Abstract
The fusion of yeast vacuoles, like other organelles, requires a Rab-family guanosine triphosphatase (Ypt7p), a Rab effector and Sec1/Munc18 (SM) complex termed HOPS (homotypic fusion and vacuole protein sorting), and soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs). The central 0-layer of the four bundled vacuolar SNAREs requires the wild-type three glutaminyl (Q) and one arginyl (R) residues for optimal fusion. Alterations of this layer dramatically increase the Km value for SNAREs to assemble trans-SNARE complexes and to fuse. We now find that added purified HOPS complex strongly suppresses the fusion of vacuoles bearing 0-layer alterations, but it has little effect on the fusion of vacuoles with wild-type SNAREs. HOPS proofreads at two levels, inhibiting the formation of trans-SNARE complexes with altered 0-layers and suppressing the ability of these mismatched 0-layer trans-SNARE complexes to support membrane fusion. HOPS proofreading also extends to other parts of the SNARE complex, because it suppresses the fusion of trans-SNARE complexes formed without the N-terminal Phox homology domain of Vam7p (Qc). Unlike some other SM proteins, HOPS proofreading does not require the Vam3p (Qa) N-terminal domain. HOPS thus proofreads SNARE domain and N-terminal domain structures and regulates the fusion capacity of trans-SNARE complexes, only allowing full function for wild-type SNARE configurations. This is the most direct evidence to date that HOPS is directly involved in the fusion event.
INTRODUCTION
Proteins are routed to their correct intracellular compartment by selective sorting into budding vesicles, targeting of these vesicles to their proper organelle, and regulated membrane fusion. Fusion entails successive steps of tethering; enrichment of key proteins and lipids to form a fusion-competent membrane microdomain (Lang et al., 2001; Wang et al., 2002; Fratti et al., 2004); assembly of proteins from each apposed membrane into fusion complexes; and lipid bilayer rearrangements, often via a hemifusion intermediate. Intracellular membrane fusion requires Rab family guanosine triphosphatases (GTPases), multisubunit Rab-effector complexes, Sec1/Munc18 (SM) proteins, and soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (Jahn et al., 2003). Rab GTPases promote tethering and the enrichment of proteins and lipids in microdomains that support fusion (Lang et al., 2001; Miaczynska and Zerial, 2002; Wang et al., 2002). Rab-effector complexes bind to the active, GTP-bound form of the Rabs and are thereby activated to further the fusion cascade. SM proteins associate with SNAREs and are required for the ensuing membrane fusion (Rizo and Südhof, 2002). Model studies, using SNARE proteins reconstituted into liposomes (Weber et al., 1998) or organelles bearing different levels of SNARE proteins (Starai et al., 2007), have shown that complexes of membrane-anchored SNAREs alone can promote either fusion with preservation of vesicular integrity and mixing of lumenal compartments (Nickel et al., 1999), membrane lysis (Dennison et al., 2006; Chen et al., 2006), or both (Starai et al., 2007). In contrast, fusion is accompanied by very little lysis when it occurs with physiological levels of SNAREs, Rab GTPase, and Rab effector (Starai et al., 2007). We are only beginning to see how SNAREs promote fusion or lysis, what regulates their function and directs it toward fusion, and how this is coordinated with Rabs, their effectors, and SM proteins.
SNARE proteins are recognized by their characteristic heptad-repeat “SNARE motifs” which assemble into four-helical coiled coils to form a SNARE complex (Fasshauer et al., 1998). Structural studies (Sutton et al., 1998) have shown that most of the amino acyl residues that face each other on the apposed surfaces of the four helices in a SNARE complex are apolar, with the notable exception of the clustered central residues of each SNARE domain, termed the 0-layer. In virtually all organisms and organelles, the 0-layer consists of three glutaminyl and one arginyl residues, forming an ionic core of the SNARE complex (Fasshauer et al., 1998). Genetic, physiological, and biochemical studies have established the importance of the 0-layer, although it remains unclear why it is required.
SM family proteins are required for the SNARE-dependent fusion of biological membranes (Toonen and Verhage, 2003; Burgoyne and Morgan, 2007). SM proteins interact with SNARE proteins in at least three different modes. Munc18-1 binds to the “closed” conformation of syntaxin, thereby preventing its assembly into SNARE complexes (Dulubova et al., 1999). Other SM proteins can bind to the extreme N terminus of their cognate syntaxins without preventing SNARE complex assembly (Dulubova et al., 2002; Yamaguchi et al., 2002). SM proteins can also bind to a preassembled SNARE complex and not the free syntaxin (Carr et al., 1999; Scott et al., 2004). Elucidating the molecular mechanisms of SNARE-dependent membrane fusion will require understanding the functional interactions between SNAREs and SM family proteins during fusion.
Vacuoles (lysosomes) from Saccharomyces cerevisiae are convenient for the study of SNARE-, SM protein-, and Rab GTPase-dependent fusion. Vacuole homotypic fusion is required for normal organelle structure in vivo (Wada et al., 1992), and it can be monitored conveniently in vitro by colorimetric assays (Haas et al., 1994; Jun et al., 2007). When purified vacuoles are incubated with ATP, their cis-SNARE complexes are disassembled early in the incubation by the combined action of Sec17p and Sec18p (Mayer et al., 1996; Ungermann et al., 1998a; Jun and Wickner, 2007a). Tethering requires the vacuolar Rab GTPase Ypt7p (Mayer and Wickner, 1997) and its hexameric effector complex, termed HOPS (homotypic fusion and vacuole protein sorting; Stroupe et al., 2006). HOPS consists of a core of four subunits, Vps 11, 16, 18, and 33, associated on the vacuole with two other subunits, Vps39p and Vps41p (Sato et al., 2000; Seals et al., 2000). After tethering, vacuoles are drawn against each other, establishing three microdomains: the disk-shaped boundary domain of tightly apposed membrane from each vacuole; the ring-shaped vertex domain at the edge of the boundary domain; and the outside domain, which does not touch the other vacuole (Wang et al., 2002). Each of the proteins (Wang et al., 2002) and lipids (Fratti et al., 2004) that are required for vacuole fusion become enriched at the vertex ring, and fusion ensues. The vertex ring-enriched proteins (Ypt7p, HOPS, SNAREs, and others) and lipids (phosphoinositides, ergosterol, and diacylglycerol) are interdependent for their vertex enrichment (Wang et al., 2003; Fratti et al., 2004). Studies with reversible inhibitory ligands of these proteins and lipids show that the formation of fusion-competent vertex ring microdomains is reversible or highly cooperative (Jun et al., 2006).
HOPS has a direct affinity for phosphoinositides and for the Vam7p SNARE (Stroupe et al., 2006). The HOPS subunit Vps39p has nucleotide exchange activity for Ypt7p (Wurmser et al., 2000), and HOPS binds specifically to the GTP-bound form of Ypt7p (Seals et al., 2000). The HOPS subunit Vps33p is a member of the SM protein family. Ypt7p and HOPS are necessary for SNAREs to pair in trans (Collins and Wickner, 2007). Vacuole fusion requires three glutaminyl (Q)-SNAREs (Vti1p, Vam3p, and Vam7p) and one arginyl (R)-SNARE (Nyv1p). Although the wild-type 3Q:1R set of SNAREs gives optimal fusion, fusion can also occur in vitro with 4Q SNAREs or even with 2Q SNAREs and 2R SNAREs when complex assembly is driven by high concentrations of SNAREs (Fratti et al., 2007). Vacuolar SNARE complexes are associated with Sec17p (α-SNAP) or HOPS, but not with both (Collins et al., 2005). Although Sec17p association allows SNARE complexes to be disassembled, the role of HOPS association in SNARE complex function is unknown.
We now show that the addition of purified HOPS to vacuole fusion reactions reduces the levels of noncanonical trans-SNARE complexes formed during fusion, and further reduces the capacity of these mismatched complexes to undergo fusion. With wild-type 3Q:1R SNAREs, additional HOPS stimulates trans-SNARE complex formation (Collins and Wickner, 2007) and fusion. However, with 4Q SNAREs, 2Q:2R SNAREs, or 3Q:1R SNAREs in rotated positions in the transcomplex, the addition of purified HOPS suppresses the level of trans-SNARE complex and even further inhibits the subsequent fusion. Furthermore, exogenous HOPS strongly suppresses vacuole fusion when trans-SNARE complexes include a mutant Vam7p SNARE lacking its phosphoinositide-binding Phox homology (PX) domain. Thus, the HOPS association with SNARE complexes proofreads the wild-type conformations of the trans-SNARE complex and regulates its capacity to lead to membrane fusion.
MATERIALS AND METHODS
Yeast Strains, Vacuole Isolation, and In Vitro Fusion Assay
Vacuoles were isolated by floatation on discontinuous Ficoll gradients (Haas et al., 1994) from S. cerevisiae strains BJ3505 (MATα ura3-52 trp1-Δ101 his3-Δ200 lys2-801 gal2 (gal3) can1 prb1-Δ1.6R pep4::HIS3) (Jones, 2002) and DKY6281 (MATα ura3-52 leu2-3112 trp1-Δ901 his3-Δ200 lys2-801 suc2-Δ9 pho8::TRP1) (Haas et al., 1994), or isogenic strains expressing Nyv1R192Qp (RFY1 and RFY2) (Fratti et al., 2007). Unless otherwise stated, the in vitro vacuole fusion reactions in this study were ATP-free fusion reactions (Thorngren et al., 2004), incubated at 27°C for 90 min. These reactions contained 3 μg of pep4Δ vacuoles (from BJ3505 derivatives) and 3 μg of pho8Δ vacuoles (from DKY6281 derivatives) in 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-KOH, pH 6.8, 200 mM sorbitol, 125 mM KCl, 5 mM MgCl2, 10 μM CoA, 815 nM purified Pbi2p (IB2), and 10 mg/ml bovine serum albumin (BSA). Mature Pho8p alkaline phosphatase activity was assayed as a measure of vacuole fusion (Haas, 1995). Fusion units (U) are micromoles of p-nitrophenylate formed per minute per microgram of pep4Δ vacuole.
Reagents
Antibodies were purified as described previously, and they were dialyzed into PS buffer (20 mM PIPES-KOH, pH 6.8, and 200 mM sorbitol) with 125 mM KCl. Anti-Vam3p (Wang et al., 2003) was used at 900 nM, and affinity-purified anti-Sec17p (Haas and Wickner, 1996) was used at 209 nM. Recombinant glutathione transferase (GST)-Vam7p, GST-Vam7Q283Rp, and GST-Vam7-SD fusion proteins were purified via glutathione-affinity chromatography and dialyzed into PS containing 125 mM KCl (Fratti et al., 2007; Fratti and Wickner, 2007).
HOPS Overproduction
A yeast strain producing a Vps33-TEV-GST fusion protein was constructed by transforming BJ2168 (MATα ura3-52 leu2-3112 trp1-Δ101 prb1-1122 pep4-3 prc1-407 gal2) (Zubenko et al., 1980) with a polymerase chain reaction (PCR) product formed from a two-round PCR amplification. A GST-TRP1 PCR product was amplified from pFA6a-GST-TRP1 (Longtine et al., 1998) by using primers 1a and 1b (Table 1), introducing a tobacco etch virus (TEV) protease site upstream of the GST coding region. This product was used as a template in a second round of PCR with primers 1b and 1c. This PCR product was transformed into BJ2168, directing the recombination of the -TEV-GST-TRP1 PCR product to the 3′ end of VPS33. This strain was named CSY14.
Table 1.
Primer sets used in this study
| Primer set | Sequence (5′ to 3′) |
|---|---|
| 1a | GGT CCA GGT GAA AAT TTG TA TTT TCA AGG TGG TCC AGG TCG GAT CCC CGG GTT AAT TAA |
| 1b | GCA CAT TTG CAT ATA CAA AAA ATT AAC AAA TCT ATC ATA TAA TAA GAA TTC GAG CTC GTT TAA AC |
| 1c | GCC GAT GGC TTG ATC AAT GGC ACA AGG ATC ATG AAC TCT ATA TCT GGT CCA GGT GAA AAT TTG TAT TTT C |
| 2 | GCG CGT CGA CTT ATC AAC GCG GAA CCA GAT CCG |
| GCG CAC TAG TAT GAA TAG ATT TTG GAA TAC TAAG | |
| 3 | GCG CAC TAG TAT GAT AAA AAC ACG TAT AGA GGA AG |
| GCG CCT CGA GCT ATT CAT TCC ATT TGG CTA ATT C | |
| 4 | GCGCGGATCCATGTCCCTGAGCTCCTGGAG |
| GCGCCTCGAGTTAAATAGTGATGTCAGAATAACTG | |
| 5 | GCG CGG ATC CAT GAA AAA CCC TAG CTT CGA CTG |
| GCG CCT CGA GCT ATA TCC TGC TCA TAG TTT CAT TTG | |
| 6 | GCA AAA TAA AAA AGC ATT TTA ACG AAG AGT ATA TAC CTA CTA TTA GAC ATT AAT GCG TAC GCT GCA GGT CGA C |
| CGC CTG ATT GTT GAT CCA AAA CAG AAT CAT TCT GAT GAT TAT CTG TAG TCA TCG ATG AAT TCT CTG TCG | |
| 7 | GAT CAG CAA AAA CCC TTC AAA ATA TCA ATT TAT ACC AAA AAT TAA GAA TTC GAG CTC GTT TAA AC |
| CGC AGT GAT ATC CGA TGA TTT CAG CGA GTG TAG CTT TTG AGC TCT TAA CAT TTT GAG ATC CGG GTT TT | |
| 8 | CGG CTG TTC AAT TAT CTT TTC AAC CGG ACG GGT CTT ATA TTG ATC AGC AAA AAC CCT TCA AAA TAT C |
| CTT TGC TAA AAC TAA TTT TTG TGA CTG TTC AGT GGG CAA AAT CGC AGT GAT ATC CGA TGA TTTC |
The p400GAL1 series of vectors, based on the pRS400 vectors (Sikorski and Hieter, 1989), were a generous gift from E. Schiebel (ZMBH, Heidelberg, Germany). VPS33-TEV-GST was amplified from CSY14 chromosomal DNA by using primer set 2 and cloned into the SpeI/SalI sites of p403GAL1. VPS18, VPS11, and VPS16 were amplified from FY834 (MATα ura3-52 leu2Δ1 lys2Δ202 trp1Δ63 his3Δ200) (Winston et al., 1995) by using primer sets 3, 4, and 5, respectively. Each of these products was cloned into p405GAL1, p406GAL1, and p404GAL1, respectively, by using the SpeI/XhoI, BamHI/XhoI, and BamHI/XhoI sites.
To construct a strain expressing all six HOPS genes under the control of the GAL1 promoter, FY834 was sequentially transformed with the following linearized plasmids: p403GAL1-VPS33-TEV-GST digested with NsiI, p405GAL1- VPS18 digested with StuI, p406GAL1-VPS11 digested with BstXI, and p404GAL1-VPS16 digested with NdeI. The GAL1 promoter from pYM-N23 (Janke et al., 2004) was amplified with primer set 6, and it was targeted to recombine upstream of VPS41 through flanking homologous DNA sequence in each primer. The GAL1 promoter was amplified from pFA6a-kanMX6-PGAL (Longtine et al., 1998) by using primer set 7, and it was targeted to recombine upstream of VPS39 by extending the flanking VPS39 homology of the initial PCR product with primer set 8. This created the strain CHY26, which overproduced all of the HOPS subunits upon growth in the presence of galactose.
A cassette from pAG32 (Goldstein and McCusker, 1999) was then used to delete the PEP4 gene from CHY26 by using the PEP4 primer sequences used in the commercial nonessential deletion library (Giaever et al., 2002), resulting in CHY31 (CHY24 pep4Δ::KanMX4).
GST-HOPS Purification
CHY31 from solid medium was grown at 30°C in CSM-his-leu-trp-ura dropout medium (MP Biomedicals, Irvine, CA), supplemented with yeast nitrogen base, 0.1% monosodium glutamate, 2% glucose, 100 μg/ml Clonat (WERNER BioAgents, Jena, Germany), and 200 μg/ml G418, adjusted to pH 6.5 with KOH, for 14 h. This culture was used to inoculate 600 ml of the medium, and growth was continued for 8 h, when OD600 reached ∼1.3. Four 6-l flasks, each with 2.5 l of YP + 2% galactose, were inoculated with 160 ml each of the starter culture and grown at 30°C for 14 h to OD600 ∼ 1.5. Each flask then received 130 ml of 50% glucose, and cells were grown for an additional 90 min (final OD600 ∼ 2.0). Cells were harvested by centrifugation (Beckman JA-10 rotor; 5000 rpm, 5 min, 23°C), resuspended with a glass rod, and mixed by inversion in 100 mM Tris-Cl, pH 9.4, 10 mM dithiothreitol (50 ml/OD · l), incubated with occasional inversion in a 30°C water bath for 10 min, and centrifuged as described above. Each pellet was resuspended in 15 ml/OD · l of 80% YP, 0.16% glucose, 0.6 M sorbitol, and 0.05 M KPi, pH 7.5, mixed with 2 mg of recombinant lyticase/OD · l, and incubated 35 min at 30°C, with inversion every 10 min. Suspensions were transferred to six ice-cold JA-14 bottles and centrifuged (5000 rpm, 5 min, 2°C). Supernatants were aspirated, and the pellet was gently resuspended in 1.1× the measured pellet weight of ice-cold 15% Ficoll, 20 mM PIPES-KOH, pH 6.8, and 200 mM sorbitol. DEAE-dextran (100 μl/OD · l of a 25 mg/ml solution in 8% Ficoll [8% Ficoll (wt/vol), 20 mM PIPES-KOH, pH 6.8, 200 mM sorbitol]) was used to resuspend the pellets, which were incubated 2 min on ice with gentle mixing every 30 s, placed at 30°C for 4 min, and then returned to ice. Cells were transferred to ice-cold 60Ti ultracentrifuge tubes, 10 ml/tube, and mixed by inversion with 14 ml of 8% Ficoll [8% Ficoll (wt/vol), 20 mM PIPES-KOH, pH 6.8, and 200 mM sorbitol] before centrifugation (50,000 rpm, 65 min, 4°C). Floated vacuoles were collected, glycerol was added to 10%, phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM, and the suspension was added dropwise directly to liquid nitrogen. The frozen droplets were stored at −80°C.
Frozen vacuoles (∼800 mg of protein) were thawed, mixed with 1.2 l of cold 20 mM HEPES-NaOH, pH 7.8, 200 mM sorbitol, and 50 mM NaCl, and centrifuged (JA-14 rotor; 10,000 rpm, 15 min, 4°C). Pellets were suspended in 800 ml of HOPS vacuole lysis buffer (HVLB: 20 mM HEPES-NaOH, pH 7.8, 400 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol, and 1.0% Triton X-100) and incubated on ice for 20 min. Insoluble material was removed by centrifugation (60Ti; 20 min, 50,000 rpm, 4°C), and the supernatant was passed through a 0.2-μm filter (Millipore, Billerica, MA). This was applied to a 2.5 × 8 cm glutathione-Sepharose 4B column (GE Healthcare, Chalfont St. Giles, United Kingdom), which had been pre-equilibrated with HVLB at 4°C. The column was washed with 3 bed volumes of HVLB, and then 3 bed volumes of low Triton X-100 HOPS buffer (LTHB: 20 mM HEPES-NaOH, pH 7.8, 400 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol, and 0.004% Triton X-100). Protein was eluted from the column with LTHB + 10 mM glutathione. Fractions containing HOPS were pooled, concentrated from ∼15 to ∼1 ml, with a final protein concentration of 0.5–1.0 mg/ml in an Amicon Ultra-15 (100,000 NMWL; Millipore) centrifugal filter device, distributed into small aliquots, and frozen in liquid nitrogen. When calculated, the molar concentrations of HOPS considered the molecular mass of HOPS to be 663 kDa, assuming a 1:1:1:1:1:1 subunit stoichiometry in the complex.
RESULTS
We assay the fusion of vacuoles that have been purified from two yeast strains. One lacks vacuolar lumenal proteases and therefore accumulates inactive proalkaline phosphatase (proPho8p), and the other has normal vacuole lumenal proteases but is deleted for the gene encoding Pho8p. Although neither vacuole population has phosphatase activity, bilayer fusion and content mixing allow the proteases to gain access to proPho8p and cleave it to active Pho8p. We measure vacuole fusion and content mixing by a colorimetric assay of this active Pho8p. The homotypic fusion of yeast vacuoles normally depends upon the formation of 3Q:1R trans-SNARE complexes (Fratti et al., 2007). Although vacuoles with either 4Q or 2Q:2R SNARE complexes can fuse, fusion requires either higher levels of SNAREs or the addition of exogenous compounds that promote lipid rearrangements (Fratti et al., 2007). Because the purified HOPS complex interacts with both the Vam7p SNARE and with certain lipids (Stroupe et al., 2006), we tested whether it might restore full fusion to vacuoles with noncanonical 0-layer residues.
HOPS Complex Inhibits Fusion of Vacuoles with a Mismatched SNARE 0-Layer
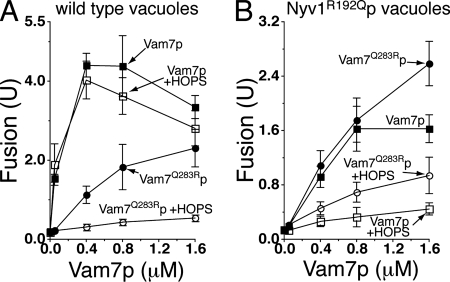
Purified vacuoles have substantial levels of unpaired, membrane-anchored Vam3p, Vti1p, and Nyv1p (Collins et al., 2005). When Vam7p, a soluble SNARE, is provided exogenously, fusion does not require ATP-dependent cis-SNARE complex disassembly (Thorngren et al., 2004; Figure 1A, filled squares). Although HOPS is required for in vitro vacuole fusion (Seals et al., 2000; Stroupe et al., 2006), isolated vacuoles bear sufficient HOPS for fusion (∼0.52 nM in the fusion reaction; Stroupe et al., 2006), and supplementation with purified HOPS complex at levels 30-fold over the endogenous vacuolar levels (15.8 nM) has little effect on fusion at any level of added Vam7p (Figure 1A, open squares). High concentrations of recombinant Vam7Q283Rp, mutated to change the 0-layer residue from Q to R, can drive fusion as a component of a 2Q:2R SNARE complex (Figure 1A, filled circles), as reported previously (Fratti et al., 2007). Unlike wild-type 3Q:1R SNARE complex fusion, however, exogenous HOPS addition caused a striking inhibition of the Vam7Q283Rp-mediated 2Q:2R fusion (Figure 1A, open circles). The HOPS SM protein subunit Vps33p, when purified from yeast cytosol, did not have proofreading activity under these conditions, even when added at a higher molar level than HOPS (data not shown). This suggests that the proofreading activity of HOPS may not reflect Vps33p function alone, although we have no independent assay to verify the activity of the isolated Vps33p.
Figure 1.
HOPS complex inhibits fusion when the SNARE 0-layer is altered. Vacuoles from either BJ3505 and DKY6281 (A) or RFY1 and RFY2 (bearing Nyv1R192Qp) (B) were assayed for fusion under standard ATP-free conditions (see Materials and Methods). Either wild-type Vam7p (squares) or mutant Vam7Q283Rp (circles) was added at the indicated concentrations, either in the absence (closed symbols) or presence (open symbols) of 15.8 nM purified HOPS complex. Results are the mean of three independent experiments ± SD.
To determine whether the HOPS complex inhibition of fusion is specific to vacuoles forming a 2Q:2R trans-SNARE complex, or whether it is a more general proofreading activity, we isolated vacuoles from strains expressing the vacuolar R-SNARE Nyv1p as a Q-SNARE (Nyv1R192Qp). The addition of Vam7Q283Rp to these vacuoles restores the potential to form a 3Q:1R SNARE complex, albeit with the position of these residues rotated with respect to the four-helical bundle, and this provided slightly greater fusion stimulation than the addition of wild-type Q-SNARE Vam7p which formed a 4Q SNARE complex (Fratti et al., 2007; Figure 1B, compare filled circles with filled squares). Adding HOPS complex suppressed the fusion of Nyv1R192Qp vacuoles supported by either Vam7p or Vam7Q283Rp (Figure 1B, open symbols). Together, HOPS complex favors the fusion activity of the wild type and properly oriented 3Q:1R SNARE complexes over any 0-layer mismatched set tested.
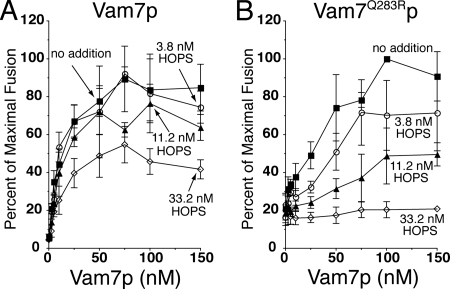
Because these assays were performed in the absence of ATP, which is normally present in the cell and in our in vitro reactions, we also tested the ability of HOPS to proofread the SNARE complex 0-layer in the presence of ATP. To render these reactions dependent on added Vam7p, Sec18p/Sec17p priming activity was blocked by α-Sec17p antibody, as reported and characterized previously (Thorngren et al., 2004). Under these conditions, fusion driven by up to 50 nM wild-type Vam7p is not affected by the addition of 3.8 or 11.2 nM HOPS, although some inhibition is seen at 33.2 nM HOPS (Figure 2A). In contrast, the fusion activity of Vam7Q283Rp is strongly reduced in a HOPS concentration-dependent manner (Figure 2B), confirming the ability of HOPS to gauge the status of the SNARE-complex 0-layer before content mixing.
Figure 2.
HOPS complex proofreads the 0-layer in the presence of ATP. Vacuoles were assayed for fusion in reactions where the priming block by α-Sec17p was bypassed by added Vam7p (Thorngren et al., 2004). Reactions (30 μl) contained vacuoles from BJ3505 and DKY6281 (3 μg each) in 20 mM PIPES-KOH, pH 6.8, 200 mM sorbitol, 150 mM KCl, 6 mM MgCl2, 10 μM CoA, 1 mM ATP, 1 mg/ml creatine kinase, 29 mM creatine phosphate, 815 nM purified IB2, and 209 nM affinity-purified α-Sec17. After 5 min on ice, HOPS and either Vam7p (A) or Vam7Q283Rp (B) was added at the indicated concentrations. Fusion was measured after 90 min at 27°C. Fusion data are from three independent reactions, standardized as a percentage of the maximum fusion signal obtained with each Vam7p addition, which was 4.78 ± 0.72 U (A) and 1.38 ± 0.33 U (B), mean ± SD.
HOPS Does Not Inhibit Fusion by Promoting Lysis
Whereas wild-type levels of SNAREs, Ypt7p, and HOPS support fusion without triggering lysis, increasing the number of trans-SNARE complexes raises the overall amount of organelle rupture, leading to fusion inhibition (Starai et al., 2007). Although levels of all four SNAREs must be elevated to cause massive lysis, detectable lysis can be triggered on wild-type vacuoles by the addition of recombinant Vam7p (Starai et al., 2007). Added HOPS can increase the number of trans-SNARE complexes formed during fusion (Collins and Wickner, 2007), although this increase does not cause additional lysis with wild-type vacuoles (Starai et al., 2007). To test whether 0-layer mismatch promotes lysis, and whether HOPS inhibits the 2Q:2R fusion driven by Vam7Q283Rp by redirecting docked vacuoles toward lysis, we assayed the release of green fluorescent protein (GFP) from the vacuole lumen during fusion.
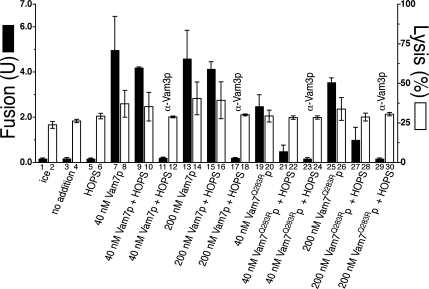
Under ATP-free conditions, the addition of Vam7p drives fusion (Figure 3, bar 5 vs. 7) as well as measurable lysis (Figure 3, bars 4 vs. 8). The addition of HOPS complex does not significantly affect this Vam7p-driven fusion or lysis (Figure 3, bars 9 and 10), and a SNARE ligand, α-Vam3p, inhibits fusion and lysis to basal levels (Figure 3, bars 11 and 12). A higher concentration of Vam7p does not drive significantly more fusion or lysis (Figure 3, bars 13–16). Vam7Q283Rp causes less fusion and lysis than the wild-type protein when used at the same concentrations (Figure 3, bars 19 and 20 and bars 25 and 26), in accordance with the finding that Vam7p must form functional transcomplexes to drive the lysis of vacuoles (Starai et al., 2007). HOPS complex inhibits the fusion caused by Vam7Q283Rp (Figure 3, bars 19 vs. 21 and 25 vs. 27), and it does not enhance Vam7Q283Rp-dependent lysis (Figure 3, bars 20 vs. 22 and 26 vs. 28). Therefore, HOPS complex does not inhibit the fusion of mismatched trans-SNARE complexes through increased membrane lysis, but rather through a 0-layer proofreading mechanism.
Figure 3.
HOPS does not promote fusion-dependent lysis with altered 0-layer SNARE complexes. Reactions (90 μl) containing 9 μg each of vacuoles from a BJ3505 derivative bearing lumenal soluble GFP (Starai et al., 2007) and from DKY6281 were incubated at 27°C under ATP-free conditions (see Materials and Methods) with the following modifications: BSA was used at 3 mg/ml, and reactions contained 0.1× protease inhibitor cocktail (50× stock: 13 μg/ml leupeptin, 25 mM 1,10-phenanthroline, 25 μg/ml pepstatin A, and 5 mM Pefabloc SC). This concentration of protease inhibitor does not inhibit the proteolytic activation of proPho8p during fusion. Where indicated, HOPS was added to 15.8 nM. After 45 min, reactions were gently mixed and placed on ice; 30 μl was withdrawn to measure fusion (black bars) via Pho8p activity, and 30 μl was assayed for vacuolar lysis (white bars) via centrifugation (5200 × g, 6 min, 4°C) and assay of GFP fluorescence in pellets and supernatants, as described previously (Starai et al., 2007).
HOPS Regulates the Assembly, and Function, of trans-SNARE Complexes
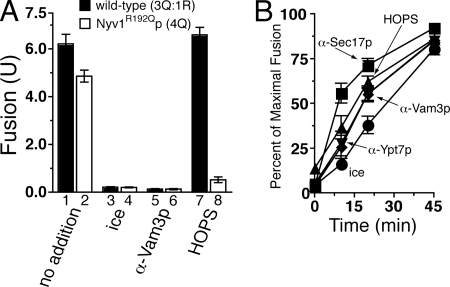
In contrast to a previous report from our group (Fratti et al., 2007)—but in agreement with other reports showing that 4Q-SNARE-mediated membrane fusion is well-tolerated (Ossig et al., 2000; Katz and Brennwald, 2000)—vacuoles bearing Nyv1R192Qp fuse with ∼78% the yield of wild-type vacuoles under our standard, ATP-containing fusion conditions (Figure 4A, bars 1 vs. 2). Adding HOPS to wild-type vacuoles does not significantly change this fusion (Figure 4A, bars 1 vs. 7), but strongly inhibits the 4Q:0R SNARE complex-mediated fusion of Nyv1R192Qp vacuoles (Figure 4A, bars 2 vs. 8). These data show that HOPS is a potent inhibitor of mismatched 0-layer fusion in a standard in vitro fusion assay.
Figure 4.
HOPS proofreads after SNARE complex disassembly and before content mixing. Vacuoles were assayed for fusion under ATP-containing conditions (20 mM PIPES-KOH, pH 6.8, 200 mM sorbitol, 150 mM KCl, 6 mM MgCl2, 10 μM CoA, 1 mM ATP, 1 mg/ml creatine kinase, 29 mM creatine phosphate, and 815 nM purified IB2). (A) Reactions (30 μl) containing either BJ3505 and DKY6281 (3 μg each, black bars) or RFY1 and RFY2 (3 μg each, white bars) were measured for fusion after 90 min at 27°C. Where indicated, HOPS was added to 20.1 nM. Results are mean ± SD. (B) Reactions (30 μl) containing RFY1 and RFY2 vacuoles were incubated at 27°C, and either the fusion inhibitors α-Sec17p (209 nM; squares), HOPS complex (20.1 nM; triangles), α-Ypt7p (467 nM; inverted triangles), or α-Vam3p (900 nM; diamonds) was added to the fusion reactions at 0-, 10-, 20-, 45-, 60-, and 90-min time points, or the reactions were removed to ice (circles) at the same intervals. After 90 min, the extent of fusion was measured and compared with the fusion levels of an uninhibited fusion reaction after 90 min (maximal fusion). Results are the mean ± SD of three independent experiments (maximal fusion was 5.32, 5.89, and 5.0 U). The first 45 min of the reaction is plotted for clarity.
By taking advantage of an assay designed to test when vacuoles become resistant to a given fusion inhibitor, we can dissect the vacuole fusion reaction into “priming,” “docking,” and “content mixing” stages (Ungermann et al., 1998b). Using this assay, we can compare the kinetics of HOPS-mediated fusion inhibition to inhibition by reagents that act at specific reaction stages. Vacuoles bearing Nyv1R192Qp become resistant to HOPS addition well after resistance to antibodies against Sec17p (Figure 4B, squares vs. triangles). HOPS resistance mirrors the resistance curves of α-Ypt7p (inverted triangles) and α-Vam3p (diamonds), and HOPS acts before content mixing (ice, circles). These data suggest that the inhibitory HOPS proofreading activity occurs after priming (defined by Sec17p function), during the trans-SNARE pairing/docking stage of vacuole fusion (defined by Ypt7p and Vam3p functions), and before membrane fusion and content mixing.
To test how HOPS may function at the trans-SNARE pairing/docking stage, we have exploited a recently developed assay of trans-SNARE complexes (Collins and Wickner, 2007) to directly measure the effects of added HOPS when the 0-layer of trans-SNARE complexes are mismatched. Under ATP-free conditions, the association of Nyv1p with Vam3p in trans and the subsequent fusion require added Vam7p (Table 2, lines 2 and 4). Adding HOPS does not significantly change the fusion yield or trans-SNARE complex formation (Table 2, line 5). Vam7Q283Rp forms fewer 2Q:2R transcomplexes compared with the wild-type 3Q:1R complex, and fusion is similarly reduced (Table 2, line 6). Added HOPS complex strongly reduces the amount of Vam7Q283Rp-containing trans-SNARE complex formed (line 7). Surprisingly, even the measurable amount of trans-SNARE complex formed with Vam7Q283Rp in the presence of HOPS is unable to drive proportional fusion (line 7). Together, these data suggest that HOPS is intimately involved in both proofreading the formation of physiological 3Q:1R 0-layer trans-SNARE complexes and in regulating the capacity of those few trans-SNARE complexes which do assemble to support fusion.
Table 2.
HOPS modulation of trans-SNARE complexes and fusion activity
| Reaction conditiona | Fusion (U) | trans-SNARE complexb | Fusion (U) trans-SNARE complex |
|---|---|---|---|
| 1. ice | 0.17 ± 0.02 | 0.4 ± 0.13 | 0.5 ± 0.17 |
| 2. no additions | 0.16 ± 0.02 | 0.9 ± 0.49 | 0.3 ± 0.19 |
| 3. HOPS | 0.16 ± 0.02 | 0.5 ± 0.08 | 0.3 ± 0.05 |
| 4. 150 nM Vam7p | 4.45 ± 0.51 | 4.1 ± 1.98 | 1.4 ± 0.81 |
| 5. 150 nM Vam7p + HOPS | 4.97 ± 0.51 | 4.3 ± 1.29 | 1.2 ± 0.32 |
| 6. 150 nM Vam7Q283Rp | 2.92 ± 0.94 | 3.1 ± 1.35 | 1.0 ± 0.39 |
| 7. 150 nM Vam7Q283Rp + HOPS | 0.62 ± 0.06 | 1.3 ± 0.18 | 0.5 ± 0.12 |
ATP-free fusion reactions (11× scale) with vacuoles from strains BJ3505 nyv1Δ CBP-VAM3 (Collins and Wickner, 2007) and DKY6281 were incubated with the indicated additions at 27°C for 45 min. Reactions were transferred to ice for 5 min, gently mixed, and a 30-μl aliquot was withdrawn to assay alkaline phosphatase activity as a measure of fusion. CBP-Vam3p affinity pull-downs were performed (Collins and Wickner, 2007). Vacuoles were sedimented (12,000 × g, 5 min, 4°C) and dissolved in ice-cold 600 μl of solubilization buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40 Alternative [Calbiochem, San Diego, CA], 10% glycerol, 0.46 μg/ml leupeptin, 3.5 μg/ml pepstatin, 2.4 μg/ml pefabloc-SC, and 1 mM PMSF). After 20 min at 4°C with nutation, insoluble material was removed by centrifugation (16,000 × g, 20 min, 4°C), and 500 μl of each vacuole extract was mixed with 50 μl of calmodulin agarose, which had been pre-equilibrated with solubilization buffer containing 2 mM CaCl2. CaCl2 was added to the vacuolar extract to final concentration of 2 mM, and the suspension was rocked for 16 h at 4°C. Beads were harvested by centrifugation (1400 × g, 2 min, 4°C) and washed five times with 500 μl of ice-cold solubilization buffer, sedimenting beads after each wash as described. Bead-bound proteins were eluted with SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer containing 5 mM EGTA, incubated 5 min at 98°C, separated by SDS-PAGE electrophoresis, and transferred to nitrocellulose. CBP-Vam3p and Nyv1p were detected by immunoblotting. Films were scanned with a computer-controlled PowerLook 1100 scanner (UMAX) and VueScan software version 8.0.4 (Hamrick Software, Phoenix, AZ). Image files were digitized, and the total pixels making up the CBP-Vam3p and Nyv1p bands were quantified using UN-SCAN-IT version 5.1 (Silk Scientific, Orem, UT). Reported values are the mean ± SD of three independent experiments.
a Standard ATP-free reactions (see Materials and Methods); HOPS added to a final concentration of 16.7 μg/ml, where noted.
b Defined as (Nyv1p pixels/CBP − Vam3p pixels) × 10.
HOPS Complex Can Proofread Other Features of the SNARE Complex
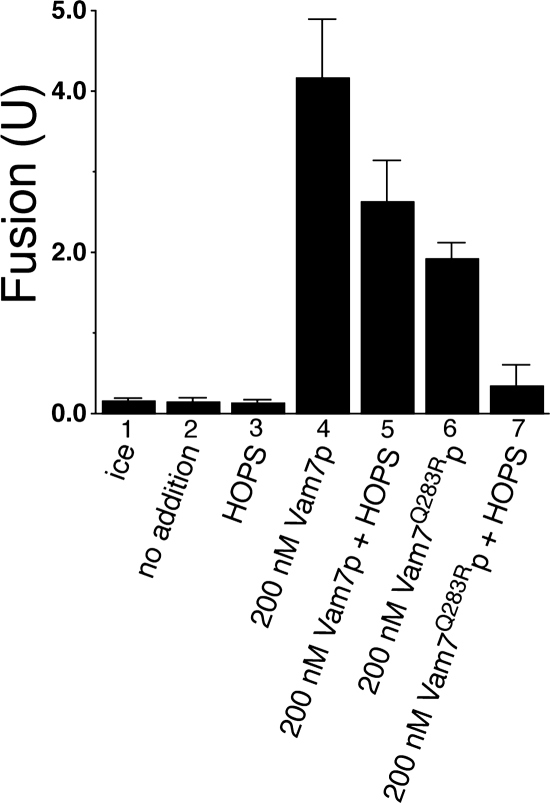
SM proteins can bind their cognate syntaxins through a conserved N-terminal “HABC” domain (Toonen and Verhage, 2003; Burgoyne and Morgan, 2007). The N-terminal domain of syntaxin 1 is essential for Munc18-1–mediated SNARE complex activation in a reconstituted system (Shen et al., 2007), and it has been reported that HOPS can associate with the syntaxin orthologue Vam3p at its N terminus (Laage and Ungermann, 2001). Therefore, we tested whether the N-terminal domain of Vam3p is required for HOPS-dependent SNARE complex 0-layer proofreading. Under ATP-free conditions, vacuoles bearing Vam3p lacking its N-terminal HABC domain (Vam3ΔN, Laage and Ungermann, 2001) can fuse when 200 nM Vam7p is provided (Figure 5, bar 4). Added HOPS slightly inhibits this fusion (bar 5). Vam7Q283R protein also supports the fusion of Vam3ΔN vacuoles (Figure 5, bar 6), but this fusion is strongly inhibited by the addition of exogenous HOPS (Figure 5, bar 7). Thus, HOPS can proofread the SNARE complex 0-layer in the absence of the N terminus of the Vam3p syntaxin.
Figure 5.
HOPS does not require the N-terminal HABC domain of Vam3p for 0-layer proofreading activity. Vacuoles from BJ3505 and DKY6281 derivatives bearing Vam3p deleted for its conserved N-terminal HABC domain (Vam3ΔN; Laage and Ungermann, 2001) were assayed for fusion under standard ATP-free conditions after 90 min at 27°C. Vam7p proteins were added at the indicated concentration, and HOPS was added to a final concentration of 25.0 nM. Results are the mean ± SD of five independent experiments.
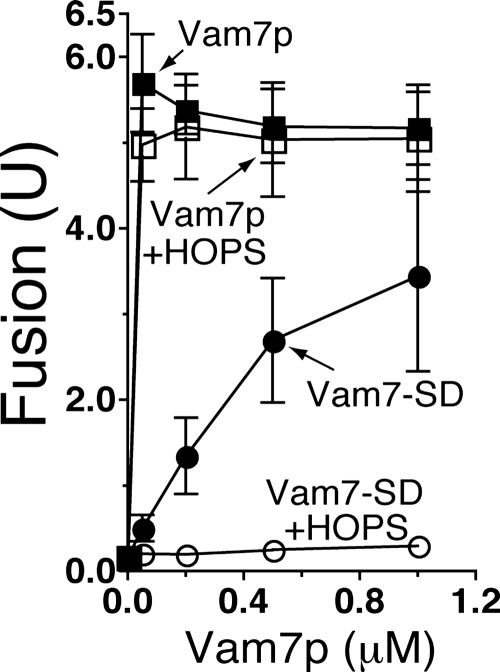
HOPS has been shown to associate with Vam7p in solution via the Vam7p phosphoinositide-binding PX domain (Stroupe et al., 2006). To test whether the HOPS proofreading of vacuole SNARE complexes extends to the Vam7p PX domain, we exploited the fact that the purified Vam7p SNARE domain, which lacks the PX domain, can support vacuole fusion in the absence of ATP (Fratti and Wickner, 2007). Vacuole fusion supported by up to 1 μM full-length, wild-type Vam7p is essentially unaffected by the addition of purified HOPS (Figure 6, compare closed squares with open squares). As reported previously, purified Vam7p SNARE domain (Vam7-SD) supports fusion with a much higher Km value compared with wild-type Vam7p (Fratti and Wickner, 2007; Figure 6, compare closed circles with closed squares). Adding HOPS to reactions containing Vam7-SD causes a striking inhibition of fusion (Figure 6, open circles), showing that the HOPS proofreading activity also gauges features of the SNARE complex in addition to the 0-layer and outside the SNARE domain coiled coils.
Figure 6.
HOPS proofreads at least one domain outside of the SNARE motif. Vacuoles from BJ3505 and DKY6281 were assayed for fusion under standard ATP-free conditions after 90-min incubation with either wild-type Vam7p (squares) or the Vam7p SNARE domain which lacks the PX domain (Vam7-SD, circles). Assays were performed in the absence (closed symbols) or presence (open symbols) of 20.1 nM purified HOPS complex. Results are the mean ± SD of three independent experiments.
DISCUSSION
SM family proteins perform two related functions, the proofreading of SNARE-dependent membrane fusion events and the activation of the SNARE complex for fusion (Peng and Gallwitz, 2002; Scott et al., 2004; Shen et al., 2007). The biochemical and genetic data regarding SM proteins clearly point to an essential role, yet the molecular interactions governing SM protein function at each distinct step are only recently being elucidated. Our current study examines the physical and functional relationship during homotypic vacuolar fusion between the HOPS complex, which includes Vps33p, an SM protein, and the SNARE complex.
We find that HOPS plays a significant role in ensuring correct trans-SNARE complex formation and function. Alterations in the buried 0-layer of the SNARE complex that cause a non3Q:1R—or even a rotated 3Q:1R—structure are significant enough to cause HOPS to inhibit assembly (Figure 1 and Table 2). Because these residues are not surface exposed (Fasshauer et al., 1998; Sutton et al., 1998), HOPS may sense a conformational change in the SNAREs resulting from these seemingly slight changes in the 0-layer, due to a dedicated and specific mode of binding to SNAREs, or the assembled SNARE complex. Binding interactions are often regulated by slight conformational differences, which are necessary to ensure interaction specificity above the background of other similar biological structures (Savir and Tlusty, 2007). In other studies (Shen et al., 2007), mutations in residues adjacent to the +1 and +5 layers in the SNARE motif, which are surface-exposed in the SNARE complex, abolish Munc18-1–mediated stimulation of liposome lipid mixing without abolishing the basal SNARE-mediated lipid mixing. In contrast, added HOPS complex inhibits the fusion activity of noncanonical 0-layer SNARE complexes which otherwise have significant fusion activity (Figures 1–6). Although the N terminus of Vam3p promotes HOPS recruitment to the vacuole (Laage and Ungermann, 2001), the 0-layer proofreading activity does not require the N-terminal HABC SM-protein binding domain of Vam3p (Figure 5), and thus HOPS recognition of the assembled SNARE complex is not simply focused on the Vam3p HABC domain. HOPS strongly suppresses the fusion capacity of trans-SNARE complexes lacking the Vam7p PX domain (Figure 6), possibly reflecting the direct affinity of this PX domain for HOPS (Stroupe et al., 2006). HOPS may use several modes of binding to the assembled SNARE complex to regulate its assembly (Jun et al., 2007b) and may continue to function as part of the trans-SNARE complex during fusion. These results support the finding that Munc18-1 can bind to synaptic SNARE complexes in the absence of the syntaxin N-terminal HABC domain (Shen et al., 2007), and they are in accord with another report describing multiple modes of SM protein:SNARE complex binding for yeast Vps45p (Carpp et al., 2006).
We have reported previously (Fratti et al., 2007; Fratti and Wickner, 2007) that the Km value for wild-type Vam7p to support vacuole fusion is far lower than the Km value for either Vam7Q283Rp or Vam7-SD. This may be caused by the proofreading function of the endogenous vacuolar HOPS. Added Vam7Q283Rp or Vam7-SD, which exceeds the HOPS proofreading capacity, may lead to fusion. Fusion, whether supported by wild-type or 2Q:2R trans-SNARE complexes (Fratti et al., 2007), is blocked by antibodies to the HOPS subunit Vps33p. Because HOPS activity is required for the formation of even mismatched trans-SNARE complexes and for the resulting membrane fusion, it is likely that SNARE complex proofreading and the support of fusion are distinct HOPS activities, each essential for physiological vacuole fusion.
SM proteins can bind to the closed conformation of free syntaxins and prevent them from entering SNARE complexes in solution (Pevsner et al., 1994; Dulubova et al., 1999; Yang et al., 2000; Misura et al., 2000). Yeast Sly1p, when prebound to the Golgi syntaxin Sed5p, can also prevent the formation of soluble 2Q:2R SNARE complexes (Peng and Gallwitz, 2002). However, without SNARE transmembrane anchors, this study could not measure the effects of the assembled 2Q:2R SNARE complexes or the SM protein on membrane fusion. Peng and Gallwitz (2004) also found that direct, high-affinity binding of Sly1p to Sed5p was, surprisingly, not required for either protein's activity in vivo. Furthermore, mutant versions of Sly1p could bind a number of nonsyntaxin SNAREs, suggesting that SM proteins could potentially interact with each of the SNAREs involved in a particular SNARE complex assembly pathway. How, then, do SM proteins prevent non3Q:1R or noncognate SNARE complexes from forming? Simple steric hindrance from a prebound and specific SM:syntaxin complex may be responsible, because each SM protein is well localized to an individual subcellular compartment, and it cannot compensate for the absence of another SM protein at another organelle (Toonen and Verhage, 2003). In contrast, SM proteins may enhance the stability of cognate SNARE complexes, thereby increasing the chances of that SNARE complex causing a fusion event. Vacuole SNAREs, like their neuronal counterparts, may initially form a fusion-incompetent, “open” transcomplex that must be converted to the fusion-competent “closed” SNARE complex. HOPS and its SM subunit Vps33p might prevent noncognate SNARE complexes from progressing from the open to the closed state, whereas SNAREs alone may freely assemble and disassemble without the ability to cause fusion. It has also been proposed that SM proteins can spatially segregate noncognate SNAREs from the active sites of fusion, concentrating the correct SNAREs and increasing the probability that appropriate SNARE complexes will form (Bethani et al., 2007). In addition, the activity of Munc18-1 on the SNARE-dependent lipid mixing of reconstituted proteoliposomes is only seen with the cognate neuronal SNAREs, and not with other SNAREs that can support a background lipid mixing rate in this assay (Shen et al., 2007). This SM protein-mediated proofreading of SNARE identity is distinct from the proofreading of the proper 0-layer and N-domain state of the SNARE complex, which we present here.
In addition to proofreading the SNARE complex, HOPS has tethering and Ypt7p/Rab GTPase nucleotide exchange functions, each of which is also required for efficient membrane fusion (Price et al., 2000; Seals et al., 2000; Wurmser et al., 2000; Stroupe et al., 2006). It is unclear whether the RabGTPase nucleotide exchange activity of HOPS is required for its proofreading activity, but neither normal vacuole fusion nor HOPS proofreading activity responds to exogenous additions of GTP (data not shown). However, SM proteins could link Rab GTPase and SNARE functions; a direct interaction between Munc18-1 and Rab3A was recently detected in bovine brain extracts, in accord with this idea (Graham et al., 2008). Therefore, the proofreading or fusion activities of SM proteins could be regulated by cognate Rab GTPase proteins, although SM proteins and HOPS can activate SNARE-dependent lipid mixing in reconstituted systems that lack Rab GTPases (Scott et al., 2004; Shen et al., 2007; Mima and Wickner, unpublished data).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM-23377. V.J.S. was supported by National Institutes of Health fellowship T32 AR07576, Autoimmunity and Connective Tissue Training Grant, and C.M.H. was supported by training grant 5T32 GM-008704, Molecular and Cell Biology at Dartmouth.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0077) on April 16, 2008.
REFERENCES
- Bethani I., Lang T., Geumann U., Sieber J. J., Jahn R., Rizzoli S. O. The specificity of SNARE pairing in biological membranes is mediated by both proof-reading and spatial segregation. EMBO J. 2007;26:3981–3992. doi: 10.1038/sj.emboj.7601820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne R. D., Morgan A. Membrane trafficking: three steps to fusion. Curr. Biol. 2007;17:R255–R258. doi: 10.1016/j.cub.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Carpp L. N., Ciufo L. F., Shanks S. G., Boyd A., Bryant N. J. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J. Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C. M., Grote E., Munson M., Hughson F. M., Novick P. J. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J. Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Arac D., Wang T.-M., Gilpin C. J., Zimmerberg J., Rizo J. SNARE-mediated lipid mixing depends on the physical state of the vesicles. Biophys. J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. M., Thorngren N. L., Fratti R. A., Wickner W. T. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J. 2005;24:1775–1786. doi: 10.1038/sj.emboj.7600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. M., Wickner W. T. trans-SNARE complex assembly and yeast vacuole membrane fusion. Proc. Natl. Acad. Sci. USA. 2007;104:8755–8760. doi: 10.1073/pnas.0702290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennison S. M., Bowen M. E., Brunger A. T., Lentz B. R. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG-mediated membrane fusion. Biophys. J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Sugita S., Hill S., Hosaka M., Fernandez I., Südhof T. C., Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi T., Gao Y., Min S. W., Huryeva I., Südhof T. C., Rizo J. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton B. R., Brunger A. T., Jahn R. Conserved features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R. A., Jun Y., Merz A. J., Margolis N., Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J. Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R. A., Collins K. M., Hickey C. M., Wickner W. Stringent 3Q:1R composition of the SNARE 0-layer can be bypassed for fusion by compensatory SNARE mutation or by lipid bilayer modification. J. Biol. Chem. 2007;282:14861–14867. doi: 10.1074/jbc.M700971200. [DOI] [PubMed] [Google Scholar]
- Fratti R. A., Wickner W. Distinct targeting and fusion functions of the PX and SNARE domains of yeast vacuolar Vam7p. J. Biol. Chem. 2007;282:13133–13138. doi: 10.1074/jbc.M700584200. [DOI] [PubMed] [Google Scholar]
- Giaever G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Graham M. E., Handley M. T., Barclay J. W., Ciufo L. F., Barrow S. L., Morgan A., Burgoyne R. D. A gain of function mutant of Munc18-1 stimulates secretory granule recruitment and exocytosis and reveals a direct interaction of Munc18-1 with Rab3. Biochem. J. 2008;402:407–416. doi: 10.1042/BJ20071094. [DOI] [PubMed] [Google Scholar]
- Haas A., Conradt B., Wickner W. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J. Cell Biol. 1994;126:87–97. doi: 10.1083/jcb.126.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci. 1995;17:283–294. [Google Scholar]
- Haas A., Wickner W. Homotypic vacuole fusion requires Sec17p (yeast alpha-SNAP) and Sec18p (yeast NSF) EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Lang T., Südhof T. C. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jones E. W. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 2002;351:127–150. doi: 10.1016/s0076-6879(02)51844-9. [DOI] [PubMed] [Google Scholar]
- Jun Y., Thorngren N., Starai V. J., Fratti R. A., Collins K., Wickner W. Reversible, cooperative reactions of yeast vacuole docking. EMBO J. 2006;25:5260–5269. doi: 10.1038/sj.emboj.7601413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y., Wickner W. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc. Natl. Acad. Sci. USA. 2007a;104:13010–13015. doi: 10.1073/pnas.0700970104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun Y., Xu H., Thorngren N., Wickner W. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. EMBO J. 2007b;26:4935–4945. doi: 10.1038/sj.emboj.7601915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Brennwald P. Testing the 3Q:1R “rule”: mutational analysis of the ionic “zero” layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol. Biol. Cell. 2000;11:3849–3858. doi: 10.1091/mbc.11.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laage R., Ungermann C. The N-terminal domain of the t-SNARE Vam3p coordinates priming and docking in yeast vacuole fusion. Mol. Biol. Cell. 2001;12:3375–3385. doi: 10.1091/mbc.12.11.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Bruns D., Wenzel D., Riedel D., Holroyd P., Thiele C., Jahn R. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mayer A., Wickner W., Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) precedes docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- Mayer A., Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J. Cell Biol. 1997;136:307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M., Zerial M. Mosaic organization of the endocytic pathway. Exp. Cell Res. 2002;272:8–14. doi: 10.1006/excr.2001.5401. [DOI] [PubMed] [Google Scholar]
- Misura K.M.S., Scheller R. H., Weis W. I. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Nickel W., Weber T., McNew J. A., Parlati F., Söllner T. H., Rothman J. E. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc. Natl. Acad. Sci. USA. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossig R., Schmitt H. D., de Groot B., Riedel D., Keränen S., Ronne H., Grubmüller H., Jahn R. Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J. 2000;19:6000–6010. doi: 10.1093/emboj/19.22.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R., Gallwitz D. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J. Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R., Gallwitz D. Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport. EMBO J. 2004;23:3939–3949. doi: 10.1038/sj.emboj.7600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J., Hsu S. C., Braun J. E., Calakos N., Ting A. E., Bennett M. K., Scheller R. H. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Price A., Seals D., Wickner W., Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J. Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Südhof T. C. SNAREs and Muc18 in synaptic vesicle fusion. Nat. Rev. Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- Sato T. K., Rehling P., Peterson M. R., Emr S. D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Savir Y., Tlusty T. Conformational proofreading: the impact of conformational changes on the specificity of molecular recognition. PLoS ONE. 2007;2:e468. doi: 10.1371/journal.pone.0000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B. L., Van Komen J. S., Irshad H., Liu S., Wilson K. A., McNew J. A. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J. Cell Biol. 2004;167:75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Tareste D. C., Paumet F., Rothman J. E., Melia T. J. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V. J., Jun Y., Wickner W. Excess vacuolar SNAREs drive lysis and Rab bypass fusion. Proc. Natl. Acad. Sci. USA. 2007;104:13551–13558. doi: 10.1073/pnas.0704741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C., Collins K. M., Fratti R. A., Wickner W. T. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Thorngren N., Collins K. M., Fratti R. A., Wickner W., Merz A. J. A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion. EMBO J. 2004;23:2765–2776. doi: 10.1038/sj.emboj.7600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen R. F., Verhage M. Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 2003;13:177–186. doi: 10.1016/s0962-8924(03)00031-x. [DOI] [PubMed] [Google Scholar]
- Ungermann C., Nichols B. J., Pelham H.R.B., Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J. Cell Biol. 1998a;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Sato K., Wickner W. Defining the functions of trans-SNARE pairs. Nature. 1998b;396:543–548. doi: 10.1038/25069. [DOI] [PubMed] [Google Scholar]
- Wada Y., Ohsumi Y., Anraku Y. Isolation and characterization of two classes of vam mutants. J. Biol. Chem. 1992;267:18655–18670. [PubMed] [Google Scholar]
- Wang L., Seeley E. S., Wickner W., Merz A. J. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- Wang L., Merz A. J., Collins K. M., Wickner W. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J. Cell Biol. 2003;160:365–374. doi: 10.1083/jcb.200209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Dulubova I., Min S. W., Chen X., Rizo J., Südhof T. C. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev. Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- Yang B., Steegmaier M., Gonzalez L. C., Scheller R. H. nSec1 binds a closed conformation of syntaxin 1A. J. Cell Biol. 2000;148:247–252. doi: 10.1083/jcb.148.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko G. S., Mitchell A. P., Jones E. W. Mapping of the proteinase b structural gene PRB1, in Saccharomyces cerevisiae and identification of nonsense alleles within the locus. Genetics. 1980;96:137–146. doi: 10.1093/genetics/96.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]