Abstract
Mutations in the SFTPC gene associated with interstitial lung disease in human patients result in misfolding, endoplasmic reticulum (ER) retention, and degradation of the encoded surfactant protein C (SP-C) proprotein. In this study, genes specifically induced in response to transient expression of two disease-associated mutations were identified by microarray analyses. Immunoglobulin heavy chain binding protein (BiP) and two heat shock protein 40 family members, endoplasmic reticulum-localized DnaJ homologues ERdj4 and ERdj5, were significantly elevated and exhibited prolonged and specific association with the misfolded proprotein; in contrast, ERdj3 interacted with BiP, but it did not associate with either wild-type or mutant SP-C. Misfolded SP-C, ERdj4, and ERdj5 coprecipitated with p97/VCP indicating that the cochaperones remain associated with the misfolded proprotein until it is dislocated to the cytosol. Knockdown of ERdj4 and ERdj5 expression increased ER retention and inhibited degradation of misfolded SP-C, but it had little effect on the wild-type protein. Transient expression of ERdj4 and ERdj5 in X-box binding protein 1−/− mouse embryonic fibroblasts substantially restored rapid degradation of mutant SP-C proprotein, whereas transfection of HPD mutants failed to rescue SP-C endoplasmic reticulum-associated protein degradation. ERdj4 and ERdj5 promote turnover of misfolded SP-C and this activity is dependent on their ability to stimulate BiP ATPase activity.
INTRODUCTION
The unfolded protein response (UPR) is a highly dynamic molecular pathway that rapidly adjusts protein synthesis, chaperone-mediated protein folding, and degradation of misfolded protein clients in response to stress. Accumulation of unfolded or misfolded protein in the endoplasmic reticulum (ER) is monitored by three membrane proteins, PERK, activating transcription factor (ATF)-6, and IRE-1, that transduce signals to the cytosolic and nuclear compartments to relieve ER stress (Zhang and Kaufman, 2004; Schroder and Kaufman, 2005). The cytosolic domain of PERK is a kinase that, when activated, phosphorylates the α-subunit of eukaryotic initiation factor 2, leading to a general, transient translational arrest, thereby preventing further accumulation of protein in the ER. Activation of ATF-6 results in cleavage of the cytosolic domain that encodes a leucine zipper transcription factor, ATF-6n; nuclear translocation of ATF-6n leads to increased expression of chaperones that enhance the folding capacity of the ER. The activated cytosolic domain of IRE-1 encodes a ribonuclease activity that removes an unspliced intron from the mRNA encoding the transcription factor XBP-1. Translation of active XBP-1 results in further up-regulation of chaperone expression and components of the ER-associated degradation (ERAD) machinery and secretory pathway. Failure to reestablish ER homeostasis results in selective translation of ATF4, leading to apoptosis.
ERAD in yeast involves multiple pathways that monitor folding of protein domains in the ER lumen (ERAD-L), membrane (ERAD-M), and cytosol (ERAD-C) (Meusser et al., 2005; Sayeed and Ng, 2005; Bukau et al., 2006). Terminally misfolded proteins are identified and targeted for dislocation to the cytosol and degradation by the proteasome. A major challenge for the quality-control machinery is discrimination between unfolded proteins that will eventually fold correctly and terminally misfolded proteins that are potentially cytotoxic and must be rapidly degraded. The quality-control checkpoints encountered by a newly synthesized protein depend upon the topology of substrate with respect to the ER membrane. The membrane protein CFTR has relatively large cytosolic domains that are monitored by at least two checkpoints located on the cytosolic side of the ER membrane (Younger et al., 2006). Lumenal proteins and membrane proteins with relatively large lumenal domains are monitored by quality-control machinery located in the ER lumen (Huyer et al., 2004; Vashist and Ng, 2004). The glycosylation status of these proteins provides cues as to the progression of folding (Molinari et al., 2003; Oda et al., 2003; Bhamidipati et al., 2005; Szathmary et al., 2005). ER-degradation enhancing alpha-mannosidase like protein (EDEM), an XBP-1 target gene encoding a lectin, detects inappropriately trimmed glycans and redirects the misfolded glycoprotein from the calnexin/calreticulin folding cycle to the degradation pathway. In yeast, the lectin Yos9p, Kar2p (immunoglobulin heavy chain binding protein [BiP]) and Hrd3p form a complex that identifies misfolded lumenal glycoproteins leading to ubiquitination by Hrd1p (Carvalho et al., 2006; Denic et al., 2006). A similar pathway was recently described for mammalian ERAD in which sel-1 suppressor of lin-12-like (SEL1L)/Hrd3-mediated interaction of OS9 and its glycosylated substrate with Hrd1 (Christianson et al., 2008).
Molecular mechanisms governing quality control of nonglycosylated proteins, particularly mammalian substrates, is less well understood. Nonglycosylated variants of CFTR and α1-antitrypsin were degraded considerably faster than the corresponding glycosylated forms (Farinha and Amaral, 2005; Mast et al., 2005); similarly, disruption of glucose trimming of tyrosinase, leading to formation of a triglycosylated form that could not interact with calnexin, resulted in rapid degradation (Svedine et al., 2004). Importantly, several nonglycosylated, soluble ERAD client proteins were recently shown to be degraded by a pathway with a molecular signature distinct from that of glycosylated substrates (Okuda-Shimizu and Hendershot, 2007). The present study was undertaken to assess quality control of a nonglycosylated, disease-associated, integral membrane protein, surfactant protein C (SP-C).
Human SP-C is synthesized as a proprotein with a single membrane-spanning domain, a 35 amino acid cytosolic domain and a lumenal domain of 133 or 139 amino acids, depending on which allele is expressed (Nogee, 2004). Proteolytic processing of the proprotein within the late endosome/multivesicular body generates the 35 amino acid mature form of SP-C, consisting of the transmembrane domain and 12 amino acids of the cytosolic domain, that is secreted into the alveolar airspaces with surfactant membranes (Johansson et al., 2004). Mutations in one allele of the SP-C gene (SFTPC) have been associated with the occurrence of interstitial lung disease (ILD) in human patients (Nogee et al., 2001, 2002). To determine whether SFTPC mutations caused lung disease, a mutation associated with ILD in human patients was expressed in type II epithelial cells of transgenic mice (Bridges et al., 2003). Elevated transgene expression was associated with dose-dependent epithelial cell death and disruption of epithelial-mesenchymal cell signaling, leading to hypoplastic lungs and neonatal death. Dysmorphogenesis occurred in the presence of two wild-type mouse Sftpc alleles, consistent with a dominant, cytotoxic effect of the transgene. Transient expression of SP-CΔexon4 in A549, human embryonic kidney (HEK)293, or MLE-15 cells resulted in retention of misfolded SP-C proprotein in the ER and activation of the unfolded protein response (UPR) and apoptosis pathways (Bridges et al., 2003; Wang et al., 2003; Mulugeta et al., 2005, 2007). Inhibition of proteasome resulted in accumulation of proprotein in the ER, indicating that misfolded SP-C was a substrate for the endoplasmic reticulum-associated degradation (ERAD) pathway.
All but one of the SFTPC mutations associated with ILD occur in the region encoding the lumenal domain, implicating lumenal chaperones in the recognition and degradation of misfolded SP-C proprotein. Using microarray analyses, we identified genes specifically induced in response to expression of two terminally misfolded variants of SP-C. We show that two BiP cochaperones, endoplasmic reticulum-localized DnaJ homologues ERdj4 and ERdj5, play an essential role in ERAD of this nonglycosylated substrate.
MATERIALS AND METHODS
Microarray Analyses and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
SP-CWT and SP-CΔexon4 cDNA constructs were described previously (Bridges et al., 2006). SP-CL188Q mutant cDNA was generated by site-directed mutagenesis of wild-type SP-C by using the QuikChange kit (Promega, Madison, WI). All SP-C variants were subcloned into the pIRES2-EGFP vector (Clontech, Mountain View, CA) for microarray analyses. Cells were transfected with either SP-CWT, SP-CΔexon4, SP-CL188Q, or pIRES2-EGFP (empty vector control) for 24 h, harvested from culture dishes with trypsin/EDTA, and sorted by fluorescence-activated cell sorting to obtain enhanced green fluorescent protein (EGFP)-positive cells. Transfection and subsequent sorting of each SP-C variant was performed in triplicate. Total RNA was isolated from the EGFP-positive cells by using an acidified guanidinium method (Chomczynski and Mackey, 1995). Total RNA was reverse transcribed into cDNA by using a T7 promoter-dT primer, amplified, and biotinylated in an in vitro transcription reaction by using T7 RNA polymerase before hybridization to the GeneChip Human Genome U133 Plus 2.0 set (contains 54,681 probe sets representing ∼47,000 transcripts and variants, including 38,500 well-characterized genes) according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). Normalization and data analyses were performed as described previously (Bridges et al., 2006). Verification of selected genes was performed by real-time PCR analysis as described previously (Bridges et al., 2006).
Genes differentially expressed between SP-CWT and SP-C mutants (SP-CΔexon4 and SP-CL188Q) were selected with a threshold t test p value ≤0.05, False Discovery Rate (FDR) ≤ 10%, fold change ≥2.0 and a minimal of 6 present call by Affymetrix algorithm in a total of 9 samples. Potential XBP-1 target genes were identified by scanning for the TGACGTGR motif (UPR element E [UPRE]) in the promoter region (−1 kb) of differentially expressed genes allowing a maximum of one mismatch (Genomatix Software, Ann Arbor, MI). Genes encoding ER proteins were identified by a combination of Gene Ontology analysis, literature search, and protein domain prediction. Gene Ontology (GO) analysis was performed using the publicly available web-based tool DAVID for Database for annotation, visualization, and integrated discovery (Dennis et al., 2003). A subset of differentially expressed genes located in or as a subcomponent of endoplasmic reticulum was identified based on the GO cellular component annotation. Protein domain prediction was performed using PROSITE and PSORT (Nakai and Horton, 1999; Sigrist et al., 2002). PROSITE is a database of protein families, domains, and functional sites as well as associated patterns and profiles to identify them. A subset of gene products containing the PROSITE ER target sequence signature [(KRHQSA)-(DENQ)-E-L] was identified using the ps_scan algorithm to search against the PROSITE database (Gattiker et al., 2002). PSORT is a program focused to the prediction of subcellular localization sites of proteins from their amino acid sequence signatures. In addition to the recognition of the ER lumenal signal (KDEL), PSORT also retrieves two kinds of ER membrane protein signals, a di-lysine motif (KKXXX) near the C terminus and a di-arginine motif (XXRR) near the N terminus. Proteins containing ER-related motifs were combined, and duplicates were removed.
cDNA Cloning, Plasmid Construction, and Antibodies
Human ERdj4-HA, ERdj5-HA, ERdj3-VSV, and BiP-Flag were cloned from HEK293 cell cDNA by using PCR with reverse transcription. ERdj4 and ERdj5 were inserted into pIRES2-EGFP vector (Clontech), BiP was inserted into pcDNA3.1 (Invitrogen, Carlsbad, CA), and ERdj3 was inserted into pCMV6-XL5 (OriGene Technologies, Rockville, MD). The hemagglutinin (HA) tag was added at a position before the C-terminal KDEL in ERdj5 as described previously (Hosoda et al., 2003). Mouse insulin 2-Flag was cloned from mouse pancreatic β-cell cDNA using PCR with reverse transcription and inserted into the pIRES2-EGFP vector. ERdj4H54Q, ERdj5H63Q, and insulin2C96Y were generated by site-directed mutagenesis, and ERdj4ΔJ, ERdj4ΔGF, and ERdj5ΔJ were made by overlap extension PCR. Mouse anti-HA antibody was purchased from Cell Signaling Technology (Danvers, MA). Mouse anti- vesicular stomatitis virus (VSV)-G antibody was purchased from Sigma-Aldrich (St. Louis, MO). Mouse anti-Flag antibody was purchased from Stratagene (La Jolla, CA). Rabbit polyclonal anti-p97/VCP was purchased from Bethyl Laboratories (Montgomery, TX).
Cell Culture and Transfection
Untransfected HEK293 cells and two HEK293 cell lines stably expressing SP-CWT or SP-CΔexon4 were cultured in Richter's CM-medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin as described previously (Bridges et al., 2006). HEK293 cells stably expressing SP-CL188Q were generated and cultured as described previously for SP-CΔexon4 (Bridges et al., 2006). XBP-1+/+ and XBP-1−/− mouse embryonic fibroblasts (MEFs) (gifts from Dr. L. Glimcher, Harvard University, Cambridge, MA) were cultured as described previously (Lee et al., 2003), and they were transfected using Nucleofector (program A-23; Amaxa, Koeln, Germany). Transient transfection of HEK293 cells and stable cell lines was performed using Lipofectamine 2000 (Invitrogen) transfection reagents, according to the manufacturer's instructions. Where indicated, cells were treated with 5 μM MG-132 for 4 h to inhibit proteasome activity, as described previously (Bridges et al., 2003; Bridges et al., 2006). Stealth RNAi molecules targeted to ERdj4 or ERdj5 were purchased from Invitrogen and transfected into HEK293 cells with Lipofectamine 2000, as described in Supplemental Figure S3. Sense RNA sequences of Stealth RNA interference (RNAi) molecules were as follows: 5′-UGA ACA GUC AGU GUA UGU AGU AACC-3′ for ERdj4 and 5′-AGU AAA GCU CGA CAU GGU GGA CACC-3′ for ERdj5. In addition, Silencer Negative Control #1SiRNA (Ambion, Applied Biosystems, Austin, TX), which has no known target in mammalian genomes, was used as the negative control.
Pulse-Chase Experiments
Cells were incubated for 30 min in methionine- and cysteine-free DMEM (Invitrogen) supplemented with 10% dialyzed fetal bovine serum. Cells were then pulse-labeled with 0.5 mCi/ml [35S]methionine/cysteine (MP Biomedicals, Irvine, CA) for 15 min and chased in complete medium (fresh DMEM containing 0.5 mM unlabeled methionine/cysteine) for 1, 2, and 4 h, as described previously (Lin et al., 1996). Cells were subjected to chemical cross-linking using dithiobis (succinimidylpropionate) (DSP; Pierce Chemical, Rockford, IL) before harvesting. The DSP reagent was dissolved in dimethyl sulfoxide at a concentration of 50 mM and added to the cells to a final concentration of 1 mM. After 30-min incubation at room temperature, the reactions were quenched by adding 1 ml of 1 M Tris, pH 7.5, and incubating for an additional 15 min. Cells were lysed and equivalent amounts of cell lysates were immunoprecipitated with the indicated antibody and protein G beads (Zymed Laboratories, South San Francisco, CA) at 4°C overnight on a rotating wheel; captured immunocomplexes were washed six times, and then they were incubated at 100°C for 4 min with Laemmli buffer, as described previously (Lin et al., 1996). Immunoprecipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) under reducing electrophoretic conditions followed by autoradiography.
Coimmunoprecipitation (CoIP) Experiments
For CoIP experiments, cross-linking and immunoprecipitation (IP) were performed as described above. In experiments where cross-linker was omitted, cells were lysed in NP-40 buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, and protease inhibitor cocktail) before immunoprecipitation. Immunoprecipitated proteins were analyzed by SDS-PAGE under reducing electrophoretic conditions followed by Western blotting with antibodies directed against proSP-C or the epitope tag (indicated in the figure legend), as described previously (Bridges et al., 2003).
RESULTS
Microarray Analysis Reveals Candidate Genes Involved in ER Quality Control of SP-C
To identify candidate genes involved in ER quality control of SP-C, HEK293 cells were transiently transfected with mutant SP-C (SP-CΔexon4 or SP-CL188Q), SP-CWT, or vector cDNAs, and global changes in gene expression were assessed by microarray analyses. Signals from 5554 of 54,681 probe sets on the human genome genechip (representing 3633 genes and expressed sequence tags [ESTs]) were specifically increased ≥2-fold in one mutant SP-C cell line and ≥1.5 in the other mutant cell line compared with SP-CWT (Supplemental Figure S1A). Of the 3633 genes/ESTs, 2834 were detected in the human promoter database, and of these, 1028 genes contained at least one UPRE (TGACGTGR) in the 5′ flanking sequence of the gene; the UPRE was overly represented in genes up-regulated by both SP-C mutants in comparison with random analyses of promoter sequences (p = 7.5 × 10−5). Within this subset of candidate UPR target genes, 43 were annotated as genes encoding ER resident/associated proteins, some of which likely play a role in detection and degradation of misfolded SP-C (Table 1). Increased expression of selected genes was confirmed by real-time PCR (Supplemental Figure S1B).
Table 1.
ER genes up-regulated in response to transient expression of misfolded SP-C in HEK293 cells
| Gene Name | Accession no. | SP-CΔexon4:SP-Cwta | SP-CL188Q:SP-Cwtb | Description | UPREc | No. probe setsd |
|---|---|---|---|---|---|---|
| BET | NM_005868 | 2.6 | 2.2 | BET1 homologue (S. cerevisiae) | 11kb | 1 |
| CDS1 | NM_001263 | 2.5 | 1.9 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 | 11kb | 2 |
| DPM1 | NM_003859 | 2.3 | 2.1 | Dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit | 11kb | 1 |
| EXTL2 | NM_001439 | 2.4 | 1.6 | Exostoses (multiple)-like 2 | 11kb | 1 |
| FKBP14 | NM_017946 | 2.1 | 2.7 | FK506 binding protein 14, 22 kDa | 11kb | 2 |
| HYOU1 | NM_006389 | 2.2 | 1.7 | Hypoxia up-regulated 1 | 11kb | 1 |
| JPH3 | NM_020655 | 2.1 | 2.0 | Junctophilin 3 | 11kb | 1 |
| KIAA1012 | NM_014939 | 2.3 | 2.1 | KIAA1012 protein | 21kb | 1 |
| PCMT1 | NM_005389 | 2.2 | 1.8 | Methyltransferase | 11kb | 1 |
| RAB1A | NM_004161 | 2.1 | 1.6 | RAB1A, member RAS oncogene family | 11kb | 1 |
| RAB2 | NM_002865 | 2.3 | 1.7 | RAB2, member RAS oncogene family | 21kb | 1 |
| RCN2 | NM_002902 | 2.2 | 1.6 | Reticulocalbin 2, EF-hand calcium binding domain | 11kb | 1 |
| SDCBP | NM_005625 | 2.3 | 1.9 | syndecan binding protein (syntenin) | 11kb | 1 |
| SEC22L3 | NM_032970 | 2.2 | 2.1 | SEC22 vesicle trafficking protein-like 3 (Saccharomyces cerevisiae) | 11kb | 1 |
| SEC24B | NM_006323 | 3.0 | 2.1 | SEC24 related gene family, member B (S. cerevisiae) | 11kb | 1 |
| SEC24D | NM_014822 | 2.6 | 2.1 | SEC24 related gene family, member D (S. cerevisiae) | 11kb | 1 |
| SERP1 | NM_014445 | 2.9 | 2.4 | Stress-associated endoplasmic reticulum protein 1 | 11kb | 3 |
| SPC18 | NM_014300 | 2.1 | 1.9 | Signal peptidase complex (18 kDa) | 11kb | 1 |
| TRA1 | NM_003299 | 2.3 | 2.3 | Tumor rejection antigen (gp96) 1 | 11kb | 3 |
| ZMPSTE2 | NM_005857 | 2.1 | 1.7 | Zinc metalloproteinase (STE24 homologue, yeast), GRP94 | 11kb | 1 |
| ATF6 | NM_007348 | 2.4 | 2.0 | Activating transcription factor 6 | 11kb | 2 |
| PIGA | NM_002641 | 3.0 | 2.4 | Phosphatidylinositol glycan, class A | 11kb | 1 |
| SEC61G | NM_014302 | 2.5 | 2.1 | Sec61 γ subunit | 11kb | 1 |
| SLC33A1 | NM_004733 | 2.2 | 2.1 | Solute carrier family 33 (acetyl-CoA transporter), member 1 | 11kb | 3 |
| p58IPK | NM_006260 | 4.3 | 3.4 | DnaJ (Hsp40) homologue, subfamily C, member 3 | 11kb, 12kb | 3 |
| ERdj4 | NM_012328 | 4.2 (3.6)e | 3.6 (4.2) | DnaJ (Hsp40) homologue, subfamily B, member 9 | 31kb, 52kb | 3 |
| ERdj5 | NM_018981 | 4.2 (3.6) | 3.6 (4.5) | DnaJ (Hsp40) homologue, subfamily C, member 10 | 31kb, 42kb | 4 |
| XBP-1 | NM_005080 | 2 (1.4) | 1.5 (1.5) | X-box binding protein 1 | 11kb | 1 |
| ERdj1 | NM_022365 | 2.0 | 1.7 | DnaJ (Hsp40) homologue, subfamily C, member 1 | 01kb, 32kb | 1 |
| ERdj3 | NM_016306 | 1.9 | 1.9 | DnaJ (Hsp40) homologue, subfamily B, member 11 | 01kb, 32kb | 1 |
| PDIR5 | NM_006810 | 2.1 | 1.6 | Protein disulfide isomerase family A, member 5 | 11kb, 22kb | 1 |
| ERdj2 | NM_007214 | 3.0 | 2.0 | SEC63-like (S. cerevisiae) | 13kb, 32kb | 2 |
| P5 | NM_005742 | 2.0 | 1.5 | Protein disulfide isomerase family A, member 6 | 01kb, 12kb | 1 |
| ERP70 | NM_004911 | 2.3 | 1.7 | Protein disulfide isomerase family A, member 4 | 01kb, 22kb | 2 |
| GRP 58 | NM_005313 | 2.2 | 1.4 | Protein disulfide isomerase family A, member 3 | 01kb, 52kb | 2 |
| HSPA5 | NM_005347 | 3.2 (4.3) | 2.8 (4.0) | Glucose-regulated protein, 78 kDa, BiP/GRP78 | 01kb, 12kb | 1 |
| Derlin1 | NM_024295 | 2.7 | 2.4 | Der-like domain family, member 1 | 41kb, 12kb | 1 |
| CANX | NM_001024649 | 2.5 | 1.9 | Calnexin | 21kb, 12kb | 1 |
| SELS | NM_018445 | 1.9 | 2.0 | Selenoprotein S, VIMP | 11kb, 12kb | 1 |
| Trapα | NM_003144 | 2.5 | 1.6 | Translocon-associated protein alpha (SSR1) | 0 | 1 |
| Trapβ | NM_003145 | 1.9 | 1.1 | Translocon-associated protein beta (SSR2) | 41kb, 22kb | 1 |
| Trapδ | NM_006280 | 2.1 | 1.4 | Translocon-associated protein delta (SSR4) | 0, 12kb | 1 |
| HERPUD1 | NM_014685 | 3.3 | 3.4 | Homocystine-inducible, ER stress inducible, ubiquitine-like domain memb | 11kb, 12kb | 1 |
| Member 1, Herp | ||||||
| SEL1L | NM_005065 | 2.6 | 2.3 | sel-1 suppressor of lin-12-like (Caenorhabditis elegans), Hrd3 | 01kb, 22kb | 1 |
a -Fold increase in cells expressing SP-CΔexon4 relative to cells expressing SP-CWT; when more than one probe set was present on the genechip, the average -fold increase was reported.
b -Fold increase in cells expressing SP-CL188Q relative to cells expressing SP-CWT; when more than one probe set was present on the genechip, the average -fold increase was reported.
c The number of times the UPRE (TGACGTGR) occurred within 1 kb (1kb) or 2 kb (2kb) of the transcription of start site.
d The number of probe sets on the genechip for each gene/EST.
e Numbers in parentheses are results of real-time PCR analyses (see Supplemental Figure S1B).
Among the ER proteins specifically induced in response to expression of mutant SP-C, BiP/GRP78 (HSPA5), an ER-localized HSP70 family member, was significantly increased 2.8- and 3.2-fold in SP-CΔexon4– and SP-CL188Q–expressing cells, respectively (Table 1). Importantly, five HSP40 family members (DnaJ proteins) involved in modulation of HSP70 ATPase activity were also significantly induced (1.7∼4.3-fold; Table 1). Two recently identified DnaJ proteins, ERdj4 and ERdj5 (Shen et al., 2002; Cunnea et al., 2003), were especially noteworthy because these were among the most highly induced genes (3.4- to 4.3-fold) in response to expression of either SP-CΔexon4 or SP-CL188Q (Table 1). ERdj4 and ERdj5 contain eight and seven putative UPREs, respectively, within 2 kb of the transcription start site suggesting that they may be targets of XBP-1, a transcription factor involved in the regulation of ERAD. Expression of XBP-1 was also increased (1.5- to 2.0-fold) in response to expression of mutant SP-C proprotein (Table 1). Two other classes of proteins previously associated with ERAD were also specifically up-regulated in response to expression of either SP-CΔexon4 or SP-CL188Q. Five members of the protein disulfide isomerase-related family of proteins were increased 1.5- to 3-fold (Table 1); these proteins have been reported to have chaperone activity, and they contain thio-redoxin motifs involved in the formation, reduction, and rearrangement of disulfide bonds. The other class of proteins was associated with the translocon/retrotranslocon; these proteins were increased 1.9- to 3.4-fold and included SEC63; SEC61g; SERP1; Derlin-1; TRAP α, β, and δ; SEL1L; Herp; and VIMP/SELS (Table 1).
ERdj4 and ERdj5 Preferentially Associate with Misfolded SP-C
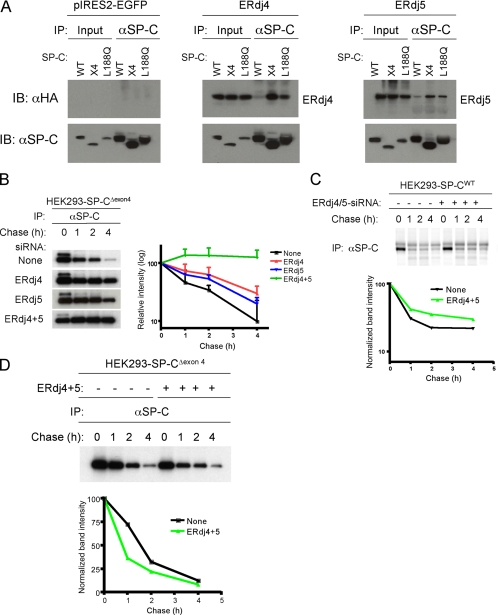
To determine whether ERdj4 and ERdj5 were specifically associated with misfolded SP-C, HEK293 cell lines stably expressing either SP-CWT or one of the two mutant SP-C proproteins (SP-CΔexon4 or SP-CL188Q) were generated (Bridges et al., 2006). Cell lines were transfected with ERdj4 or ERdj5, and 48 h later, they were treated with the proteasome inhibitor MG-132 to stabilize misfolded SP-C. After cross-linking with DSP, cells were harvested, and SP-C was immunoprecipitated from the cell lysates with an antibody directed against the cytosolic domain of SP-C that detects both wild-type and mutant protein, followed by SDS-PAGE, under reducing electrophoretic conditions, and Western blotting with HA antibody to detect ERdj proteins. Much more ERdj4 was associated with SP-CΔexon4 than with SP-CWT (Figure 1A and Table 2). Preferential association was confirmed by performing the immunoprecipitation experiment in reverse, which showed that SP-CΔexon4 but little or no SP-CWT precipitated with ERdj4 (Supplemental Figure S2Aa). Likewise, ERdj5 associated with more SP-CΔexon4 than SP-CWT (Figure 1A, Table 2, and Supplemental Figures S2Ac and S5). Similar results were obtained with another SP-C mutant, SP-CL188Q (Figure 1B and Table 2), indicating that both ERdj4 and ERdj5 preferentially associated with misfolded SP-C proprotein. Preferential association of ERdj4 or ERdj5 with mutant SP-C was not due to diminished expression of the chaperones in HEK293 cells stably expressing SP-CWT (Figure 1). In contrast to ERdj4 and ERdj5, association of ERdj3 (increased 1.9-fold in mutant SP-C–expressing cells; Table 1), with either SP-CΔexon4 or SP-CWT was not detected (Supplemental Figure S5). Together, the preferential and/or prolonged binding of ERdj4 and ERdj5 to misfolded substrates suggests that specific HSP40 family members are able to distinguish between correctly folded and misfolded SP-C, stably associating directly or indirectly with the misfolded form.
Figure 1.
ERdj4 and ERdj5 are required for degradation of SP-CΔexon4. (A) HEK293 cell lines stably expressing SP-CWT, SP-CΔexon4 (x4) or SP-CL188Q were transfected with plasmids encoding ERdj4 (middle), ERdj5 (right), or vector (left). Forty-eight hours after transfection, cells were treated with the proteasome inhibitor MG-132 to stabilize misfolded SP-C. After cross-linking with DSP, cells were harvested and equal amounts of cell lysates were subjected to IP with SP-C proprotein antibody followed by reducing SDS-PAGE, and sequential Western blotting (IB) with HA or SP-C antibodies. (B) HEK293 cells stably expressing SP-CΔexon4 were not transfected (top row) or transfected with siRNA for ERdj4, ERdj5, or both ERdj4 and ERdj5. Twenty-four hours later, cells were labeled with [35S]Met/Cys for 15 min, chased for the indicated period, and immunoprecipitated with SP-C antibody (left). The radioactivity of each SP-CΔexon4 band was analyzed by phosphorimaging analyses and normalized to the value at 0 chase time; averages of three independent experiments were plotted with SD values (right). (C) HEK293 cells stably expressing SP-CWT were transfected with siRNA for both ERdj4 and ERdj5 (lanes 5–8), or they were not transfected with siRNA (lanes 1–4). Pulse-chase, immunoprecipitation for SP-CWT proprotein, and phosphorimaging analyses were performed as described in B. (D) HEK293 cells stably expressing SP-CΔexon4 were cotransfected with plasmids encoding ERdj4 and ERdj5 (lanes 5–8), or they were left untransfected (lanes 1–4). Pulse-chase, immunoprecipitation, and phosphorimaging analyses were performed as described in B. The radioactivity of each SP-CΔexon4 band was normalized to the value at 0 chase time (bottom); pulse-chase studies represent the average of three independent experiments ± SD values.
Table 2.
Quantitative immunoprecipitation analyses of ERAD substrates with ERdj4 and ERdj5a
| Substrate | ERdj4 WT:mutant | ERdj5 WT:mutant |
|---|---|---|
| SP-CΔexon4 | 1:40.50 ± 25.48 | 1:15.99 ± 11.27 |
| SP-CL188Q | 1:30.80 ± 21.35 | 1:29.26 ± 30.33 |
| Insulin 2C96Y | 1:1.18 ± 0.27 | 1:2.70 ± 0.73 |
a HEK293 cells were transfected and subjected to immunoprecipitation analyses as described in Figure 5A. Band intensity was quantified by ImageQuant (GE Healthcare, Chalfont St. Giles, United Kingdom) analyses. The value of each WT protein was arbitrarily set to 1. The data shown are the means ± SD from at least three independent experiments.
ERdj4 and ERdj5 Are Required for Degradation of Misfolded SP-C
The prolonged association of ERdj4 and ERdj5 with mutant SP-C suggested that they play an important role in recognition and targeting of SP-C for degradation. This hypothesis was tested by knocking down ERdj4 and ERdj5 in HEK293 cells stably expressing SP-CΔexon4. Small interfering RNA (siRNA)-mediated knockdown of an ERdj4 reporter molecule in transiently transfected HEK293 cells was >90% (Supplemental Figure S3A–C), and it did not alter concentration of mRNAs encoding endogenous ERdj5, ERdj3, or actin (Supplemental Figure S3D). In the absence of siRNA, SP-CΔexon4, proprotein was barely detectable by Western blotting due to rapid degradation via the ERAD pathway (Figure 3A; Bridges et al., 2003, 2006). Pulse-chase analyses indicated that SP-CΔexon4 was degraded with a half-life of 1.7 h (Figure 1B) and that degradation was significantly slowed in the presence of ERdj4 or ERdj5 siRNAs. Knockdown of both ERdj4 and ERdj5 completely blocked the degradation of misfolded SP-C during the 4-h chase period (Figure 1B). Unlike SP-CΔexon4, turnover of SP-CWT was only slightly affected by knockdown of both ERdj4 and ERdj5 genes (Figure 1C). These results suggest that ERdj4 and ERdj5 at most play a minor role in the biosynthesis of SP-CWT.
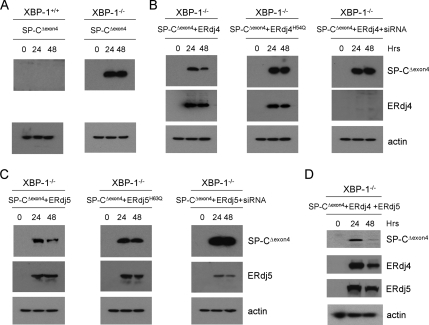
Figure 3.
ERdj4 and ERdj5 restore degradation of SP-CΔexon4 in XBP-1−/− MEFs. (A) XBP-1+/+ and XBP-1−/− MEFs were transiently transfected with plasmid encoding SP-CΔexon4 followed by Western blotting of cell lysates with SP-C and actin antibodies at 0, 24, and 48 h posttransfection. (B) XBP-1−/− MEFs were cotransfected with plasmids encoding SP-CΔexon4 and ERdj4 (left), ERdj4H54Q (middle) or ERdj4 plus ERdj4 siRNA (right). Cell lysates were prepared at 0, 24, and 48 h posttransfection, and then they were analyzed by Western blotting with SP-C, HA (ERdj4), and actin antibodies. (C) XBP-1−/− MEFs were cotransfected with plasmids encoding SP-CΔexon4 and ERdj5 (left), ERdj5H63Q (middle), or ERdj5 plus ERdj5 siRNA (right). Cell lysates were prepared at 0, 24, and 48 h posttransfection, and then they were analyzed by Western blotting with SP-C, HA (ERdj5) and actin antibodies. (D) XBP-1−/− MEFs were cotransfected with plasmids encoding SP-CΔexon4, ERdj4, and ERdj5. Cell lysates were prepared at 0, 24, and 48 h posttransfection, and then they were analyzed by Western blotting with SP-C, HA (ERdj4 and ERdj5), and actin antibodies. Within each set of experiments (A–D), samples were run on one gel to ensure identical exposure times. Actin was used to adjust exposure times across experiments.
Knockdown of ERdj4 did not affect binding of ERdj5 to SP-CΔexon4 (Supplemental Figure S2Da); likewise, knockdown of ERdj5 had no obvious effect on the association of ERdj4 with SP-CΔexon4 (Supplemental Figure S2Db). Overexpression of both ERdj4 and ERdj5 decreased the half-life of SP-CΔexon4 from 1.7 to 0.75 h (Figure 1D). Collectively, these data suggest that ERdj4 and ERdj5 associate with SP-C independently of each other and that both chaperones are required for degradation of misfolded SP-C. A similar finding was first reported for the yeast ER-localized J-domain–containing proteins Jemlp and Scjlp (Nishikawa et al., 2001); deletion of both Jem1p and Scj1p or compromised BiP function resulted in accumulation of ERAD substrates within the lumen of the ER.
BiP Is Required for Binding of ERdj5 but Not ERdj4 to Misfolded SP-C
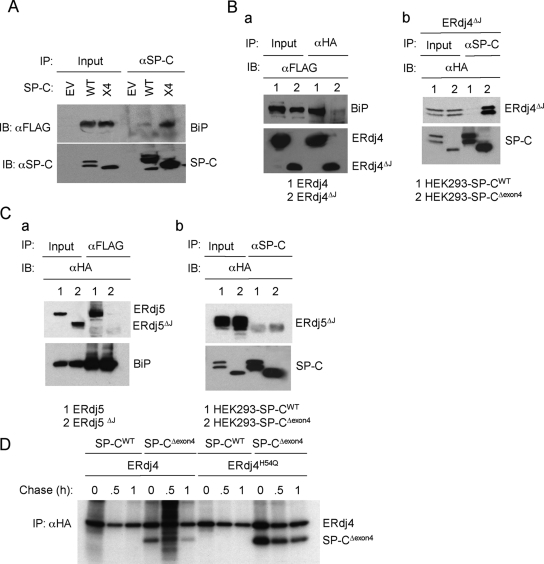
To investigate the interaction of BiP with SP-C, cell lines were transfected with BiP, treated with MG-132 and cross-linked in situ with DSP. Immunoprecipitation of cell lysates for SP-C followed by Western blotting for BiP (FLAG antibody) readily detected SP-CΔexon4 in association with BiP, both in the presence (unpublished data) and absence of DSP (Figure 2A); in contrast, much less BiP coimmunoprecipitated with SP-CWT, likely reflecting the transient nature of the chaperone-wild type protein interaction. Consistent with this finding, immunoprecipitation for BiP recovered SP-CΔexon4, but it did not recover any detectable SP-CWT, suggesting that the pool of BiP bound to mutant SP-C was much larger than that associated with the wild-type (WT) protein (unpublished data). Similar results were obtained with SP-CL188Q, confirming prolonged association of BiP with mutant SP-C (unpublished data).
Figure 2.
BiP is required for binding of ERdj5 but not ERdj4 to misfolded SP-C. (A) Cell lines stably expressing WT or SP-CΔexon4 (x4) were transfected with plasmids encoding BiP and treated with MG-132; no cross-linking agent was added in this experiment. Cell lysates were immunoprecipitated with SP-C proprotein antibody followed by Western blotting with FLAG antibody to detect BiP. EV, HEK293 cells transfected with empty vector (pIRES2-EGFP). (B) a, HEK293 cells were cotransfected with plasmids expressing ERdj4 or ERdj4ΔJ and BiP. After cross-linking, cell lysates were immunoprecipitated with HA antibody and analyzed by Western blotting with FLAG antibody to detect BiP or HA antibody to detect ERdj4. b, HEK293 cells stably expressing SP-CWT (lanes 1) or SP-CΔexon4 (lanes 2) were transfected with plasmids encoding ERdj4ΔJ, treated with MG-132, and cross-linked. Cell lysates were immunoprecipitated with SP-C antibody and analyzed by Western blotting with HA antibody to detect ERdj4ΔJ. (C) a, HEK293 cells were cotransfected with plasmids encoding ERdj5 or ERdj5ΔJ and BiP. After cross-linking with DSP, cell lysates were immunoprecipitated with FLAG antibody (BiP) and analyzed by Western blotting with HA antibody to detect ERdj5 or FLAG antibody to detect BiP. b, HEK293 cells stably expressing SP-CWT (lanes 1) or SP-CΔexon4 (lanes 2) were transfected with plasmids encoding ERdj5ΔJ, treated with MG-132, and cross-linked. Cell lysates were immunoprecipitated with SP-C antibody and analyzed by Western blotting with HA antibody to detect ERdj5ΔJ. (D) HEK293 cells stably expressing SP-CWT or SP-CΔexon4 were transfected with plasmids encoding ERdj4 or ERdj4H54Q. Forty-eight hours after transfection, cells were pulse-labeled with [35S]Met/Cys for 15 min, and then they were chased for the indicated times. After cross-linking with DSP, cell lysates were immunoprecipitated with HA antibody followed by reducing SDS-PAGE, and autoradiography.
The ATPase activity of BiP is required for substrate binding, and it is modulated by DnaJ cochaperones that interact with the ATPase domain of BiP through a conserved ∼70-amino acid J domain (Gething, 1999). To determine whether interactions of ERdj4 and ERdj5 with misfolded SP-C were BiP-dependent, J domain deletion variants of ERdj4 (ERdj4ΔJ) and ERdj5 (ERdj5ΔJ) were constructed and analyzed. As expected, ERdj4ΔJ did not coimmunoprecipitate with BiP (Supplemental Figure 2Ba), confirming that the J domain is required for interaction of ERdj4 with BiP. Unexpectedly, ERdj4ΔJ specifically coprecipitated with SP-CΔexon4 (Figure 2Bb); an ERdj4 HPD mutant also stably associated with SP-CΔexon4 but not SP-CWT (Figure 2D). In addition to the J domain, ERdj4 contains a glycine/phenylalanine-rich (G/F) region of unknown function. Deletion of the G/F domain resulted in an unstable protein that was degraded by ERAD (Supplemental Figure S2Ba), and it did not associate with SP-C (Supplemental Figure S2Bb) or BiP (Supplemental Figure S2Bc). It therefore remains unclear whether ERdj4 directly binds mutant SP-C through its G/F domain or through a partner protein. Deletion of the J domain of ERdj5 also abolished interaction with BiP (Supplemental Figure 2Ca); however, unlike ERdj4ΔJ, ERdj5ΔJ did not precipitate with SP-CΔexon4, indicating that BiP is an obligate partner for association of ERdj5 with misfolded SP-C (Supplemental Figure 2Cb). Both ERdj4 and ERdj5 associated with BiP in the absence of misfolded SP-C (Supplemental Figure 2Ba and 2Ca). ERdj3 also associated with BiP in the presence or absence of misfolded SP-C (unpublished data), but it did not associate with SP-C (Supplemental Figure S5).
ERdj4 and ERdj5 Restore SP-C Degradation in XBP-1−/− MEFs
XBP-1–deficient MEFs cells were transiently transfected with SP-CΔexon4, followed by Western blotting with the anti-SP-C antibody. In wild-type MEFs, no mutant SP-C proprotein was detected because it was rapidly degraded (Figure 3A). However, in XBP-1−/− MEFs, SP-CΔexon4 accumulated, and it was readily detected after transfection, suggesting that XBP-1 target genes are required for efficient degradation of misfolded SP-C. The concentration of SP-CΔexon4 slowly decreased over time in culture (up to 96 h), consistent with dilution of the misfolded protein during successive cell division (unpublished data). The 5′-flanking region of genes encoding ERdj4 and ERdj5 contain multiple UPREs, and mRNA transcripts corresponding to these genes were not detected in XBP-1 MEFs (unpublished data; Lee et al., 2003). To determine whether ERdj4 and/or ERdj5 could restore degradation of misfolded SP-C in XBP-1−/− MEFs, cells were cotransfected with SP-CΔexon4 and ERdj4 and/or ERdj5. The degradation of misfolded SP-C was accelerated in the presence of ERdj4 expression (Figure 3B) and to a lesser extent in the presence of ERdj5 (Figure 3C). Expression of both ERdj4 and ERdj5 nearly completely restored the degradation of mutant SP-C (Figure 3D). Importantly, ERdj4 and ERdj5 HPD mutants were less effective in restoring SP-C degradation (Figure 3, B and C, middle column), suggesting that turnover of mutant proprotein was linked to stimulation of BiP ATPase activity; additional data (Figure 2D and Supplemental Figure S2A and S2C) support this hypothesis. Finally, siRNA-mediated inhibition of ERdj4 and ERdj5 expression completely blocked rescue of SP-C ERAD. Collectively, these results are consistent with the hypothesis that ERdj4 and ERdj5 are two major XBP-1 target genes required for ERAD of misfolded SP-C.
ERdj4 and ERdj5 Form a Complex with Misfolded SP-C and Cytosolic p97
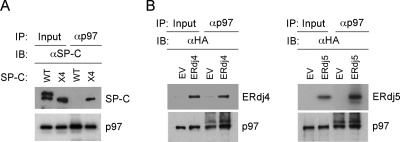
Misfolded SP-C is ultimately degraded by the proteasome and must therefore be transported out of the ER lumen to the cytosol. The cytosolic AAA-ATPase p97/VCP and its cofactors Ufd1, Npl4, and p47 bind ubiquitinated proteins as they emerge from the ER, and they contribute to substrate dislocation (Meusser et al., 2005). SP-CΔexon4 contains two lysine residues (K6 and K34) in its cytosolic domain and three lysines (K63, K153, and K160) in its lumenal C-terminal domain. Immunoprecipitation of HEK293 cells cotransfected with SP-CΔexon4 lysine>arginine mutants and ubiquitin-HA, followed by Western blotting for HA, indicated that both cytosolic and lumenal lysines were ubiquitinated (Supplemental Figure S4). Cross-linking and coprecipitation experiments indicated that SP-CΔexon4 but not SP-CWT associated with endogenous p97/VCP (Figure 4A). ERdj4 and ERdj5 also coprecipitated with endogenous p97/VCP, both in the presence (unpublished data) and absence of DSP (Figure 4B). These results suggest that ERdj4 and ERdj5 remain associated with misfolded SP-C until it is dislocated from the ER.
Figure 4.
Association of p97 with ERdj4, ERdj5, and SP-C. (A) HEK293-SP-CWT and HEK293-SP-CΔexon4 (x4) stable cell lines were treated with MG-132 and cross-linked with DSP. Cell lysates were immunoprecipitated with p97 antibody and analyzed by Western blotting with SP-C antibody. (B) HEK293 cells were transfected with plasmids encoding ERdj4 or ERdj5; no cross-linking reagent was added in this experiment. Cell lysates were immunoprecipitated with p97 antibody, and then they were analyzed by Western blotting with HA antibody.
Association of ERdj4 and ERdj5 with Other ERAD Substrates
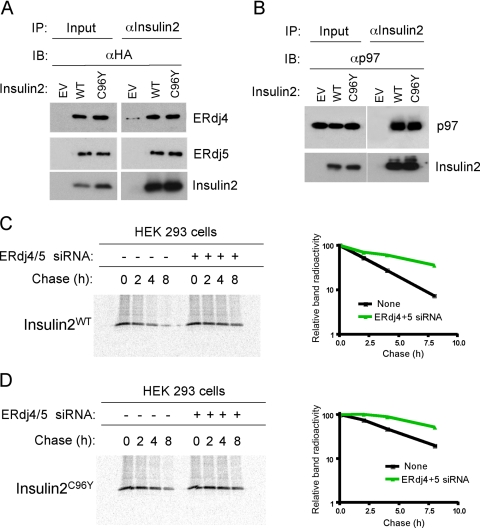
To determine whether ERdj4 and ERdj5 formed chaperone complexes with another disease-associated mutant protein, HEK293 cells were cotransfected with ERdj4 or ERdj5 and mutant insulin 2 (insulin 2C96Y) or WT insulin 2. Transfection of WT or mutant insulin 2 modestly, but consistently, increased expression of ERdj4 and ERdj5 mRNAs (unpublished data). In contrast to mutant SP-C, ERdj4 and ERdj5 formed complexes with both WT and mutant insulin 2, a nonglycosylated, secreted protein (Figure 5A and Table 2). Because WT connexin 32 and proteolipid protein were reported previously to be degraded in a proteasome-dependent manner (Vanslyke et al., 2000; Kramer-Albers et al., 2006), we determined whether WT insulin2 was part of a complex destined for degradation. Both WT and mutant insulin 2 coprecipitated with p97, suggesting that the WT protein was unstable and degraded when expressed transiently in HEK293 cells (Figure 4B). Degradation was not simply related to protein overexpression, because very little WT SP-C coprecipitated with ERdj4, ERdj5, or p97 (Table 2 and Figure 1A). Thus, ERdj4 and ERdj5 preferentially formed complexes with proteins destined for retrotranslocation and degradation. We next determined whether ERdj4 and ERdj5 were required for degradation of mutant and WT insulin 2. HEK cells were first transfected with siRNAs for ERdj4 and ERdj5 followed by expression vectors encoding WT or mutant insulin 2 and substrate disappearance from cell lysates assessed by pulse-chase experiments. siRNA-mediated inhibition of ERdj4 and ERdj5 expression (confirmed by real-time PCR; unpublished data) inhibited degradation of WT and mutant insulin 2 (Figure 5, C and D), although ERAD was not completely inhibited as for mutant SP-C (Figure 1B).
Figure 5.
ERdj4 and ERdj5 associate with WT and mutant insulin 2. (A) HEK293 cells were transfected with empty vector (EV) or cotransfected with plasmid expressing ERdj4 or ERdj5 and plasmid encoding insulin 2 or mutant insulin 2 (insulin 2C96Y). Cells were treated with MG-132 followed by cross-linking with DSP 48 h after transfection. Cell lysates were immunoprecipitated with insulin 2 antibody followed by Western blotting with HA antibody to detect ERdj4 and ERdj5. Quantitative analyses from three independent experiments are shown in Table 2. (B) HEK293 cells were cotransfected with plasmids encoding ERdj4 or ERdj5 and insulin 2 or insulin 2C96Y. After 48 h, cells were treated with MG-132 followed by cross-linking. Cell lysates were immunoprecipitated with insulin 2 antibody followed by Western blotting with p97 antibody. (C and D) HEK293 cells were cotransfected with plasmids encoding insulin2WT (C) or insulin2C96Y (D). Pulse-chase analyses were performed as described in Figure 1B. Cell lysates were immunoprecipitated with insulin 2 antibody, subjected to SDS-PAGE, and band intensity was quantitated by phosphorimaging analyses.
DISCUSSION
In the current study, a surprisingly large number of genes (7.8%; Supplemental Figure S1A) were significantly induced in response to transient expression of two different SP-C mutants associated with interstitial lung disease; in contrast, expression of only 11 genes was suppressed. This finding is remarkably similar to the results of a study in yeast, in which tunicamycin- or DTT-induced ER stress increased expression of ∼6% of the yeast genome with very little gene repression (Travers et al., 2000), and a recent study in which XBP-1 was reported to be an activator of expression (Acosta-Alvear et al., 2007). To identify genes specifically involved in the recognition and degradation of misfolded SP-C, we searched for potential XBP-1 target genes, because this transcription factor modulates expression of several key genes in the ERAD pathway. Analysis of the 5′-flanking sequence of genes induced by mutant SP-C identified >1000 genes with at least one TGACGTGR motif (UPRE) associated with XBP-1 binding (Clauss et al., 1996; Yoshida et al., 2001; Kanemoto et al., 2005; Acosta-Alvear et al., 2007; Shen and Hendershot, 2007); interrogation with other recently identified XBP-1 binding motifs would likely increase this pool (Acosta-Alvear et al., 2007). Genes encoding ER proteins were identified by database annotation, and the number of candidate genes was subsequently increased by literature search and use of algorithms predicting ER localization. This approach identified several genes that were previously shown to encode XBP-1 targets, including p58IPK/DNAJC3, ERdj4/DNAJB9, ERdj5/DNAJC10, ERdj3/HEDJ, EDEM, PDIA4/ERP70, PDIA6/PDI-P5, PDIR, and RAMP4/SERP1 (Lee et al., 2003; Shaffer et al., 2004). Thus, ectopic expression of a lung-specific, mutant, nonglycosylated, membrane protein (SP-CΔexon4 or SP-CL188Q) in embryonic kidney cells induced a subset of genes that was also associated with ER stress induced by tunicamycin or DTT.
Among ER-localized HSP40 family members, ERdj4 and ERdj5 were increased to the greatest extent in response to mutant SP-C expression. Interestingly, p58IPK, a DnaJ protein recently reported to be localized to the ER lumen (Rutkowski et al., 2007), was also robustly increased in cells expressing mutant SP-C (Table 1), suggesting that it may also be involved in ERAD of SP-C. BiP, ERdj4, and ERdj5 coprecipitated with misfolded SP-C and remained associated with the substrate until it was retrotranslocated to the cytosol. Inhibition of ERdj4 and ERdj5 expression resulted in ER retention and accumulation of misfolded SP-C. Inhibition of ERdj4 and ERdj5 did produce a minor increase in the amount of wild-type proprotein; however, this effect is consistent with ERAD-mediated turnover of a small amount of unfolded/misfolded wild-type protein, as reported for many newly synthesized proteins (Schubert et al., 2000; Rutkowski et al., 2006). Furthermore, in pulse-chase experiments, ERdj4 immunoprecipitated with mutant SP-C but not the wild-type proprotein. Expression of another ER-localized DnaJ homologue, ERdj3, was also modestly increased (1.9×) in cells expressing mutant SP-C (Table 1). ERdj3 was shown previously to be robustly increased in response to treatment with the ER stress-inducing agents tunicamycin or thapsigargin, associated with both unfolded and mutant protein substrates (Shen and Hendershot, 2005), and it was identified as a direct target of spliced XBP-1 (Kanemoto et al., 2005; Shen and Hendershot, 2007). However, ERdj3 did not associate with either WT or mutant SP-C (Supplemental Figure S5). Together, these results suggest that BiP and its cochaperones, ERdj4 and ERdj5, are part of a quality control complex that selectively detects and targets misfolded SP-C for elimination via ERAD. The observation that ERdj4 and ERdj5 associated with BiP and p97 independently of misfolded SP-C suggests that this complex may be “primed” for rapid association with misfolded/unfolded substrates; alternatively, Bip/ERdj/p97 association may be driven in part by ER stress arising from transient overexpression of ERdj4 and ERdj5 (Supplemental Figure S3E).
ERdj4 is type II DnaJ protein that contains a short cytosolic domain, a single membrane spanning domain and a lumenal juxtamembrane J domain followed by a C-terminal glycine/phenylalanine-rich region (Shen et al., 2002). In the present study, deletion of the J domain abrogated interaction of ERdj4 with BiP but not with misfolded SP-C; in fact, interaction of misfolded SP-C with ERdj4 was consistently enhanced by deletion of the J domain or mutation of the HPD motif. This finding is similar to that reported for interaction of ERdj3 with unfolded substrates (Shen and Hendershot, 2005) and suggests that the C-terminal domain containing the Gly/Phe-rich region may bind unfolded/misfolded substrate directly. This hypothesis is also supported by the observation that yeast Sis1p directly binds substrate through its C-terminal domain and suggests that type II DnaJ proteins may have chaperone activity independently of BiP (Lu and Cyr, 1998). Although ERdj3 was also increased in cells expressing mutant SP-C, knockdown of ERdj4 resulted in accumulation of misfolded protein, indicating that endogenous ERdj3 could not compensate for loss of ERdj4.
ERdj5 (DNAJC10/JPDI) is an ER-resident lumenal protein with a canonical C-terminal KDEL retrieval motif, a J domain, and four thioredoxin-like domains (Cunnea et al., 2003; Hosoda et al., 2003). Deletion of the J domain prevented interaction of ERdj5 with both BiP and misfolded SP-C, suggesting that association of ERdj5 with substrate was mediated by BiP. Surprisingly, mutation of the HPD motif significantly enhanced association of ERdj5 with misfolded SP-C (Supplemental Figure S2A). Further analyses revealed that, in contrast to results of a previous study (Hosoda et al., 2003), the HPD mutation did not alter association of ERdj5 (or ERdj4) with BiP (Supplemental Figure S2C). Therefore, stable complexes of misfolded SP-C, BiP, and ERdj5 formed in the absence of the catalytic activity of ERdj5. Complex formation would presumably bring the thioredoxin-like domains of ERdj5 into contact with misfolded SP-C. In this context ERdj5 may function similarly to PDI, promoting the formation, reduction or isomerization of disulfide bonds. PDI was reported to play an important role in disulfide bond reduction (unfoldase activity) before dislocation of misfolded protein (Tsai et al., 2001). The failure of two previous studies to detect ERdj5 oxidoreductase activity in vitro (Cunnea et al., 2003; Hosoda et al., 2003) may reflect the substrate specificity of ERdj5 or the requirement of a partner protein(s), such as BiP, for unfoldase activity. Alternatively, ERdj5 may act as a redox sensor to identify proteins with unpaired/mispaired cysteines and target them for elimination via ERAD.
In addition to mutant SP-C, another disease-linked mutant protein, insulin 2C96Y, formed complexes with ERdj4, ERdj5, and p97. Unexpectedly, ERdj4 and ERdj5 also formed a complex with WT insulin 2 and p97. Knockdown of ERdj4 and ERdj5 slowed degradation of both WT and mutant insulin 2. Thus, ERdj4 and ERdj5 are likely part of a larger lumenal quality control complex that targets unstable/misfolded proteins for dislocation and degradation by proteasome.
Okuda-Shimizu and Hendershot (2007) recently showed that three separate BiP-bound, nonglycosylated ERAD substrates coprecipitated with derlin-1, Herp, Hrdl, and p97. Herp played an important role in facilitating retrotranslocation and degradation of these proteins; in contrast, Herp was not required for ERAD of misfolded α1-antitrypsin, consistent with separate retrotranslocation pathways for the glycosylated and nonglycosylated ERAD substrates used in this study. Interestingly, expression of Herp was elevated 3.3- and 3.4-fold in cells expressing SP-CΔexon4 and SP-CL188Q, respectively (Table 1). Although it is not yet clear whether Herp is required for retrotranslocation of SP-C, degradation was clearly dependent on association with ERdj4 and ERdj5. A similar requirement for DnaJ association was described previously for BiP-bound ERAD substrates (Nishikawa et al., 2001). It is possible that the major function of ERdj4 and ERdj5 is simply to facilitate solubility of misfolded proteins by modulating the activity of BiP. However, the apparent direct interaction of ERdj4 with misfolded SP-C and the presence of multiple thioredoxin-like domains in ERdj5 suggest a larger role in quality control that could include recognition and/or targeting of misfolded SP-C to the retrotranslocation apparatus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. L. H. Glimcher for XBP-1−/− MEFs. This research was supported by the National Heart, Lung, and Blood Institute grants HL-61646 and HL-56387.
Abbreviations used:
- ATF
activating transcription factor
- BiP
immunoglobulin heavy chain binding protein
- CFTR
cystic fibrosis transmembrane conductance regulator
- DSP
dithiobis (succinimidylpropionate)
- EDEM
endoplasmic reticulum-degradation enhancing alpha-mannosidase like protein
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- ERdj
endoplasmic reticulum-localized DnaJ homologue
- HA
hemagglutinin
- HEK
human embryonic kidney
- HSP
heat shock protein
- IP
immunoprecipitation
- MEF
mouse embryonic fibroblast
- SEL1L
sel-1 suppressor of lin-12-like
- SP-C
surfactant protein C
- UPR
unfolded protein response
- UPRE
unfolded protein response element
- WT
wild type
- XBP-1
X-box binding protein 1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0674) on April 9, 2008.
REFERENCES
- Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., Lennon C. J., Kluger Y., Dynlacht B. D. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A., Denic V., Quan E. M., Weissman J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Bridges J. P., Xu Y., Na C. L., Wong H. R., Weaver T. E. Adaptation and increased susceptibility to infection associated with constitutive expression of misfolded SP-C. J. Cell Biol. 2006;172:395–407. doi: 10.1083/jcb.200508016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges J. P., Wert S. E., Nogee L. M., Weaver T. E. Expression of a human surfactant protein C mutation associated with interstitial lung disease disrupts lung development in transgenic mice. J. Biol. Chem. 2003;278:52739–52746. doi: 10.1074/jbc.M309599200. [DOI] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Mackey K. Modification of the TRI Reagent(TM) procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss I. M., Chu M., Zhao J. L., Glimcher L. H. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnea P. M., et al. ERdJ5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 2003;278:1059–1066. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Farinha C. M., Amaral M. D. Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol. Cell. Biol. 2005;25:5242–5252. doi: 10.1128/MCB.25.12.5242-5252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A., Gasteiger E., Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- Gething M. J. Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- Hosoda A., Kimata Y., Tsuru A., Kohno K. JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J. Biol. Chem. 2003;278:2669–2676. doi: 10.1074/jbc.M208346200. [DOI] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Johansson J., Weaver T. E., Tjernberg L. O. Proteolytic generation and aggregation of peptides from transmembrane regions: lung surfactant protein C and amyloid beta-peptide. Cell Mol. Life Sci. 2004;61:326–335. doi: 10.1007/s00018-003-3274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemoto S., Kondo S., Ogata M., Murakami T., Urano F., Imaizumi K. XBP1 activates the transcription of its target genes via an ACGT core sequence under ER stress. Biochem. Biophys. Res. Commun. 2005;331:1146–1153. doi: 10.1016/j.bbrc.2005.04.039. [DOI] [PubMed] [Google Scholar]
- Kramer-Albers E. M., Gehrig-Burger K., Thiele C., Trotter J., Nave K. A. Perturbed interactions of mutant proteolipid protein/DM20 with cholesterol and lipid rafts in oligodendroglia: implications for dysmyelination in spastic paraplegia. J. Neurosci. 2006;26:11743–11752. doi: 10.1523/JNEUROSCI.3581-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. H., Iwakoshi N. N., Glimcher L. H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Phillips K. S., Wilder M. R., Weaver T. E. Structural requirements for intracellular transport of pulmonary surfactant protein B (SP-B) Biochim. Biophys. Acta. 1996;1312:177–185. doi: 10.1016/0167-4889(95)00201-4. [DOI] [PubMed] [Google Scholar]
- Lu Z., Cyr D. M. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Mast S. W., Diekman K., Karaveg K., Davis A., Sifers R. N., Moremen K. W. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 2005;15:421–436. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Mulugeta S., Maguire J. A., Newitt J. L., Russo S. J., Kotorashvili A., Beers M. F. Misfolded BRICHOS SP-C mutant proteins induce apoptosis via caspase-4- and cytochrome c-related mechanisms. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;293:L720–L729. doi: 10.1152/ajplung.00025.2007. [DOI] [PubMed] [Google Scholar]
- Mulugeta S., Nguyen V., Russo S. J., Muniswamy M., Beers M. F. A surfactant protein C precursor protein BRICHOS domain mutation causes endoplasmic reticulum stress, proteasome dysfunction, and caspase 3 activation. Am. J. Respir. Cell. Mol. Biol. 2005;32:521–530. doi: 10.1165/rcmb.2005-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Horton P. PSORT: a program for detecting protein trafficking signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogee L. M. Alterations in SP-B and SP-C expression in neonatal lung disease. Annu. Rev. Physiol. 2004;66:601–623. doi: 10.1146/annurev.physiol.66.032102.134711. [DOI] [PubMed] [Google Scholar]
- Nogee L. M., Dunbar A. E., Wert S., Askin F., Hamvas A., Whitsett J. A. mutation in the surfactant protein C gene associated with interstitial lung disease. Chest. 2002;121:20S–21S. doi: 10.1378/chest.121.3_suppl.20s. [DOI] [PubMed] [Google Scholar]
- Nogee L. M., Dunbar A. E., Wert S. E., Askin F., Hamvas A., Whitsett J. A. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N. Engl. J. Med. 2001;344:573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- Oda Y., Hosokawa N., Wada I., Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Okuda-Shimizu Y., Hendershot L. M. Characterization of an ERAD Pathway for Nonglycosylated BiP Substrates, which Require Herp. Mol. Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Akha A.A.S., Raden D., Kaufman R. J. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:2024–2041. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., Kang S. W., Goodman A. G., Garrison J. L., Taunton J., Katze M. G., Kaufman R. J., Hegde R. S. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayeed A., Ng D.T.W. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schubert U., Anton L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Shaffer A. L., et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shen Y., Hendershot L. M. ERdj3, a stress-inducible ER DnaJ homologue, serves as a co-factor for BiP interactions with unfolded substrates. Mol. Biol. Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Hendershot L. M. Identification of ERdj3 and OBF-1/BOB-1/OCA-B as direct targets of XBP-1 during plasma cell differentiation. J. Immunol. 2007;179:2969–2978. doi: 10.4049/jimmunol.179.5.2969. [DOI] [PubMed] [Google Scholar]
- Shen Y., Meunier L., Hendershot L. M. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- Sigrist C. J., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinformatics. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- Svedine S., Wang T., Halaban R., Hebert D. N. Carbohydrates act as sorting determinants in ER-associated degradation of tyrosinase. J. Cell Sci. 2004;117:2937–2949. doi: 10.1242/jcs.01154. [DOI] [PubMed] [Google Scholar]
- Szathmary R., Bielmann R., Nita-Lazar M., Burda P., Jakob C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER–associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Vanslyke J. K., Deschenes S. M., Musil L. S. Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol. Biol. Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. J., Mulugeta S., Russo S. J., Beers M. F. Deletion of exon 4 from human surfactant protein C results in aggresome formation and generation of a dominant negative. J. Cell Sci. 2003;116:683–692. doi: 10.1242/jcs.00267. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zhang K. Z., Kaufman R. J. Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 2004;279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.