory role during unperturbed cell cycle.
We propose a simple control mechanism as a model, in which active Cds1p regulates the release of Flp1p out of the nucleolus. According to our results, Rad24p may be involved in this shuttle mechanism. Nuclear Flp1p would in its turn directly or indirectly regulate full activation of Cds1p in response to replication stress (see model in Supplemental Figure 11). Furthermore, by means of Cds1p-dependent Flp1p phosphorylation, an adequate checkpoint activation could be achieved.
The present study suggests that elements capable of regulating Cds1p protein levels and Cds1p-associated kinase activity may be targets of Flp1p. Among these feasible targets, it would be reasonable to include uncharacterized fission yeast Ssa1p and Ssa2p chaperone homologues (Wood et al., 2002) that may eventually play a role in maintaining Cds1p levels. Swi1p, Swi3p, and Sap1p are chromatin interacting proteins that have been involved in the activation of Cds1p in response to HU or MMS (Noguchi et al., 2003, 2004; Matsumoto et al., 2005; Sommariva et al., 2005; Noguchi and Noguchi, 2007). Swi1p, a Tof1p homologue, and Swi3p prevent DNA replication fork collapse, and they are required for the proficient activation of the kinase Cds1p upon activation of the checkpoint to replicative stress (Noguchi et al., 2003, 2004; Sommariva et al., 2005). Significantly, Hsk1p/Dfp1p complex is needed to activate Cds1p in response to replication stress and DNA damage (Takeda et al., 2001; Fung et al., 2002). Hsk1p and Dfp1p, Cdc7p and Dbf4p fission yeast homologues, have a well-characterized role in regulating the initiation of DNA replication and, as Mus81p, Swi1p, Swi3p, and Mcm4p, are key targets of Cds1p in fulfilling the essential role of the checkpoint kinase in maintaining DNA replication fork stability when responding to DNA damage and replication fork stalling (Brown and Kelly, 1999; Snaith et al., 2000; Noguchi et al., 2003, 2004; Kai et al., 2005; Matsumoto et al., 2005; Sommariva et al., 2005; Bailis et al., 2008). Analysis of these potential targets as well as identification of further Flp1p substrates and the study of a possible role of Flp1p in regulating recovery from checkpoint response will be the subject of future studies.
Our findings show that fission yeast Cdc14p-family phosphatase Flp1p, in addition to its role in the exit from mitosis, has a role in the checkpoint response to replication stress. Moreover, we propose that Cds1p regulates its own-full activation in response to checkpoint induction through Flp1p. It will be of general interest to understand whether the same is true for homologues of Flp1p and Cds1p in higher eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the B05 laboratory at the Centro de Investigación del Cáncer for helpful discussions, and we especially appreciate the technical assistance of Sonia Andrés. We also thank S. Moreno and M. Sacristán for critically reading this manuscript. We are grateful to P. Russell (The Scripps Research Institute), S. Moreno (Centro de Investigación del Cáncer), E. Noguchi (School of Medicine, Drexel University), M. J. O'Connell (Mount Sinai School of Medicine), and P. Pérez (Instituto de Microbiología-Bioquímica) for fission yeast strains and methods. We also are grateful to the Yeast Genetic Resource Centre (Japan) for the Gar2-RFP strain. A.B. received financial support for this research work through Programa General de Conocimiento (BFU2006–05878/BMC and GEN2003-20243-C08-04), Instituto de Salud “Carlos III” (Ingenio 2010/Consolider RD06/0020/0024), and Junta de Castilla y Leon (CSI08A05) grants from the Spanish Science and Technology Ministry and Junta de Castilla y León. H.D.-C. was supported by a predoctoral fellowship from the Spanish Science Ministry.
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- DiFMUP
6,8-difluoro-4-methylumbelliferyl phosphate
- GFP
green fluorescent protein
- GST
glutathione transferase
- Ha
hemagglutinin
- HU
hydroxyurea
- MBP
myelin basic protein
- MMS
methyl methanesulfonate
- SIN
septation initiation network
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0737) on April 2, 2008.
REFERENCES
- Baber-Furnari B. A., Rhind N., Boddy M. N., Shanahan P., Lopez-Girona A., Russell P. Regulation of mitotic inhibitor Mik1 helps to enforce the DNA damage checkpoint. Mol. Biol. Cell. 2000;11:1–11. doi: 10.1091/mbc.11.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis J. M., Luche D. D., Hunter T., Forsburg S. L. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote S-phase genome stability. Mol. Cell. Biol. 2008;28:1724–1738. doi: 10.1128/MCB.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Furnari B., Mondesert O., Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Russell P. DNA replication checkpoint. Curr. Biol. 2001;11:R953–R956. doi: 10.1016/s0960-9822(01)00572-3. [DOI] [PubMed] [Google Scholar]
- Brondello J. M., Boddy M. N., Furnari B., Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell. Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. W., Kelly T. J. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc. Natl. Acad. Sci. USA. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. T., Peli-Gulli M. P., Simanis V., McCollum D. S. pombe FEAR protein orthologs are not required for release of Clp1/Flp1 phosphatase from the nucleolus during mitosis. J. Cell Sci. 2006;119:4462–4466. doi: 10.1242/jcs.03220. [DOI] [PubMed] [Google Scholar]
- Chen L., Liu T. H., Walworth N. C. Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev. 1999;13:675–685. doi: 10.1101/gad.13.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z., Li J., Eshaghi M., Peng X., Krishna R., Karaturi M., Liu J. Modulation of cell cycle-specific gene expressions at the onset of S phase contributes to the robust DNA replication checkpoint response in fission yeast. Mol. Biol. Cell. 2007;18:1756–1767. doi: 10.1091/mbc.E06-10-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Preciado V., Ufano S., Bueno A. Limiting amounts of budding yeast Rad53 S-phase checkpoint activity results in increased resistance to DNA alkylation damage. Nucleic Acids Res. 2006;34:5852–5862. doi: 10.1093/nar/gkl741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueille N., Salimova E., Esteban V., Blanco M., Moreno S., Bueno A., Simanis V. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 2001;114:2649–2664. doi: 10.1242/jcs.114.14.2649. [DOI] [PubMed] [Google Scholar]
- Du L. L., Nakamura T. M., Moser B. A., Russell P. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 2003;23:6150–6158. doi: 10.1128/MCB.23.17.6150-6158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaway S., Liu H. Y., Walworth N. C. Interaction of 14-3-3 protein with Chk1 affects localization and checkpoint function. J. Cell Sci. 2005;118:39–50. doi: 10.1242/jcs.01570. [DOI] [PubMed] [Google Scholar]
- Esteban V., Blanco M., Cueille N., Simanis V., Moreno S., Bueno A. A role for the Cdc14-family phosphatase Flp1p at the end of the cell cycle in controlling the rapid degradation of the mitotic inducer Cdc25p in fission yeast. J. Cell Sci. 2004;117:2461–2468. doi: 10.1242/jcs.01107. [DOI] [PubMed] [Google Scholar]
- Esteban V., Vazquez-Novelle M. D., Calvo E., Bueno A., Sacristan M. P. Human Cdc14A Reverses CDK1 Phosphorylation of Cdc25A on Serines 115 and 320. Cell Cycle. 2006;5:2894–2898. doi: 10.4161/cc.5.24.3566. [DOI] [PubMed] [Google Scholar]
- Forbes K. C., Humphrey T., Enoch T. Suppressors of cdc25p overexpression identify two pathways that influence the G2/M checkpoint in fission yeast. Genetics. 1998;150:1361–1375. doi: 10.1093/genetics/150.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J. C., al-Khodairy F., Fotou E., Sheldrick K. S., Griffiths D. J., Carr A. M. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science. 1994;265:533–535. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- Fung A. D., Ou J., Bueler S., Brown G. W. A conserved domain of Schizosaccharomyces pombe dfp1+ is uniquely required for chromosome stability following alkylation damage during S phase. Mol. Cell. Biol. 2002;22:4477–4490. doi: 10.1128/MCB.22.13.4477-4490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B., Blasina A., Boddy M. N., McGowan C. H., Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari B., Rhind N., Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Gulli M. P., Girard J. P., Zabetakis D., Lapeyre B., Melese T., Caizergues-Ferre M. Gar2 is a nucleolar protein from Schizosaccharomyces pombe required for 18S rRNA and 40S ribosomal subunit accumulation. Nucleic Acids Res. 1995;23:1912–1918. doi: 10.1093/nar/23.11.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M., Boddy M. N., Russell P., Wang T. S. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev. 2005;19:919–932. doi: 10.1101/gad.1304305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommajosyula N., Rhind N. Cdc2 Tyrosine phosphorylation is not required for the S-phase DNA damage checkpoint in fission Yeast. Cell Cycle. 2006;5:2495–2500. doi: 10.4161/cc.5.21.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Wang Y. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol. Cell. Biol. 2007;27:5067–5078. doi: 10.1128/MCB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H. D., Griffiths D. J., Edwards R. J., Christensen P. U., Murray J. M., Osman F., Walworth N., Carr A. M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein R., Mortensen U. H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A., Furnari B., Mondesert O., Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A., Kanoh J., Russell P. Nuclear exclusion of Cdc25 is not required for the DNA damage checkpoint in fission yeast. Curr. Biol. 2001;11:50–54. doi: 10.1016/s0960-9822(00)00026-9. [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Ogino K., Noguchi E., Russell P., Masai H. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J. Biol. Chem. 2005;280:42536–42542. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- McGlynn P., Lloyd R. G. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002a;18:413–419. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- McGlynn P., Lloyd R. G. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002b;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- McGlynn P., Lloyd R. G. Replicating past lesions in DNA. Mol. Cell. 2002c;10:700–701. doi: 10.1016/s1097-2765(02)00687-1. [DOI] [PubMed] [Google Scholar]
- Meister P., Poidevin M., Francesconi S., Tratner I., Zarzov P., Baldacci G. Nuclear factories for signalling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 2003;31:5064–5073. doi: 10.1093/nar/gkg719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Taddei A., Vernis L., Poidevin M., Gasser S. M., Baldacci G. Temporal separation of replication and recombination requires the intra-S checkpoint. J. Cell Biol. 2005;168:537–544. doi: 10.1083/jcb.200410006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Karagiannis J., Sevugan M., Singh P., Balasubramanian M. K. The 14-3-3 protein rad24p modulates function of the cdc14p family phosphatase clp1p/flp1p in fission yeast. Curr. Biol. 2005;15:1376–1383. doi: 10.1016/j.cub.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami H., Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- Noguchi C., Noguchi E. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics. 2007;175:553–566. doi: 10.1534/genetics.106.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E., Noguchi C., Du L. L., Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E., Noguchi C., McDonald W. H., Yates J. R., 3rd, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 2004;24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- O'Connell M. J., Raleigh J. M., Verkade H. M., Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill T., Giarratani L., Chen P., Iyer L., Lee C. H., Bobiak M., Kanai F., Zhou B. B., Chung J. H., Rathbun G. A. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 2002;277:16102–16115. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- Peng C. Y., Graves P. R., Thoma R. S., Wu Z., Shaw A. S., Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Rhind N., Furnari B., Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- Rhind N., Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998a;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Russell P. Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol. 1998b;18:3782–3787. doi: 10.1128/mcb.18.7.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Russell P. Roles of the mitotic inhibitors Wee1 and Mik1 in the G(2) DNA damage and replication checkpoints. Mol. Cell. Biol. 2001;21:1499–1508. doi: 10.1128/MCB.21.5.1499-1508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 1997;283:506–520. doi: 10.1016/s0076-6879(97)83040-6. [DOI] [PubMed] [Google Scholar]
- Sicard H., Faubladier M., Noaillac-Depeyre J., Léger-Silvestre I., Gas N., Caizergues-Ferrer M. The role of the Schizosaccharomyces pombe gar2 protein in nucleolar structure and function depends on the concerted action of its highly charged N terminus and its RNA-binding domains. Mol. Biol. Cell. 1998;9:2011–2023. doi: 10.1091/mbc.9.8.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith H. A., Brown G. W., Forsburg S. L. Schizosaccharomyces pombe Hsk1 is a potential Cds1p target required for genome integrity. Mol. Cell. Biol. 2000;20:7922–7932. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommariva E., Pellny T. K., Karahan N., Kumar S., Huberman J. A., Dalgaard J. Z. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol. Cell. Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Ogino K., Tatebayashi K., Ikeda H., Arai K., Masai H. Regulation of checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1. Mol. Biol. Cell. 2001;12:1257–1274. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., McCollum D. Distinct nuclear and cytoplasmic functions of the S. pombe Cdc14-like phosphatase Clp1p/Flp1p and a role for nuclear shuttling in its regulation. Curr. Biol. 2005;15:1384–1389. doi: 10.1016/j.cub.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Trautmann S., Rajagopalan S., McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell. 2004;7:755–762. doi: 10.1016/j.devcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B. A., Jorgensen P., Tyers M., Gould K. L., McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Vazquez-Novelle M. D., Esteban V., Bueno A., Sacristan M. P. Functional homology among human and fission yeast Cdc14 phosphatases. J. Biol. Chem. 2005;280:29144–29150. doi: 10.1074/jbc.M413328200. [DOI] [PubMed] [Google Scholar]
- Walworth N., Davey S., Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- Wolfe B. A., Gould K. L. Fission yeast Clp1p phosphatase affects G2/M transition and mitotic exit through Cdc25p inactivation. EMBO J. 2004;23:919–929. doi: 10.1038/sj.emboj.7600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B. A., McDonald W. H., Yates J. R., 3rd, Gould K. L. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell. 2006;11:423–430. doi: 10.1016/j.devcel.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Wood V., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Xu Y. J., Davenport M., Kelly T. J. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev. 2006;20:990–1003. doi: 10.1101/gad.1406706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.