Abstract
The Cajal body (CB) is a nuclear structure closely associated with import and biogenesis of small nuclear ribonucleoprotein particles (snRNPs). Here, we tested whether CBs also contain mature snRNPs and whether CB integrity depends on the ongoing snRNP splicing cycle. Sm proteins tagged with photoactivatable and color-maturing variants of fluorescent proteins were used to monitor snRNP behavior in living cells over time; mature snRNPs accumulated in CBs, traveled from one CB to another, and they were not preferentially replaced by newly imported snRNPs. To test whether CB integrity depends on the snRNP splicing cycle, two human orthologues of yeast proteins involved in distinct steps in spliceosome disassembly after splicing, hPrp22 and hNtr1, were depleted by small interfering RNA treatment. Surprisingly, depletion of either protein led to the accumulation of U4/U6 snRNPs in CBs, suggesting that reassembly of the U4/U6·U5 tri-snRNP was delayed. Accordingly, a relative decrease in U5 snRNPs compared with U4/U6 snRNPs was observed in CBs, as well as in nuclear extracts of treated cells. Together, the data show that particular phases of the spliceosome cycle are compartmentalized in living cells, with reassembly of the tri-snRNP occurring in CBs.
INTRODUCTION
Pre-mRNA splicing is a two-step transesterification reaction catalyzed by the spliceosome, a large complex assembled from preformed subcomplexes, called spliceosomal small nuclear ribonucleoprotein particles (snRNPs) and hundreds of additional proteins (Jurica and Moore, 2003). In turn, the five major spliceosomal snRNPs, U1, U2, U4, U5, and U6, each consist of a single small nuclear RNA (snRNA) and specific set of proteins. Among the shared protein components of snRNPs are the seven Sm proteins, which are assembled as a stable, heteroheptameric ring on the RNA polymerase II-transcribed snRNAs: U1, U2, U4, and U5. After transcription, these snRNAs are transported to the cytoplasm, where the Sm ring is assembled on snRNAs by the SMN complex. Subsequently, the 5′ ends of the snRNAs are hypermethylated to generate the trimethyl-guanosine cap, which together with SMN, promotes snRNP nuclear import (reviewed in Will and Luhrmann, 2001; Matera and Shpargel, 2006; Tycowski et al., 2006). Because snRNPs do not shuttle between the nucleus and cytoplasm (Änkö and Neugebauer, unpublished data), Sm ring assembly seems to occur early and only once in the lifetime of each snRNP. A related heteroheptameric ring, consisting of seven “like-Sm” (LSm) proteins, is assembled on U6, an RNA polymerase III transcript, which is thought to remain in the nucleus for all assembly steps (Achsel et al., 1999; Mayes et al., 1999; Kiss, 2004; Listerman et al., 2007).
Once back in the cell nucleus, snRNPs first accumulate in CBs before distributing throughout the nucleoplasm, where splicing occurs (Sleeman and Lamond, 1999; Sleeman et al., 2001; Neugebauer, 2002). This suggests a role for CBs in nuclear steps of snRNP maturation, a prediction borne out by the following set of observations. First, posttranscriptional modifications of the snRNAs themselves occur in CBs after snRNP reimport from the cytoplasm (Darzacq et al., 2002; Kiss, 2002; Jady et al., 2003). These modifications, including pseudouridinylation and 2′-O-methylation, are guided by small Cajal body-specific RNAs. Second, CBs are the site of complex assembly steps that involve RNA–RNA annealing and the sequential addition of proteins. For example, the U4/U6 snRNP is formed when the U4 and U6 snRNAs anneal, a step catalyzed by U6-specific LSm proteins and SART3 (also named hPrp24 or p110) (Ghetti et al., 1995; Raghunathan and Guthrie, 1998; Achsel et al., 1999; Bell et al., 2002). Subsequently, the U4/U6·U5 tri-snRNP assembles when U5 snRNP associates by protein–protein interactions with the U4/U6 snRNP (Makarova et al., 2002; Schaffert et al., 2004). Both U4/U6 and U4/U6·U5 tri-snRNP assembly occur in CBs (Schaffert et al., 2004; Stanek and Neugebauer, 2004). Recently, mathematical modeling of U4/U6 snRNP formation in the cell nucleus revealed that accumulation of U4 and U6 snRNPs in CBs should greatly increase the efficiency of U4/U6 assembly (Klingauf et al., 2006). An additional role of CBs in U2 snRNP formation (Nesic et al., 2004) further points to CBs as the key site of nuclear steps in snRNP assembly. The observation that depletion of coilin, a protein required for snRNP concentration in CBs, impairs cell proliferation (Lemm et al., 2006) is consistent with the proposal that snRNP assembly is inefficient in the absence of CBs.
snRNPs must not only assemble de novo but also may regenerate after splicing to complete the so-called spliceosome cycle. During spliceosome assembly and activation, snRNPs undergo structural rearrangements, including U4/U6 snRNA unwinding and release of the U4 snRNP from the spliceosome (Staley and Guthrie, 1998). After splicing, mRNA is released from the spliceosome by the DEAH-box helicase hPrp22/HRH1 and snRNPs remain associated with the excised intron lariat (Company et al., 1991; Ohno and Shimura, 1996). In Saccharomyces cerevisiae, a complex of three proteins (Prp43/Ntr1/Ntr2) was shown to be essential for release of individual snRNPs from the lariat (Arenas and Abelson, 1997; Martin et al., 2002; Tsai et al., 2005; Boon et al., 2006; Tanaka et al., 2007; Tsai et al., 2007). If these released snRNPs are to participate in subsequent rounds of splicing, they have to be reassembled into the active U4/U6·U5 tri-snRNP. Several studies provide genetic and biochemical evidence for snRNP reassembly (Raghunathan and Guthrie, 1998; Bell et al., 2002; Verdone et al., 2004; Chen et al., 2006). Although snRNPs are highly expressed, the long half-lives of snRNAs suggests that they likely recycle and function again (Yu et al., 1999).
In the present study, we address the hypothesis that snRNPs cycle more than once through CBs. We show in living cells that CBs contain mostly mature snRNPs, which are capable of exchanging with nucleoplasm and visiting multiple CBs. Targeted knockdown of proteins involved in spliceosome recycling, hPrp22, and the human homologue of the recently identified yeast Ntr1, led to a dramatic accumulation of the U4/U6 snRNP in CBs. These data demonstrate that the CB is a vital way station in the spliceosomal cycle.
MATERIALS AND METHODS
Cells and Antibodies
HeLa cells were cultured in DMEM supplemented with 10% fetal calf serum, penicillin, and streptomycin (Invitrogen, Carlsbad, CA). To create a stable HeLa cell line expressing human Prp8 tagged with green fluorescent protein (GFP) at the C terminus and expressed under the control of its own promoter, a bacterial artificial chromosome (BAC) harboring the human Prp8 gene was obtained from the BACPAC Resources Center (http://bacpac.chori.org). Neo/Kanr-dsRed and EGFP-IRES-Neo cassettes were polymerase chain reaction (PCR) amplified with primers carrying 50 nucleotides of homology to the targeting sequence. Recombineering of the BACs was performed as described previously (Zhang et al., 2000) (Gene Bridges, Dresden, Germany); and after transduction, neo-resistant cells were sorted by fluorescence-activated cell sorting (FACS) to obtain single colonies. Immunoprecipitation using anti-GFP antibodies showed that hPrp8-GFP is properly incorporated into the U5 snRNP and the tri-snRNP (data not shown).
The following antibodies were used: rabbit anti-SART3/p110 antibodies (Stanek et al., 2003); monoclonal antibody (mAb) anti-coilin (5P10) (Almeida et al., 1998), kindly provided by M. Carmo-Fonseca (Institute of Molecular Medicine, Lisbon, Portugal); rabbit antibodies against LSm4 (Achsel et al., 1999); hPrp31 (U4/U6–61K) (Makarova et al., 2002); hPrp4 (U4/U6–60K) (Lauber et al., 1997); and hSnu114 (U5–116K) (Fabrizio et al., 1997), kindly provided by R. Lührmann (Max Planck Institute, Göttingen, Germany). Monoclonal antibodies against U2B” and U1-70K were purchased from Progen (Heidelberg, Germany). Rabbit anti-mouse Ntr1 was raised against a peptide, LQNEFNPNRQRHWQ (amino acids 32–45; Zymed Laboratories, South San Francisco, CA), and was provided by Michael Paine (University of Southern California, Los Angeles, CA).
Protein Tagging
SART3-cyan fluorescent protein (CFP), coilin-CFP, and hPrp31-CFP were described previously (Stanek and Neugebauer, 2004). SMN-yellow fluorescent protein (YFP) was kindly provided by M. Dundr (Chicago Medical School, North Chicago, IL; Dundr et al., 2004) and SmB-YFP and SmD1-GFP by A. Lamond (University of Dundee, United Kingdom; Sleeman and Lamond, 1999). SmB was subcloned into ECFP-C1 (Clontech, Mountain View, CA), photoactivatable (PA)-GFP-C1 (Patterson and Lippincott-Schwartz, 2004) and E5-red fluorescent protein (RFP)-C1 by using HindIII/KpnI sites. SmD1 was recloned into ECFP-C1, PA-GFP-C1, and E5-RFP-C1 by using BamHI/PstI sites. E5-RFP-C1 vector was created by replacing GFP sequence in GFP-C1 plasmid (Clontech) with E5-RFP sequence from pTimer-1 plasmid (Clontech) by using AgeI/BglII restriction sites. SART3-HcDiRed construct was created by cloning SART3 sequence into the HcDiRed-N1 vector, which originated from the H2B-HcDiRed-N1 plasmid obtained from J. Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany; Gerlich et al., 2003).
Live Cell Imaging
Cells were plated on glass bottomed Petri dishes (MatTek, Ashland, MA), and after 20–24 h, they were transfected with appropriate DNA constructs by using FuGENE 6 (Roche Diagnostics, Mannheim, Germany). The cells were imaged 22–24 h after transfection by using either a Zeiss 510 microscope equipped with water immersion objective (63× 1.2 numerical aperture [NA]) or a Leica SP2 confocal microscope equipped with water immersion objective (63× 1.2 NA) and an environmental chamber controlling CO2 level and temperature. PA-GFP was activated by short pulses of 405-nm laser line, and images of activated PA-GFP (excitation with 488-nm laser line) and either CFP (excitation with 458-nm laser line) or HcDiRed (excitation with 594-nm laser line) were acquired at 15-s intervals in activation of one CB or every 2 min in activation of the whole nucleus. The raw images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). For publication, fluorescent levels were linearly adjusted using Adobe Photoshop (Adobe Systems, Mountain View, CA).
For E5-RFP experiments, cells were transfected with SmB-E5-RFP or SmD1-E5-RFP, fixed at different times after transfection with 4% paraformaldehyde/piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) and embedded in glycerol containing 4′,6-diamidino-2-phenylindole and 2.5% 1,4-diazabicyclo[2.2.2]octane (DABCO; Sigma Chemie, Deisenhofen, Germany) as an anti-fade reagent. Alternatively, cells were treated for 2 h before fixation with 30 ng/ml leptomycin B (LC Laboratories Woburn, MA). Images were collected using the DeltaVision microscope system (Applied Precision, Issaquah, WA) coupled with Olympus IX70 microscope equipped with oil immersion objective (60× 1.4 NA) by using the same settings for each sample. Stacks of 25 z-sections with 200-nm z-step were collected per sample and subjected to mathematical deconvolution by using measured point spread function (SoftWorx; Applied Precision). Mean intensities in green and red channel were quantified using SoftWorx.
Fluorescence Resonance Energy Transfer (FRET) Measurement
Cells were transfected with fluorescent protein-tagged constructs using FuGENE 6, grown for 24–26 h, and fixed in 4% paraformaldehyde/PIPES (Sigma-Aldrich) for 10 min at room temperature. After rinsing with Mg-phosphate-buffered saline (PBS) (PBS supplemented with 10 mM Mg2+) and water, cells were embedded in glycerol containing DABCO. FRET was measured by acceptor photobleaching method as described previously (Stanek and Neugebauer, 2004) by using the Leica SP2 confocal microscope. Intensities of CFP (excited by 405-nm laser set to 5–10% of maximum power) and YFP (excited by 514-nm laser line set to 2% of maximum power) were measured. Then, YFP was bleached in a region of interest by three to five intensive (30% maximum power) pulses of 514-nm laser line and CFP and YFP fluorescence measured again. Apparent FRET efficiency calculated according to the equation FRETefficiency[%] = (CFPafter − CFPbefore) × 100/CFPafter. Unbleached regions of the same cell were used as a negative control. Ten cells were measured per each FRET pair, and average and SE were calculated.
Small Interfering RNA (siRNA) Transfection
Preannealed siRNA duplexes were obtained either from Ambion (Austin, TX) or QIAGEN (Hilden, Germany). Three independent siRNA duplexes were used against hNtr1, and five duplexes were used to target hPrp22. The sequences of sense siRNAs were as follows: from Ambion: hNtr1-27-5′-CCUGUUAAGCAGGACGACUtt, hNtr1-28-5′-GCAGGACGACUUUCCUAAGtt, hNtr1-29-5′-GGAUUAGCAAGAAGCUCACtt; hPrp22-55-5′-GCUUUAAUGCCCAGCGCAGtt; hPrp22-56-5′-GGAAUAAAGUGAAGUCUAGtt; and hPrp22-57-5′-CCCAAAUAGACGGCGAAAUtt; from Qiagen: hPrp22-3-5′-GGGACAGGACAAAGAAGAAtt and hPrp22-4-5′-CAGAGAAGUGGGAGAUCAAtt.
The negative control 1 siRNA from Ambion was used as a negative control. Oligofectamine (Invitrogen) was used for siRNA transfection. Cells were incubated 48 h before further treatment. Within this incubation period we did not observe any extensive cell death with respect to the treatment with the negative control siRNA.
Reverse Transcription (RT)-PCR
Total RNA was isolated 48 h after siRNA transfection by using TRIzol reagent (Invitrogen). cDNA was synthesized using a gene-specific reverse primer and SuperScript III (Invitrogen). Taq polymerase was used to amplify cDNA (25 cycles). Controls without RT reaction were performed to verify that there was no residual DNA contamination. The following primers were used for RT-PCR and quantitative PCR: hPrp22-For, CAAGAGGTGGGCTACACCAT; hPrp22-Rev, 5′-TGATCGCGTACTGAGTGAGG; hNtr1-For, 5′-TGTCTTCACTCCTGGCTCCT; hNtr1-Rev, 5′-AAGCCACTTGGGGAAGAAGT; 18S-For, 5′-TTGTTGGTTTTCGGAACTGAG; 18S-Rev, 5′-GCAAATGCTTCGGCTCTGGTC; c-myc-mRNA-For, 5′-GCGACTCTGAGGAGGAACAAGAAG; c-myc-mRNA-Rev, 5′-ACTCTGACCTTTTGCCAGGAGC; c-myc-pre-mRNA-For, 5′-TGCTCCCTTTATTCCCCCAC; c-myc-pre-mRNA-Rev, 5′-GGTCATAGTTCCTGTTGGTGAAGC; LDHA mRNA-For, 5′-AGAACACCAAAGATTGTCTCTGGC; LDHA mRNA-Rev, 5′-TTTCCCCCATCAGGTAACGG; LDHA pre-mRNA-For, 5′-CCTTTCAACTCTCTTTTGGCAACC; LDHA pre-mRNA-Rev, 5′-AATCTTATTCTGGGGGGTCTGTTC; Tubulin mRNA-For, 5′-GCTGCTTTGTGGAGTGGATTCC; Tubulin mRNA-Rev, 5′-CCGTGTTGTTGCCAATGAAGG; Tubulin pre-mRNA-For, 5′-GACCTTCCTCCTGCTTTCAGTTC; and Tubulin pre-mRNA-Rev, 5′-TCTGCTTGTGTTCCCAGTTGC.
Quantitative PCR was done as described previously (Listerman et al., 2006), and ratio of pre-mRNA to mRNA was calculated for each siRNA treatment according to RsiRNA = 2(Ctpre-mRNA − CtmRNA), normalized to NC siRNA treated cells (Rn = RsiRNA/RncsiRNA), and plotted.
Glycerol Gradient Ultracentrifugation
Nuclear extracts were prepared according to Dignam et al. (1983), diluted in gradient buffer (20 mM HEPES/KOH, pH 8.0, 150 mM NaCl, 1.5 mM MgCl2, and 0.5 mM dithiothreitol), and fractionated in a linear 10–30% glycerol gradient by centrifugation at 32,000 rpm for 17 h by using the SW-41 rotor (Beckman Coulter, Fullerton, CA). Individual fractions (700 μl) were collected, and RNA was extracted from each fraction with phenol:chloroform:isoamylalcohol, separated on 10% urea-polyacrylamide gel electrophoresis (PAGE), and silver stained. In parallel, proteins were precipitated from the phenol phase by acetone, dissolved in SDS-PAGE sample buffer, and analyzed by immunoblotting.
Indirect Immunofluorescence
Forty-eight hours after the siRNA transfection the cells were fixed in 4% paraformaldehyde/PIPES for 10 min, permeabilized for 5 min with 0.2% Triton X-100 (Sigma Chemie), and incubated with appropriate primary antibodies. Secondary anti-rabbit antibodies conjugated with fluorescein isothiocyanate (FITC) and anti-mouse antibodies conjugated with tetramethylrhodamine B isothiocyanate (TRITC) (Jackson ImmunoResearch Laboratories, West Grove, PA) were used. Images were collected using a DeltaVision microscope system and subjected to mathematical deconvolution as described above. Mean fluorescence intensities in CBs and the nucleoplasm were determined in individual optical sections by using ImageJ as described previously (Stanek and Neugebauer, 2004).
In Situ Hybridization
Digoxigenin-labeled DNA probes directed against human U2, U4, and U5 snRNAs were obtained by PCR as described previously (Bell et al., 2002) by using pSP65U2H, pSPU4b, pSP64U5 (Black and Pinto, 1989) as templates. Forty-eight hours after siRNA transfection cells were fixed in 4% paraformaldehyde/PIPES for 10 min, permeabilized with 0.5% Triton X-100 for 5 min, and incubated with anti-SART3 antibodies as a marker of CBs followed by incubation with secondary antibody conjugated with FITC (Jackson ImmunoResearch Laboratories). Cells were again fixed in 4% paraformaldehyde/PIPES for 5 min, quenched for 5 min in 0.1 M glycine/0.2 M Tris, pH 7.4, and incubated with digoxigenin-labeled probe in 2× SSC/50% formamide/10% dextran sulfate/1% BSA for 60 min at 37°C. After washing in 2× SSC/50% formamide, 2× SSC and 1× SSC, the probe was detected by mouse anti-digoxigenin antibody (Roche Diagnostics) followed by incubation with goat anti-mouse antibody coupled with TRITC (Jackson ImmunoResearch Laboratories). Images were collected using a DeltaVision microscope system, and fluorescence intensities in CBs and the nucleoplasm were determined as describe above.
RESULTS
Accumulation of Mature snRNPs in Cajal Bodies
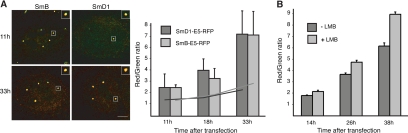
It is not known whether the pool of snRNPs in CBs consists of only newly imported and incompletely assembled snRNPs, or whether mature snRNPs that have already participated in splicing accumulate in CBs as well. We probed the relative “age” of snRNPs in CBs, by tagging SmB and SmD1 proteins with a “fluorescent timer,” a mutant of red fluorescent protein drFP583 (E5-RFP) that changes its fluorescence emission properties during maturation, converting from green-to-red emission within the course of 3 h (Terskikh et al., 2000). Changes in the red-to-green ratio over time are therefore indicative of relative shifts in the age of the molecules present in a given subcellular location. Because there is no evidence of Sm protein exchange once the Sm ring has been assembled on snRNA (Wang and Meier, 2004; Shpargel and Matera, 2005), we assume that Sm proteins remain stably associated with nuclear snRNPs after assembly and import; thus, Sm protein dynamics likely reflect those of mature snRNPs. In addition, fluorescently tagged Sm proteins efficiently incorporate into snRNP particles, and their expression is comparable with endogenous Sm proteins (Supplemental Figure 1). HeLa cells expressing SmB- or SmD1-E5-RFP were fixed at different times after transfection, and E5-RFP fluorescence intensities in the green and red spectra were measured (Figure 1A). Note that both Sm-E5-RFP constructs localized properly in CBs. Surprisingly, both green and red forms of E5-RFP were clearly present in CBs at the earliest time point (11 h) that CBs were detectable, indicating that a portion of E5-RFP had already matured to the red form before fluorescence reached the detection threshold. Absolute intensities of green fluorescence did not change over the time, indicating a constant influx of new snRNPs that must reside for a consistent period in the CB. An increase in red fluorescence and the red-to-green fluorescence ratio was observed, and over the period of 22 h it increased ∼2 times in the nucleoplasm and 3 times in CBs. If CBs selectively recruited only newly imported snRNPs, we would have expected the red-to-green ratio to remain the same or even decrease in CBs, despite the fact that more of the red variant accumulates in the nucleus.
Figure 1.
Mature snRNPs accumulate in Cajal bodies. To determine the age of snRNPs in CBs, SmB and SmD1 proteins that are stable components of snRNPs were tagged with E5-RFP. During maturation, E5-RFP changes its fluorescence from green to red in a time course of a couple hours. (A) HeLa cells were transfected with SmB-E5-RFP or SmD1-E5-RFP and fixed at different times after transfection. Ratio of red to green was measured in CBs (bars are SD) and in the nucleoplasm (lines). Red fluorescence in CBs increased ∼3 times over 22 h, indicating that mature snRNPs accumulate in CBs. (B) Cells were transiently transfected with SmB-E5-RFP, and 2 h before fixation they were treated with leptomycin B (LMB) that inhibits biogenesis of new snRNPs by blocking export of new snRNA from the nucleus. A 45% increase of red-to-green ratio after 38 h indicates import inhibition of the green (new) variant of SmB-E5-RFP. Bars are SE.
To test whether Sm-E5-RFP proteins are imported to the cell nucleus within snRNP particles, cells were treated for 2 h before fixation with leptomycin B that efficiently inhibits export of newly synthesized snRNAs to the cytoplasm and consequently also import of newly formed snRNPs. A 45% increase in the red-to-green ratio observed 38 h after transfection (Figure 1B) indicates impairment of green SmB-E5-RFP import, indicating that SmB-E5-RFP is indeed imported together with snRNAs beginning the biogenesis pathway in the cytoplasm. The finding that CBs accumulate more old snRNPs in CBs compared with the nucleoplasm indicates that the pool of snRNPs concentrated in CB contains not only fresh snRNPs but also relatively older, presumably mature snRNPs.
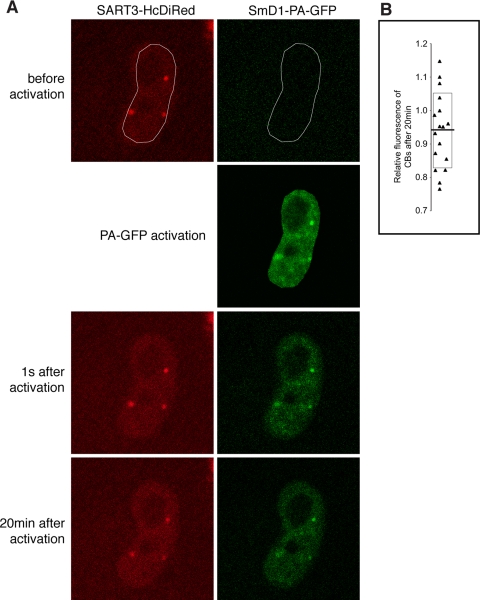
A complementary experiment was suggested by the fact that, although they are highly concentrated in CBs, snRNPs exchange rapidly with the surrounding nucleoplasm and only reside in CBs for few seconds on average (Dundr et al., 2004; Sleeman, 2007). We tagged Sm proteins with PA-GFP to determine whether the snRNPs that exit CBs at any given moment are replaced by new or old snRNPs. SmB- or SmD1-PA-GFP was expressed and the entire nucleus was photoactivated (Figure 2). If CBs contain exclusively new (nonactivated) snRNPs imported from the cytoplasm, fluorescence would be lost from these CBs over time. On the contrary, SmB-GFP fluorescence CBs remained high 20 min after photoactivation (Figure 2), 100 times longer than the residence time of snRNPs in CBs. Measurement of fluorescence intensities in CBs and nucleoplasm revealed a 0–25% decrease (average 6%) in fluorescence intensity in CBs relative to nucleoplasm. These data show that the exchanging population of snRNPs in CBs consists largely of “older” nuclear snRNPs.
Figure 2.
snRNPs newly imported from the cytoplasm represent only a minor fraction in CBs. (A) The SmD1 protein was tagged with PA-GFP and coexpressed in HeLa cells with SART3-HcDiRed as a marker of CBs. To distinguish between nuclear and cytoplasmic pool of snRNPs, nuclear snRNPs were specifically activated by short pulse of 405-nm laser line. Images were taken every 2 min for total 20 min. (B) Quantification of CB fluorescence. To avoid photobleaching effects, the ratio of fluorescent signals CB:nucleoplasm were determined immediately after activation and 20 min later. Values for individual CBs are plotted. Mean value is indicated by a solid line SD by a box. A small decrease in CB fluorescence indicates that within 20 min, snRNPs imported from the cytoplasm represent only small fraction of snRNPs in CBs.
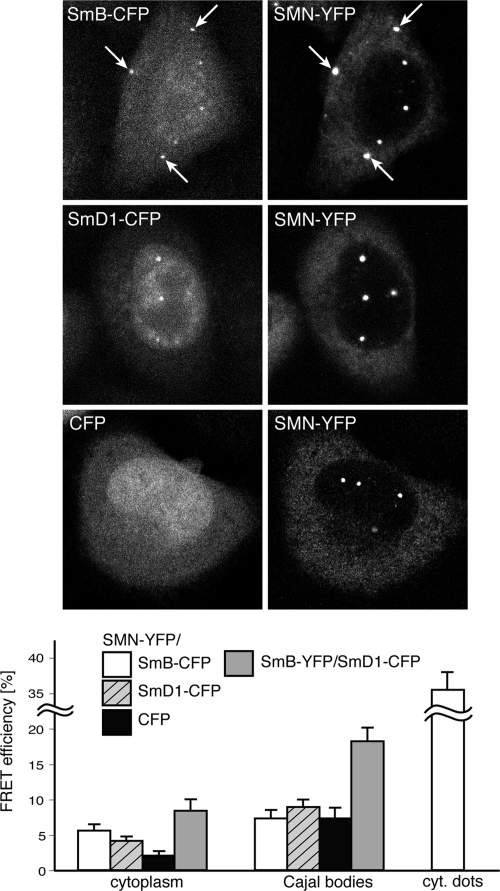
To further examine the assembly status of snRNPs in CBs and in the cytoplasm, we probed snRNP interactions with the SMN protein, which is localized both in the cytoplasm and in CBs. The SMN protein is a part of the SMN complex, which ensures proper assembly of Sm rings on snRNPs in the cytoplasm (Terns and Terns, 2001; Meister et al., 2002; Paushkin et al., 2002; Matera and Shpargel, 2006). The significance of SMN accumulation in CBs is not known. Because SMN is required for the import of newly assembled snRNPs (Narayanan et al., 2002, 2004), it is hypothesized that SMN is coimported into CBs with new snRNPs (Matera and Shpargel, 2006). Interactions between snRNPs and the SMN protein were measured by FRET. Because Sm proteins and SMN are present in the cytoplasm and in CBs, a FRET signal could indicate the presence of snRNP–SMN complexes, changes in their structure and composition in the different cellular compartments, or both. SmD1-CFP or SmB-CFP were coexpressed with SMN-YFP in HeLa cells, and FRET was measured by the acceptor photobleaching method in the cytoplasm and CBs of the same cell (Figure 3). As a negative control, SMN-YFP was coexpressed with CFP only. As expected, FRET between SMN and the Sm proteins was detected in the cytoplasm, but no FRET signal above the negative control was observed in CBs. Overall higher FRET values in CBs are likely caused by local accumulation of SMN. In some cells, Sm proteins and SMN accumulated in cytoplasmic bodies, presumably due to coexpression of exogenous SMN and Sm proteins (Shpargel et al., 2003); these aggregates resembled recently identified U bodies (Liu and Gall, 2007) and contained similar amounts of fluorescent proteins as nuclear CBs. High FRET signals detected in these cytoplasmic inclusions indicate accumulation of SmB–SMN complexes. In addition, FRET between SmB-YFP and SmD1-CFP were robust in the cytoplasm as well as in CBs. Preassembled Sm subcomplexes are used during assembly of the Sm-ring; SmB and SmD1 proteins are not part of the same Sm subcomplexes, but they lay next to each other in the ring (Raker et al., 1996; Kambach et al., 1999). Thus, detection of FRET between these two components represents a good marker for Sm-ring assembly and further indicates that fluorescently tagged Sm proteins are correctly incorporated into mature snRNPs. These results show that snRNPs in CBs differ from snRNPs localized in the cytoplasm; snRNPs in CBs either do not interact with SMN or snRNP-SMN complexes in CBs have an altered structure that does not support FRET detection.
Figure 3.
The SMN protein interacts with Sm proteins in the cytoplasm. To compare SMN–snRNP complexes in CBs and the cytoplasm, SMN-YFP was coexpressed with SmD1-CFP, SmB-CFP, or CFP alone as a negative control, and FRET was measured in the cytoplasm and in CBs by acceptor photobleaching method. The SMN-Sm FRET signal was two- to threefold higher over negative control in the cytoplasm but not in CBs. The SmB–SmD1 pair used as a positive control exhibited high FRET signal both in the cytoplasm and in CBs. In some cells, we observed cytoplasmic accumulation of SmB and SMN proteins (arrows) and high FRET signal was measured in these cytoplasmic inclusions. Ten measurements for each pair are averaged and are shown in the graph with SE bars.
Cycling of snRNPs between CBs
Because Sm proteins exchange rapidly between CBs and the nucleoplasm (Dundr et al., 2004; Sleeman, 2007), our finding that CBs contain predominantly mature snRNPs implies that snRNPs cycle constantly between CBs and the nucleoplasm, visiting CBs repeatedly. To test this directly, SmD1 or SmB proteins tagged with PA-GFP were coexpressed with CFP-tagged markers of CBs (SART3, coilin). The Sm-PA-GFP proteins were activated by a short pulse of 405 nm laser in one CB and the movement of activated molecules monitored every 15 s over a 5-min time period (Figure 4, Supplemental Videos). Activated SmB- and SmD1-PA-GFP proteins moved from the activated CB, diffused throughout the nucleoplasm and accumulated in another (nonactivated) CB within the time course of the experiment. This behavior was not affected by coexpression of any of the CB markers used and no movement of activated molecules was observed in fixed cells (Supplemental Figure 2). These data show directly that snRNPs repeatedly visit CBs.
Figure 4.
snRNPs cycle between CBs. To observe movement of snRNPs between CBs and the nucleoplasm, SmB-PA-GFP was coexpressed with SART3-CFP and SmD1-PA-GFP with coilin-CFP. Sm-PA-GFPs were specifically activated in one CB (circle) by short pulse of 405-nm laser, and movement of activated molecules was observed for 5 min (also see Supplemental Videos). Activated molecules moved throughout the whole nucleoplasm and accumulated in other CBs in the same nucleus (arrows). The detection system was adjusted to detect very low signals of PA-GFP, but by using this setup we also detected cell autofluorescence in the cytoplasm (stars).
Accumulation of U4/U6 snRNPs in CBs after Inhibition of Spliceosome Disassembly
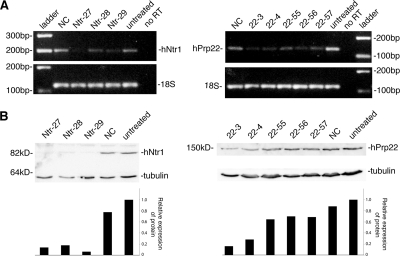
Previous reports showed that ongoing snRNP biogenesis is necessary for structural integrity of CBs (Shpargel and Matera, 2005; Girard et al., 2006; Lemm et al., 2006). However, our data show that the CB contains mainly mature snRNPs. We therefore aimed to test whether an ongoing supply of mature snRNPs is important for maintaining the CB structure as well. Two proteins implicated in spliceosome disassembly were depleted, with the expectation that inhibition of snRNP recycling after splicing would reduce the supply of mature snRNPs. Three different siRNA duplexes were used to target the human homologue of Ntr1 (also named TFIP11; Wen et al., 2005) and five duplexes for hPrp22. After 48 h, total RNA and proteins were isolated and mRNA and protein levels tested (Figure 5).
Figure 5.
siRNA targeted depletion of hPrp22 and hNtr1. (A) Five different siRNAs against hPrp22 and three against hNtr1 protein were used. Cells were treated with siRNAs for 48 h, and mRNA levels of hPrp22 and hNtr1 were determined by RT-PCR. RT-PCR of 18S rRNA served as a loading control. (B) Extract from cells treated for 48 h with siRNAs was loaded on gel, and hNtr1 and hPrp22 protein levels were determined. Anti-tubulin antibody used as a loading control.
To address the effects of hNtr1 and hPrp22 knockdown on CB structure, CBs were immunodetected after the siRNA treatment with anti-coilin and anti-SART3 antibodies. Coilin is a universal marker of CBs; SART3 interacts with the U4/U6 snRNP and its presence in the CB is sensitive to transcription/splicing inhibition (Stanek et al., 2003). Surprisingly, CBs remained intact and accumulated SART3 after either treatment (Figure 6A; data not shown). In fact, the detection of SART3 fluorescent signal in CBs was enhanced by treatment, and an elevated accumulation in CBs of other U4/U6 snRNP components was also observed upon depletion of hPrp22 or hNtr1 (Figure 6B). The CB accumulation of U4/U6-specific markers was not due to a decrease in nucleoplasmic fluorescence, which remained mostly unaffected by the siRNA treatment. Partial effects on the U2-specific U2B” protein were observed after hPrp22 but not hNtr1 knockdown, and no significant changes in CB localization of the U2 snRNA or the U1-specific U1–70K protein were detected. In contrast, CB localization of the three tested U5 snRNP components—hSnu114, hPrp8-GFP, and the U5 snRNA—decreased after siRNA treatment. The hPrp8-GFP protein was expressed from a human BAC under the control of the endogenous promoter and was properly incorporated into the U5 and tri-snRNPs (data not shown).
Figure 6.
U4/U6-specific markers accumulate in CBs after hPrp22 and hNtr1 knockdown. (A) Cells treated with siRNA against hPrp22 (22–3) and hNtr1 (Ntr-27) for 48 h were fixed, and localization of snRNP-specific proteins was determined by antibody staining. To avoid distortion of CB morphology due to changes in intensity, the intensities of the images shown were adjusted to an equal maximum. This results in apparent fluorescent reduction in the nucleoplasm after siRNA treatments, which was not observed at raw images. NC, negative control siRNA. hSnu114, green; coilin, red. (B) Quantification of fluorescence is shown in the graph. Fluorescence ratio CB:nucleoplasm was calculated for each CB, and average with SE bars is shown (number of measured CBs indicated inside bars). *p < 0.0001 as determined by Student's t test with respect to cells treated with control siRNA.
Depletion of hPrp22 or hNtr1 caused the dramatic and specific accumulation of U4/U6 snRNP components in CBs. To determine whether bona fide U4/U6 snRNP particles had formed in CBs, we used FRET to measure interactions that are specific for the SART3·U4/U6 complex, a transient intermediate en route to tri-snRNP formation that was previously localized to CBs (Stanek and Neugebauer, 2004). Cells were incubated for 24h with the indicated siRNA, cotransfected with hPrp31-CFP and SART3-YFP, incubated additional 24 h and fixed. FRET was measured as described previously in the nucleoplasm and CBs by acceptor photobleaching (Stanek and Neugebauer, 2004). No significant changes were observed in CBs after treatment with anti-hPrp22 or anti-hNtr1 siRNAs (Figure 7), showing that U4/U6 components accumulating in CBs are assembled into the U4/U6 snRNP. These data indicate that inhibition of spliceosome recycling leads to specific accumulation of the U4/U6 snRNPs in CBs. To test whether U4/U6 snRNP accumulation in CBs resulted from an inhibition of splicing, three different genes (c-myc, LDHA, and tubulin) were tested for splicing efficiency by RT-quantitative PCR. Partial increases in pre-mRNA-to-mRNA ratios were detected after treatment with anti-hPrp22 siRNAs, but no substantial inhibition of splicing was observed after hNtr1 depletion (Supplemental Figure 3) indicating that U4/U6 snRNP concentration in CBs is not likely a general result of splicing inhibition and a lack of expression of necessary snRNP components.
Figure 7.
Assembled U4/U6 snRNP accumulates in Cajal bodies after hPrp22 and hNtr1 knockdown. (A) Cells treated with siRNA against hPrp22 (22-3) were transfected with SART3-YFP and hPrp31-CFP, and FRET was measured by acceptor photobleaching in the nucleoplasm and CBs. (B) Quantification of SART3-YFP/hPrp31-CFP FRET measurements in cells treated with control (NC), hPrp22-3, or hNtr1-27 siRNAs. Average values of 10 measurements with SE bars are shown.
Enhanced accumulation of the U4/U6 snRNPs in CBs had been previously observed after inhibition of U4/U6·U5 snRNP assembly (Schaffert et al., 2004). If depletion of hNtr1 and hPrp22 inhibits spliceosome recycling and thus reduces the amount of the free U5 snRNP, assembly of the functional tri-snRNP might be delayed. To assess the influence of hPrp22 or hNtr1 knockdown on snRNP assembly and recycling, nuclear extract was prepared from cells treated for 48 h with anti-hPrp22–3 or anti-hNtr1–27 siRNAs and snRNPs analyzed by glycerol gradient ultracentrifugation. RNA and proteins were isolated from individual fractions and U5-specific hSnu114 and U4/U6-associated hPrp4 proteins detected by immunoblotting (Figure 8). To determine the position of individual snRNP complexes in the gradient snRNAs from each fraction were resolved on denaturing gels and silver stained (data not shown). hPrp4 sedimented in two distinct peaks that reflect U4/U6 snRNPs (fractions 6–8) and U4/U6·U5 tri-snRNPs (fractions 11–13). The U5 marker hSnu114 fractionated with the tri-snRNP and with the mono-U5 snRNP (fractions 8–10). In addition to the mono-U5 and the tri-snRNP, hSnu114 accumulated in faster sedimenting complexes (fractions 15 and 16) in anti-hPrp22 siRNA treated cells, which might correspond to post-spliceosomal complexes (Makarov et al., 2002) that were not recycled properly after hPrp22 knockdown. Importantly, decreases in the mono-U5 snRNP relative to the U5 in the tri-snRNP fractions were observed in both hPrp22 and hNtr1 siRNA-treated cells, consistent with the reduced U5 snRNP concentration in CBs after both knockdowns (Figure 6).
Figure 8.
Mono-U5 snRNP is reduced after hPrp22 and hNtr1 knockdowns. HeLa cells were treated with 22-3 or Ntr-27 siRNA for 48 h, and nuclear extracts were centrifuged on 10–30% glycerol gradients. Parallel RNA gels were used for determination of snRNP complexes position in gradients. Proteins from individual fractions were isolated, and hSnu114 (marker of the U5 snRNP) and hPrp4 (marker of the U4/U6 snRNP) were detected. In siRNA-treated cells, the level of the free U5 snRNP decreased (fractions 8–10).
DISCUSSION
It was previously shown that U2, U4/U6, and U4/U6·U5 tri-snRNP assembly steps occur in CBs (Nesic et al., 2004; Schaffert et al., 2004; Stanek and Neugebauer, 2004); however, it was not known whether the assembly events reflected de novo snRNP assembly or reassembly after splicing. In this study, we present data showing that in addition to de novo snRNP assembly, CBs could serve as the snRNP recycling center. These conclusions are based on three independent lines of evidence. First, we show by several experimental means, that the snRNPs concentrated in CBs include a substantial “mature” snRNP pool. Second, we show directly that snRNPs visit multiple CBs within the same nucleus and do so frequently; thus, there is no reason to suspect that CBs preferentially contain newly imported snRNPs. Third, targeted depletion of two factors required for spliceosome disassembly and snRNP release, hPrp22 and hNtr1, leads to an accumulation of U4/U6 snRNPs in CBs. We argue below that this accumulation likely reflects a failure of U5 snRNP recovery from spliceosomes; in the absence of sufficient U5 snRNP flux through CBs, tri-snRNP formation is blocked. These observations provide novel insights into how phases of the spliceosome cycle are compartmentalized in living cells.
Experiments establishing the relative maturity of snRNPs in CBs relied on three different fluorescence microscopy techniques. The E5-RFP fluorescent protein tag, which changes emission spectra from green to red as it matures, and PA-GFP were both used to tag Sm proteins (B and D1; see Figures 1 and 2) and to monitor snRNP movements. Both approaches proved that the snRNP pool in CBs largely consists of mature snRNPs that have visited CBs already multiple times. In agreement with these data, snRNP movements between CBs were observed, in which snRNPs photoactivated in one CB reappeared within a time course of minutes in another distant CB. A similar movement of snRNPs between CBs was described recently (Sleeman, 2007).
In the third approach, we sought to compare snRNPs in CBs and in the cytoplasm by measuring FRET signals between Sm proteins and SMN, which plays a role in the cytoplasmic phase of snRNP biogenesis. SMN is coimported with new snRNPs to the nucleus, and it is highly concentrated in CBs; because SMN binds coilin, it has been proposed that SMN delivers the newly imported snRNPs to CBs (Stanek and Neugebauer, 2006). However, although we detected SMN-snRNP interactions in the cytoplasm, we did not observe any significant interaction in CBs. This indicates that either 1) the SMN–snRNP complex falls apart immediately after the import of snRNPs, 2) that a conformational change occurs within the CB that is unfavorable for FRET, or 3) newly imported snRNP–SMN complexes form only a small fraction of snRNPs and SMN in CBs, and these are below the detection limit of the FRET assay. At present, we cannot distinguish among these possibilities; if snRNP–SMN complexes are present in CBs, they must differ at least conformationally from the complexes found in the cytoplasm. Perhaps SMN protein accumulates in the CB as a result of binding coilin after snRNP import and dissociation in the CB. Presumably, SMN eventually returns to the cytoplasm for further rounds of Sm ring assembly.
The conclusion that CBs contain not only new snRNPs imported from the cytoplasm but also mature snRNPs agrees with findings that the concentration of snRNPs in CBs is transcription dependent (Carmo-Fonseca et al., 1992; Blencowe et al., 1993; Stanek et al., 2003); in the absence of new intron-containing transcripts (i.e., under conditions of transcriptional blockade), the splicing process provides fewer “used” snRNPs for recycling. This highlights a paradox emerging in the field, because it has been proposed that snRNP biogenesis and import from the cytoplasm is required for snRNP accumulation in CBs and for integrity of CBs themselves (Carvalho et al., 1999; Shpargel and Matera, 2005; Girard et al., 2006; Lemm et al., 2006). If only a small proportion of snRNPs in CBs are newly imported, how can this fraction be required for CB integrity? The simplest explanation is that experimental reduction of snRNP biogenesis at various stages has long-term effects on the overall concentration of snRNPs in the nucleus, not just an acute effect on import. A reasonable proposal stemming from our present study and consistent with prior work of others is that CB integrity depends on the cellular level of splicing activity and the absolute concentration of nuclear snRNPs (Carmo-Fonseca et al., 1992; Blencowe et al., 1993; Sleeman et al., 2001; Stanek et al., 2003).
Our data show that mature snRNPs repeatedly visit CBs. To test whether this cycling through CBs correlates with snRNP regeneration after splicing, proteins involved in spliceosome disassembly were depleted, and localization of distinct snRNPs in CBs was examined. Surprisingly, depletion of two tested proteins involved in spliceosome disassembly resulted in accumulation of U4/U6 snRNPs in CBs. Because transcription/splicing inhibition by α-amanitin leads to opposite effects—the snRNPs and SART3 leave CBs (Carmo-Fonseca et al., 1992; Blencowe et al., 1993; Sleeman et al., 2001; Stanek et al., 2003)—it seems unlikely that U4/U6 snRNP accumulates in CBs as a result of splicing inhibition. In addition, splicing of c-fos and c-myc pre-mRNAs was only slightly reduced after siRNA treatment (Supplemental Figure 3). Instead, the phenotype more closely resembles the situation after inhibition of U4/U6·U5 snRNP formation, when accumulation of U4/U6 snRNPs in CBs was also observed (Schaffert et al., 2004). Why does inhibition of spliceosome disassembly and inhibition of tri-snRNP assembly have the same phenotype? According to current models of spliceosome recycling, inhibition of this process should trap U5 and U6 snRNPs in the late spliceosome (Will and Luhrmann, 2006) and thus decrease levels of free U5 and U6 snRNPs in the nucleoplasm. In contrast, the U4 snRNP leaves the spliceosome at an earlier step, just as the tri-snRNP joins the assembling spliceosome (Makarov et al., 2002; Chan et al., 2003). Thus, the level of free U4 mono-snRNP in the nucleoplasm should be unaffected by Prp22 or Ntr1 depletion. Early studies showed that there is two- to threefold excess of the U6 snRNP over the U5 and U4 snRNPs (Tycowski et al., 2006), making it unlikely that U6 snRNP levels are limiting. In contrast to U4 and U6; however, levels of free U5 snRNP are likely decreased and formation of the U4/U6·U5 tri-snRNP inhibited, as was shown after Ntr1 depletion in yeast (Boon et al., 2006). Consistent with this, we observed a decrease in U5 snRNP levels both in CBs and nuclear extracts after knockdown. Thus, inhibition of spliceosome disassembly leads to a similar situation as inhibition of tri-snRNP assembly—accumulation of the U4/U6 snRNPs in CBs. Apparently, the supply of new U5 snRNPs from the cytoplasm is not sufficient to keep up with tri-snRNP assembly, underscoring the importance of the recycled U5 snRNP for assembly and regeneration of tri-snRNPs.
Together, these observations suggest that snRNP reassembly after splicing may obey similar rules to de novo snRNP assembly, even though the assembly of new snRNPs includes additional steps that need not be repeated (e.g., posttranscriptional RNA modification). This implies that snRNP assembly in CBs at any stage of their life cycle must be independent of snRNA posttranscriptional modifications or any other steps in snRNP biogenesis. Finally, it has been shown by mathematical modeling that CBs increase the rate of U4/U6 snRNP assembly, by providing a local environment with elevated snRNP concentrations (Klingauf et al., 2006). Thus, the localization of snRNPs to CBs likely promotes the assembly of new as well as regenerating snRNPs by the same mechanism, because snRNPs from either source will meet elevated concentrations of their potential partners in the CB.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Reinhard Lührmann, Marc Schneider (Max Planck Institute, Göttingen, Germany), Maria Carmo-Fonseca, Angus Lamond, Jan Ellenberg, Michael Paine (University of Southern California, Los Angeles, CA), Petr Draber (Institute of Molecular Genetics, Academy of Sciences of the Czech Republic), and Miroslav Dundr for gifts of reagents. We thank Angus Lamond, Jason Swedlow, and Jean Beggs for helpful suggestions and Magdalena Strzelecka, Marta Pabis, and Arnold Kiss for comments on the manuscript. We are grateful to Jason Swedlow, Sam Swift, Jan Peychl, and Ondrej Sebesta for help with live cell imaging and Christian Merz and Reinhard Lührmann for help with the gradient centrifugation. This work was supported by the Max Planck Society and a grant from the DFG (to K.N.), a Partner group grant from the Max Planck Gesellschaft, the Grant Agency of the Czech Republic (301/05/0601), and the institutional project AV0Z50520514 awarded by the Academy of Sciences of the Czech republic (to D.S.). M.B. was supported by the grant GACR 204/07/0133.
Abbreviations used:
- CB
Cajal body
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein particle.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1259) on March 26, 2008.
REFERENCES
- Achsel T., Brahms H., Kastner B., Bachi A., Wilm M., Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F., Saffrich R., Ansorge W., Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J. E., Abelson J. N. Prp 43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Pinto A. L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol. Cell. Biol. 1989;9:3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Carmo-Fonseca M., Behrens S. E., Luhrmann R., Lamond A. I. Interaction of the human autoantigen p150 with splicing snRNPs. J. Cell Sci. 1993;105:685–697. doi: 10.1242/jcs.105.3.685. [DOI] [PubMed] [Google Scholar]
- Boon K. L., Auchynnikava T., Edwalds-Gilbert G., Barrass J. D., Droop A. P., Dez C., Beggs J. D. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell. Biol. 2006;26:6016–6023. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Pepperkok R., Carvalho M. T., Lamond A. I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M. T., Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Chen C. H., Kao D. I., Chan S. P., Kao T. C., Lin J. Y., Cheng S. C. Functional links between the Prp19-associated complex, U4/U6 biogenesis, and spliceosome recycling. RNA. 2006;12:765–774. doi: 10.1261/rna.2292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Darzacq X., Jady B. E., Verheggen C., Kiss A. M., Bertrand E., Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hebert M. D., Karpova T. S., Stanek D., Xu H., Shpargel K. B., Meier U. T., Neugebauer K. M., Matera A. G., Misteli T. In vivo kinetics of Cajal body components. J. Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Laggerbauer B., Lauber J., Lane W. S., Luhrmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Beaudouin J., Kalbfuss B., Daigle N., Eils R., Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Ghetti A., Company M., Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–145. [PMC free article] [PubMed] [Google Scholar]
- Girard C., Neel H., Bertrand E., Bordonne R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res. 2006;34:2925–2932. doi: 10.1093/nar/gkl374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady B. E., Darzacq X., Tucker K. E., Matera A. G., Bertrand E., Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 2003;22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M. S., Moore M. J. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke S., Young R., Avis J. M., de la Fortelle E., Raker V. A., Luhrmann R., Li J., Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Klingauf M., Stanek D., Neugebauer K. M. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol. Biol. Cell. 2006;17:4972–4981. doi: 10.1091/mbc.E06-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber J., Plessel G., Prehn S., Will C. L., Fabrizio P., Groning K., Lane W. S., Luhrmann R. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–941. [PMC free article] [PubMed] [Google Scholar]
- Lemm I., Girard C., Kuhn A. N., Watkins N. J., Schneider M., Bordonne R., Luhrmann R. Ongoing U snRNP Biogenesis Is Required for the Integrity of Cajal Bodies. Mol. Biol. Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I., Bledau A. S., Grishina I., Neugebauer K. M. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I., Sapra A. K., Neugebauer K. M. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Liu J. L., Gall J. G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. USA. 2007;104:11655–11659. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Makarova O. V., Makarov E. M., Liu S., Vornlocher H. P., Luhrmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6center dotU5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–1157. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Schneider S., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- Matera A. G., Shpargel K. B. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 2006;18:317–324. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Mayes A. E., Verdone L., Legrain P., Beggs J. D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Eggert C., Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Narayanan U., Achsel T., Luhrmann R., Matera A. G. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Narayanan U., Ospina J. K., Frey M. R., Hebert M. D., Matera A. G. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum. Mol. Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D., Tanackovic G., Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- Neugebauer K. M. On the importance of being co-transcriptional. J. Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Ohno M., Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- Patterson G. H., Lippincott-Schwartz J. Selective photolabeling of proteins using photoactivatable GFP. Methods. 2004;32:445–450. doi: 10.1016/j.ymeth.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- Raghunathan P. L., Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- Raker V. A., Plessel G., Luhrmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Schaffert N., Hossbach M., Heintzmann R., Achsel T., Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K. B., Matera A. G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K. B., Ospina J. K., Tucker K. E., Matera A. G., Hebert M. D. Control of Cajal body number is mediated by the coilin C-terminus. J. Cell Sci. 2003;116:303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J. Cell Sci. 2007;120:1540–1550. doi: 10.1242/jcs.001529. [DOI] [PubMed] [Google Scholar]
- Sleeman J. E., Ajuh P., Lamond A. I. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- Sleeman J. E., Lamond A. I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Staley J. P., Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stanek D., Neugebauer K. M. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek D., Neugebauer K. M. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- Stanek D., Rader S. D., Klingauf M., Neugebauer K. M. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Aronova A., Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M. P., Terns R. M. Macromolecular complexes: SMN–the master assembler. Curr. Biol. 2001;11:R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- Terskikh A., et al. “Fluorescent timer”: protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- Tsai R. T., Fu R. H., Yeh F. L., Tseng C. K., Lin Y. C., Huang Y. H., Cheng S. C. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. T., Tseng C. K., Lee P. J., Chen H. C., Fu R. H., Chang K. J., Yeh F. L., Cheng S. C. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K. T., Kolev N. G., Conrad N. K., Fok V., Steitz J. A. The ever-growing world of small nuclear ribonucleoproteins. In: Gesteland R. F., Cech T. R., Atkins J. F., editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 327–368. [Google Scholar]
- Verdone L., Galardi S., Page D., Beggs J. D. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 2004;14:1487–1491. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Wang C., Meier U. T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Lei Y. P., Zhou Y. L., Okamoto C. T., Snead M. L., Paine M. L. Structural organization and cellular localization of tuftelin-interacting protein 11 (TFIP11) Cell Mol. Life Sci. 2005;62:1038–1046. doi: 10.1007/s00018-005-4547-z. [DOI] [PubMed] [Google Scholar]
- Will C. L., Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Will C. L., Luhrmann R. Spliceosome structure and function. In: Gesteland R. F., Cech T. R., Atkins J. F., editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 369–400. [Google Scholar]
- Yu Y. T., Sharl E. C., Smith C. M., Steitz J. A. The growing world of small nuclear ribonucleoproteins. In: Gesteland C., Atkins, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 487–524. [Google Scholar]
- Zhang Y., Muyrers J. P., Testa G., Stewart A. F. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.