Abstract
The AAA+ ATPase VPS4 plays an essential role in multivesicular body biogenesis and is thought to act by disassembling ESCRT-III complexes. VPS4 oligomerization and ATPase activity are promoted by binding to LIP5. LIP5 also binds to the ESCRT-III like protein CHMP5/hVps60, but how this affects its function remains unclear. Here we confirm that LIP5 binds tightly to CHMP5, but also find that it binds well to additional ESCRT-III proteins including CHMP1B, CHMP2A/hVps2–1, and CHMP3/hVps24 but not CHMP4A/hSnf7–1 or CHMP6/hVps20. LIP5 binds to a different region within CHMP5 than within the other ESCRT-III proteins. In CHMP1B and CHMP2A, its binding site encompasses sequences at the proteins' extreme C-termini that overlap with “MIT interacting motifs” (MIMs) known to bind to VPS4. We find unexpected evidence of a second conserved binding site for VPS4 in CHMP2A and CHMP1B, suggesting that LIP5 and VPS4 may bind simultaneously to these proteins despite the overlap in their primary binding sites. Finally, LIP5 binds preferentially to soluble CHMP5 but instead to polymerized CHMP2A, suggesting that the newly defined interactions between LIP5 and ESCRT-III proteins may be regulated by ESCRT-III conformation. These studies point to a role for direct binding between LIP5 and ESCRT-III proteins that is likely to complement LIP5's previously described ability to regulate VPS4 activity.
INTRODUCTION
Multivesicular bodies (MVBs) are a subset of late endosomes morphologically characterized by the presence of intralumenal vesicles (ILVs; Gruenberg, 2004; Piper and Katzmann, 2007). Signaling receptors destined for degradation as well as certain lysosomal proteins are sorted into ILVs en route to the lysosome (Katzmann et al., 2002). Protein machinery involved in MVB biogenesis was discovered in studies of protein sorting to the vacuole in Saccharomyces cerevisiae. Functional loss of what are termed the class E Vps (vacuolar protein sorting) proteins prevents delivery of cargo into the vacuole. Cargo accumulates instead on the limiting membrane of the vacuole and in an adjacent abnormal compartment referred to as the “class E compartment” (Raymond et al., 1992). Eighteen class E Vps proteins have been identified in yeast, and these proteins are highly conserved throughout evolution (Babst, 2005; Hurley and Emr, 2006; Saksena et al., 2007). Interestingly, several mammalian class E Vps proteins are also involved in viral budding and cytokinesis, demonstrating a conserved role in topologically similar membrane budding and fission reactions (Demirov and Freed, 2004; Morita and Sundquist, 2004; Bieniasz, 2006; Carlton and Martin-Serrano, 2007; Morita et al., 2007).
A majority of class E Vps proteins are components of four complexes that include Vps27/Hse1 (sometimes referred to as ESCRT-0), ESCRT-I, ESCRT-II, and ESCRT-III, where ESCRT is an acronym for endosomal sorting complex required for transport. These complexes are recruited (possibly sequentially) to endosomal membranes where they function in sorting cargo and generating ILVs. The AAA+ ATPase Vps4 is recruited by ESCRT-III to disassemble and recycle the ESCRT machinery (Hurley and Emr, 2006; Saksena et al., 2007; Williams and Urbe, 2007).
ESCRT-III components are small (200–250 amino acid) structurally related proteins. All have basic N-terminal and acidic C-terminal halves and are thought to share a common set of six α-helices (Muziol et al., 2006; Shim et al., 2007). There are six ESCRT-III–related proteins in yeast (Vps2, Vps24, Vps20, and Snf7, core members; and Did2/Vps46 and Vps60, proposed regulatory members), and these are extended to eleven proteins in humans (Kranz et al., 2001; Babst et al., 2002; Saksena et al., 2007). Mammalian ESCRT-III proteins are referred to either as orthologues of their yeast counterparts or as CHMPs (charged multivesicular body proteins). To standardize our discussion of the large group of mammalian ESCRT-III proteins, we will primarily use the CHMP nomenclature in this article.
Unlike ESCRT-I and -II that are stable heteropolymeric complexes, ESCRT-III proteins are monomers in the cytoplasm and only assemble into complex on the endosomal membrane (Babst et al., 2002). In current models, ESCRT-III proteins are maintained in a metastable “closed” conformation in the cytoplasm and “open” when they bind to the membrane and assemble into polymers (Hurley and Emr, 2006; Saksena et al., 2007; Williams and Urbe, 2007; Shim et al., 2007). These polymers may deform the membrane and participate in forming ILVs (Hanson et al., 2008). Previously we defined ∼40 amino acids at the extreme C-terminus of each core ESCRT-III protein as an autoregulatory domain that controls transition between these states (Shim et al., 2007). These 40 amino acids include a short C-terminal α-helix and a linker that connects it to the rest of the protein.
ESCRT-III does not spontaneously disassemble, but instead requires energy input from the AAA+ (ATPases associated with a variety of cellular activities) protein Vps4 (Babst et al., 1998), of which there are two isoforms in mammalian cells, VPS4A and VPS4B/SKD1. We will use VPS4 to refer generically to the different forms of this enzyme. VPS4 has recently been shown to bind via its N-terminal microtubule interacting and trafficking (MIT) domain to a short motif present in a subset of ESCRT-III proteins, including CHMP1 (Did2 in yeast), CHMP2 (Vps2 in yeast), and CHMP3 (Vps24 in yeast; Obita et al., 2007; Stuchell-Brereton et al., 2007). This VPS4 binding motif is in a short C-terminal α-helix and is referred to as the MIT domain–interacting motif (MIM; Obita et al., 2007). The C-termini of the remaining ESCRT-III proteins (CHMP4 [Snf7 in yeast], CHMP5 [Vps60 in yeast], and CHMP6 [Vps20 in yeast]) do not contain the conserved MIM despite the fact that some of them have previously been shown to bind to VPS4 (von Schwedler et al., 2003; Yeo et al., 2003; Xiao et al., 2007). How VPS4 interacts with these proteins remains to be determined.
Although VPS4 activity is essential for MVB biogenesis, little is known about how it works. The AAA+ domain of VPS4 is similar to other AAA+ domains with the exception of an inserted β-sheet motif (referred to as the β domain) and a C-terminal α-helix (Scott et al., 2005a; Xiao et al., 2007). Like other AAA+ ATPases, VPS4 is thought to function as an oligomeric ring. VPS4 is primarily a monomer or dimer in the cytoplasm, and its assembly into a ring is enhanced by interaction of its β domain with the cofactor LIP5 (Vta1 in yeast; Scott et al., 2005a; Azmi et al., 2006; Vajjhala et al., 2006; Hartmann et al., 2008; Yu et al., 2008).
LIP5/Vta1 is a ∼300 amino acid long highly charged protein. Deletion of Vta1 in yeast leads to defects in cargo sorting and vacuolar morphology (Shiflett et al., 2004) and knockdown of LIP5 in mammalian cells significantly impairs receptor down-regulation and viral budding (Ward et al., 2005). A conserved domain at the C-terminus of LIP5 (the “VSL (Vta1/SBP1/LIP5) domain”) mediates LIP5 dimerization and interaction with Vps4 (Azmi et al., 2006).
In addition to binding to VPS4, LIP5/Vta1 has been found to interact with CHMP5/Vps60, one of the proposed regulatory ESCRT-III proteins (Shiflett et al., 2004; Ward et al., 2005; Azmi et al., 2006; Rue et al., 2007). This interaction is robust and has been documented in many systems. Less well explored connections between Vta1 and a few other ESCRT-III–related proteins have been reported, primarily in yeast. In particular, Vta1 binds to Did2/Vps46 (yeast ortholog of CHMP1; Lottridge et al., 2006) and the name Vta1 (Vps twenty [Vps20]–associated 1) was originally derived from a connection between Vps20 and Vta1, although this interaction has not been reproduced (Yeo et al., 2003; Azmi et al., 2006).
In the present study, we directly examine the ability of LIP5 to bind each of the six classes of ESCRT-III–related proteins in order to determine whether ternary interactions between LIP5, VPS4, and ESCRT-III might play a role in ESCRT-III disassembly. We confirm that LIP5 binds to CHMP5, but also find that it binds to CHMP1B, CHMP2A, and CHMP3 but not to CHMP4A or CHMP6. Mapping the binding sites reveals that LIP5 binds to the extreme C-terminal region of CHMP1B and CHMP2A and instead to an internal sequence in CHMP5. Complexes of LIP5 with CHMP5 are preferentially soluble, whereas those between LIP5 and CHMP2A are polymeric and insoluble. The C-terminal binding site for LIP5 in CHMP1B and CHMP2A overlaps with the previously defined “MIT interacting motif” or MIM responsible for recruiting Vps4. Surprisingly, we find evidence of a second binding site for VPS4 within these ESCRT-III proteins that may allow them to simultaneously interact with VPS4 and LIP5. These studies suggest that LIP5 is deeply intertwined with ESCRT-III and VPS4 in the pathway leading to multivesicular body formation.
MATERIALS AND METHODS
Plasmids
The following ESCRT-III and VPS4 constructs have been previously described: pGEX4T-1 CHMP4A residues 1-222; pHO4d VPS4B(E235Q)-His6/myc; pEGFP C1 VPS4B(E235Q); pcDNA3.1 FLAG-CHMP4A 1-222, 1-181, 1-147; and pGEX4T-1-CHMP2A 1-222, 1-180, and 1-144 (Lin et al., 2005; Shim et al., 2007). ESCRT-III and VPS4 constructs prepared for this study include pGEX4T-1-CHMP6 1-201; pGEX4T-1-CHMP3 1-222; pGEX4T-1-CHMP2A 1-219, 1-216, 1-203, and 1-193; pGEX4T-1-CHMP1B 1-199, 1-181, 106-199, 106-181, and 169-199; pGEX4T-1-CHMP5 1-219, 121-149, 121-158, 121-175, 121-219, 149-175, and 149-183; pcDNA3.1-FLAG-CHMP2A 1-219, 1-206, and 1-193; pcDNA3.1-FLAG-CHMP1B 1-199, 1-181, 1-168, and 1-136; pcDNA3.1-FLAG-CHMP5 1-219; pcDNA4TO-CHMP5 1-219 His6myc; pET28a-VPS4A MIT domain (1-75); and pGEX4T-1-VPS4B(E235Q) and pGEX4T-1-VPS4B(E235Q, ΔGAI deletion of 390-396). cDNAs used to create these constructs were either from the Mammalian Genome Collection (human CHMP1B, CHMP2B, and CHMP5; IMAGE ID: 6165059, 3460712, and 4094210, respectively) or previously described (CHMP2A, CHMP3, CHMP4A, and CHMP6; Lin et al., 2005; Shim et al., 2007). For insertion into pGEX4T-1, pcDNA3.1-FLAG, pcDNA4/TO-His6myc or pET28a, BamHI and XhoI sites were added to the fragments as they were amplified by PCR. QuickChange (Stratagene, La Jolla, CA) site-directed mutagenesis was used to introduce point mutations into CHMP2A and CHMP1B as indicated in the text.
pEGFP C1-LIP5 was a kind gift from Dr. Jerry Kaplan (University of Utah, Salt Lake City, UT). For bacterial expression of His6-LIP5, PCR amplified cDNA was inserted into pET28a between NdeI and XhoI sites. GFP-LIP5 ΔN contains residues 76-307 in pEGFP C1 between BglII and HindIII sites. All constructs were sequenced using ABI big dye reagents at the Nucleic Acid Chemistry Laboratory (Washington University, St. Louis, MO).
Protein Expression and Purification
BL21(DE3) Escherichia coli transformed with the indicated constructs were grown at 37°C to a 600 nm optical density of ∼1, transferred to room temperature, and brought to 0.4 μM IPTG for 3 h to induce expression. Pelleted bacteria were resuspended in buffer A (20 mM Tris, 250 mM NaCl, 5% glycerol, 1 mM DTT, 1 mM PMSF, pH 7.4) and lysed by sonication. Bacterial lysates were centrifuged at 66,000 × g for 20 min. Clarified lysates were bound to glutathione Sepharose (Amersham Biosciences, Piscataway, NJ) or Ni2+-NTA agarose (Qiagen, Valencia, CA) for 1 h at 4°C. Unbound material was removed by washes in buffer A, and proteins were eluted in buffer A containing either glutathione (50 mM) or imidazole (160 mM). Purified protein was quantitated using Bradford reagent with BSA as a standard. Proteins were snap-frozen in liquid nitrogen and stored at −80°C.
GST Pulldown Assays
Where indicated, GST proteins immobilized on beads were combined with clarified bacterial lysate containing His6-LIP5. This lysate was prepared from BL21(DE3) E. coli expressing pET28a-LIP5 grown as above. Bacteria were lysed in buffer B (30 mM HEPES, 120 mM NaCl, 5% glycerol, 1 mM PMSF, pH 7.4), brought to 0.5% Triton X-100, and centrifuged at 66,000 × g for 20 min. Clarified bacterial lysate was incubated with immobilized glutathione S-transferase (GST) fusion proteins for 1 h at 4°C in buffer B. Beads were washed, and bound material was analyzed by SDS-PAGE and staining with Coomassie brilliant blue. For the experiment shown in Figure 6B, GST proteins were combined with mammalian cell lysate containing green fluorescent protein (GFP)-LIP5. This was prepared from HEK293T cells transfected with pEGFP C1-LIP5 solubilized in buffer B containing 0.5% Triton X-100 and centrifuged at 15,000 × g for 15 min. After the binding reaction, bound and unbound material was detected by SDS-PAGE and immunoblotting with rabbit anti-GFP antibody (Lin et al., 2005; 1:2500).
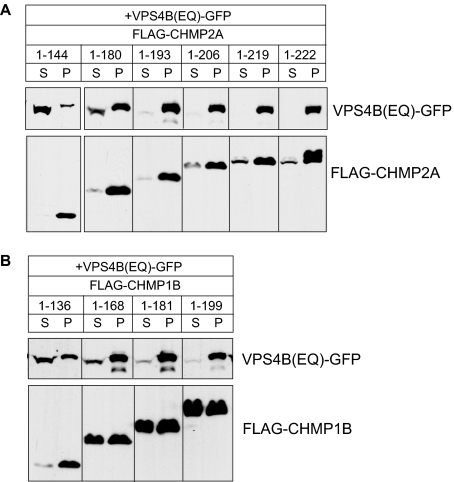
Figure 6.
Effects of C-terminal deletions suggest existence of secondary binding site for VPS4B in CHMP2A and CHMP1B. (A) HEK293T cells cotransfected with VPS4B(E235Q)-GFP and indicated FLAG-CHMP2A constructs were solubilized in 1% Triton X-100 and centrifuged. The resulting distribution of VPS4B(E235Q) and CHMP2A between supernatant (S) and pellet (P) was visualized by immunoblotting. (B) Same experiment but with FLAG-CHMP1B constructs.
To assess competition between LIP5 and the VPS4A MIT domain, GST proteins immobilized on glutathione Sepharose were blocked in buffer C (20 mm Tris, pH 7.8, 100 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 5 mM DTT, 5% glycerol) also containing 1% casein for 1 h. Beads were incubated with His6-LIP5 (0.4–12.8 μM) with or without 300 μM His6-VPS4A MIT domain (CHMP2A) and with or without 500 μM MIT domain (CHMP1B) for 1 h at 4°C. Control experiments were carried out with or without 300 μM ribonuclease A. Beads were then washed three times in buffer C. Bound proteins were analyzed by SDS-PAGE. Gels were stained with colloidal Coomassie (G Biosciences, St. Louis, MO) and visualized using an Odyssey Infrared imager (LiCor Biosciences, Lincoln, NE). Bands were quantified with Odyssey 2.1 software and were within the experimentally determined linear range of detection.
Solid-Phase LIP5-binding Assay
Microtiter plate wells containing immobilized antibody against GST (Pierce, Rockford, IL, or EMD Biosciences, Gibbstown, NM) were incubated with 100 μl of the indicated GST-ESCRT-III or GST-VPS4B protein at 10 μg/ml for 1 h in buffer D (20 mm Tris, pH 7.4, 150 mM NaCl, pH7.4; estimated to be ∼60–100 times the binding capacity of the plate as per manufacturer data). For VPS4 binding assays, buffer D contained 100 mM KOAc in place of NaCl, 5 mM MgCl2, and 1 mM ATP. Plates were washed three times in buffer D with 0.05% Tween-20 and blocked for 1 h in buffer containing 1% casein (Pierce plates) or 1% casein + 0.5% BSA (EMD plates). Plates were then washed, and 100 μl of His6-LIP5 at the indicated concentration in buffer D was added and incubated for 1 h. Plates were again washed with buffer D, and 100 μl of a 1:2000 dilution of NTA-horseradish peroxidase (HRP; Qiagen) was bound for 1 h. After washing, 100 μl of TMB-Ultra (Pierce) was added to wells for ∼3 min. Absorbance was read at 652 nm on a Bio-Tek plate reader. Background signal arising from nonspecifically bound His6-LIP5 (measured in parallel wells containing no GST protein or GST alone) was subtracted from each value. The background was concentration dependent, and in a typical assay ranged from ABS of 0.1 for 4 nM His6-LIP5-0.23 for 3 μM His6-LIP5. Corrected absorbance data were analyzed using Prism (GraphPad, San Diego CA) to define an EC50 by nonlinear regression analysis using the formula ABS = (ABSmax * X)/(EC50 + X).
Tissue Culture and Transfection
HEK293T cells were grown in DMEM (Invitrogen-BRL, Gaithersburg, MD) containing 5% fetal bovine serum (Invitrogen-BRL), 5% supplemented calf serum (Hyclone Laboratories, Logan, UT) and 2 mM glutamine. Cells were transfected with the indicated plasmid(s) using Lipofectamine 2000 (Invitrogen, Carlsbad CA) following the manufacturer's instructions and then used for experiments 18–24 h after transfection.
Sedimentation Assay
Sedimentation assays were performed as described previously (Shim et al., 2007). Briefly, transiently transfected HEK293T cells in 6-cm dishes were solubilized in 1% Triton X-100 and centrifuged at 10,000 × g for 15 min at 4°C. Pellets were resuspended to the same volume as supernatant in lysis buffer, and equal volumes of the fractions were analyzed by immunoblotting with rabbit anti-FLAG antibody (Sigma, St. Louis MO, 1:2500) or rabbit anti-GFP.
Immunoprecipitation
Transiently transfected HEK293T cells in 6-cm dishes were solubilized in 500 μl buffer E (0.5% Triton X-100, 30 mM HEPES, 120 mM NaCl, 1 mM PMSF and complete protease inhibitor; Roche Diagnostics, Alameda, CA). Insoluble material was removed by centrifugation at 15,000 × g for 15 min. Soluble lysate was incubated with 20 μl of protein A Sepharose CL-4B (GE Healthcare, Piscataway, NJ) for 25 min to remove nonspecifically interacting material, then with 6 μl of rabbit anti-GFP antibody for 2 h, and finally with 30 μl of protein A Sepharose for 1 h, all at 4°C. Bound protein and lysate were analyzed by immunoblotting using mouse monoclonal anti-myc (Developmental Studies Hybridoma Bank, Iowa City, IA, 1:2500) and rabbit anti-GFP.
RESULTS
LIP5 Binds Tightly to Several ESCRT-III Proteins in Addition to CHMP5
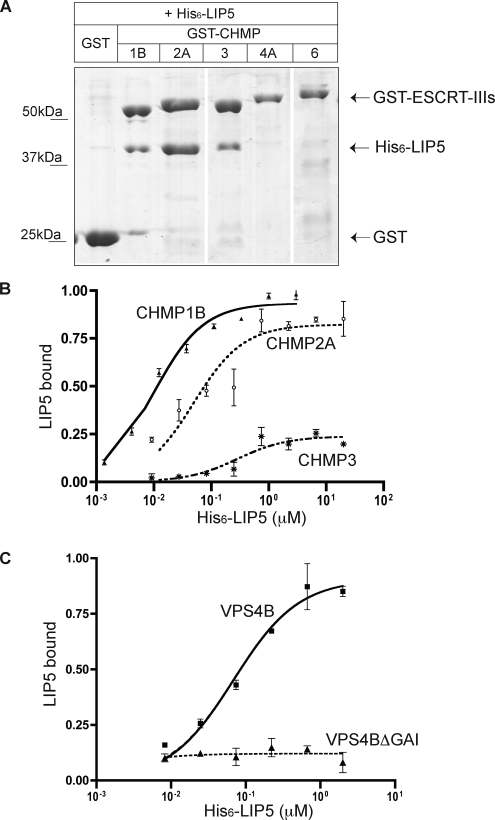
The AAA+ ATPase VPS4 plays a key role in MVB biogenesis (Babst et al., 1997, 1998; Babst et al., ; Bishop and Woodman, 2000; Fujita et al., 2003), but precisely what it does and how this is regulated remains unclear. To gain new insight into this reaction, we explored connections between ESCRT-III and a known cofactor of VPS4, LIP5. A previous study demonstrated that LIP5 bound efficiently but apparently uniquely to the ESCRT-III like protein CHMP5 (Ward et al., 2005); this interaction was also found in a reciprocal immunoprecipitation of proteins that bind to CHMP5 (Ma et al., 2007). In yeast, Vta1p, the LIP5 orthologue, binds both to Vps60p (CHMP5 ortholog; Shiflett et al., 2004; Azmi et al., 2006; Rue et al., 2007) and to Did2p/Vps46p (CHMP1 ortholog; Lottridge et al., 2006). On this basis, we asked whether LIP5 also interacts with other human ESCRT-III proteins. We expressed ESCRT-III proteins representing each of the ESCRT-III subfamilies as GST-fusion proteins in E. coli and carried out in vitro binding experiments. In a survey GST pulldown, we found that CHMP1B, CHMP2A/hVps2-1, and CHMP3/hVps24 all bound to His6-LIP5, whereas CHMP4A/hSnf7–1 and CHMP6/hVps20 did not (Figure 1A).
Figure 1.
LIP5 binds to ESCRT-III proteins. (A) Interaction of LIP5 with a subset of ESCRT-III proteins. GST and GST-ESCRT-III proteins immobilized on glutathione-Sepharose beads were incubated with E. coli lysate containing His6-LIP5. Bound material was separated on a SDS-PAGE gel and stained with Coomassie blue. Where necessary, lanes were rearranged as indicated by white lines. Immunoblotting with an anti-His6 antibody confirmed that no His6-LIP5 bound to GST-CHMP4A or GST-CHMP6 (not shown). (B) Solid phase assay of LIP5 binding to GST-CHMP1B, 2A, and 3. His6-LIP5 bound to immobilized GST-CHMP proteins was detected with NTA-HRP and TMB colorimetric substrate. EC50 values determined by nonlinear regression analysis ranged from 10 to 20 nM for CHMP1B (▴, solid line), from 49–60 nM for CHMP2A (○, dotted line), and from 0.3–1 μM for CHMP3 (*, alternating dashed line) in several independent experiments. Error bars, the SD from one experiment run in duplicate. Absorbance data were normalized to the Bmax for CHMP1B. (C) Binding of His6-LIP5 to GST-VPS4B(E235Q) and GST-VPS4B(E235Q, ΔGAI) β domain mutant. EC50 for VPS4B (E235Q) (■, solid line) was 60 nM. LIP5 binding to VPS4B(E235Q, ΔGAI) did not change as a function of LIP5 added (▴, dotted line). Error bars, the SD from one experiment run in duplicate, and the absorbance data were normalized to the Bmax for VPS4B(E235Q).
To quantitatively compare binding of LIP5 to these different proteins, we immobilized each GST-CHMP fusion protein on microtiter plates using anti-GST antibodies and measured binding of His6-LIP5 across a range of concentrations using Ni2+-NTA conjugated to HRP and a colored substrate to detect bound His6-LIP5. EC50 values for LIP5 binding ranged from 10 to 20 nM for binding to GST-CHMP1B to 0.3–1 μM for binding to GST-CHMP3 (Figure 1B). There was no binding above background to immobilized GST, GST-CHMP4A, or GST-CHMP6 (data not shown).
For comparison, we also quantitated the interaction of LIP5 with VPS4B. These two proteins have previously been shown to bind each other with an EC50 of 53 nM in surface plasmon resonance experiments (Fujita et al., 2004). We found that His6-LIP5 bound to immobilized GST-VPS4B(E235Q) with an EC50 of 60 nM (Figure 1C) and that this binding was abolished by a short deletion within the VPS4B β domain (Δ390–396) known to block interaction of yeast Vps4 and Vta1 (Vajjhala et al., 2006; Figure 1C). We conclude that LIP5 binds with submicromolar affinity to both a subset of ESCRT-III proteins and to VPS4B. As will be described below, we confirmed in parallel studies that LIP5 also binds to CHMP5 with comparable or even higher affinity (see Figure 8). The interaction of LIP5 with CHMP1B might have been anticipated based on earlier studies in yeast (Lottridge et al., 2006), but the association of LIP5 with the core ESCRT-III proteins CHMP2A and CHMP3 was unexpected and raises the possibility of a more intimate relationship between LIP5 and ESCRT-III than previously appreciated.
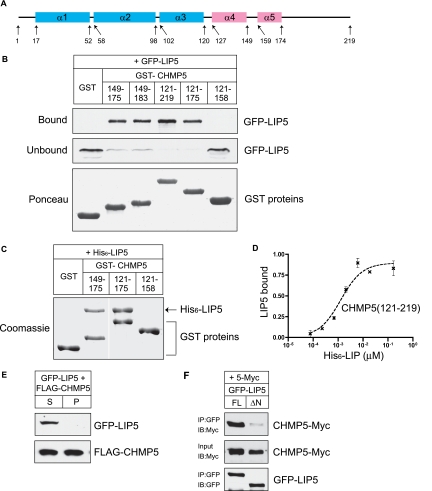
Figure 8.
Unique properties of CHMP5-LIP5 complex: LIP5 binds to CHMP5 α5 preferentially in the soluble fraction. (A) Predicted secondary structure of CHMP5. (B) GST and GST-CHMP5 proteins immobilized on beads were incubated with HEK293T cell lysate containing GFP-LIP5. Bound and unbound fractions were analyzed by immunoblotting with anti-GFP antibody. GST proteins were visualized by staining the immunoblot with Ponceau red. (C) GST and GST-CHMP5 proteins on beads were incubated with E. coli lysate containing His6-LIP5, and the bound material was analyzed by staining with Coomassie blue. Where necessary, lanes were rearranged as indicated by white lines. (D) Binding of His6-LIP5 to immobilized GST-CHMP5(121-219), detected, and analyzed as in Figure 1B. EC50 values ranged from 1 to 2 nM. Error bars, SD from one experiment performed in duplicate. (E) LIP5 does not cosediment with CHMP5. HEK293T cells cotransfected with GFP-LIP5 and FLAG-CHMP5 were solubilizated in 1% Triton X-100 and centrifuged. The resulting supernatant (S) and pellet (P) were analyzed by immunoblotting. (F) Coimmunoprecipitation of CHMP5-myc with LIP5-GFP from cotransfected HEK293T cells. LIP5-GFP or LIP5ΔN-GFP was immunoprecipitated from the soluble lysate of cotransfected cells. Bound proteins and lysate were analyzed by immunoblotting.
LIP5 Binding to CHMP2A and CHMP1B Is Mediated by C-Terminal Sequences
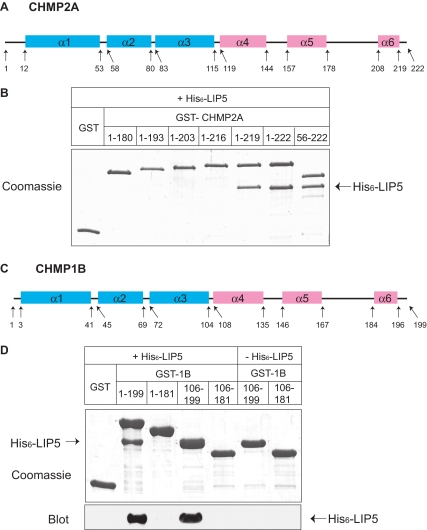
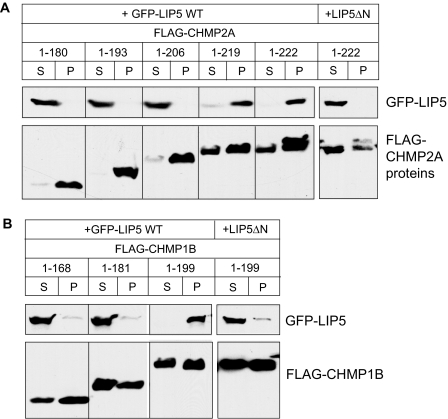
To understand how LIP5 binds to ESCRT-III proteins, we began by looking for its binding site in the core ESCRT-III protein CHMP2A. We examined interaction between LIP5 and a series of CHMP2A deletion proteins that lack one or more of the protein's predicted six α-helices (Figure 2A), as previously described (Shim et al., 2007). Because even the shortest deletion from the C-terminus (leaving an α1–α5 protein, residues 1-180) abolished binding, we generated a series of smaller deletions from the C-terminus to determine whether binding required α6 or sequences within the long linker between α5 and α6. GST-CHMP2A fusion proteins were purified from E. coli and combined with His6-LIP5 to assess their interaction (Figure 2B). Removing three amino acids from the C-terminus of CHMP2A (leaving residues 1-219) was not expected to significantly affect α6 and did not change binding of LIP5. On the other hand, removing six or more amino acids (thus perturbing or removing α6) abolished interaction of CHMP2A with LIP5. Deleting α1 from CHMP2A (leaving residues 56-222) did not perturb LIP5 binding, demonstrating that the interaction is independent of CHMP2A's N-terminus.
Figure 2.
C-terminal sequences in CHMP2A and CHMP1B are required for LIP5 binding. (A) Predicted CHMP2A secondary structure obtained using a neural network based algorithm (http://www.compbio.dundee.ac.uk/∼www-jpred/). Pink and blue boxes correspond to predicted α-helices with pI higher than 8 and lower than 6, respectively. (B) Effects of deleting N- and C-terminal sequences from CHMP2A on LIP5 binding. GST and GST-CHMP2A proteins with the indicated sequences immobilized on beads were incubated with E. coli lysate containing His6-LIP5. Bound material was analyzed by staining with Coomassie blue. (C) Predicted CHMP1B secondary structure. (D) Effects of deleting N- and C-terminal sequences from CHMP1B on LIP5 binding. GST and GST-CHMP1B proteins with the indicated sequences immobilized on beads were incubated with E. coli lysate containing His6-LIP5. Bound material was analyzed by staining with Coomassie blue (top panel) and by immunoblotting with an anti-His6 antibody (bottom panel). Immunoblotting was needed because His6-LIP5 migrates similarly to GST-CHMP1B(106-199).
In further experiments, we found the same requirement for extreme C-terminal sequences for interaction of CHMP1B with LIP5 (Figure 2C). Removing the predicted α6 from CHMP1B's C-terminus disrupted LIP5 binding, whereas deleting the N-terminal half of the protein had no effect (Figure 2D). These experiments demonstrate that sequences within α6 are needed for CHMP2A and CHMP1B to bind LIP5.
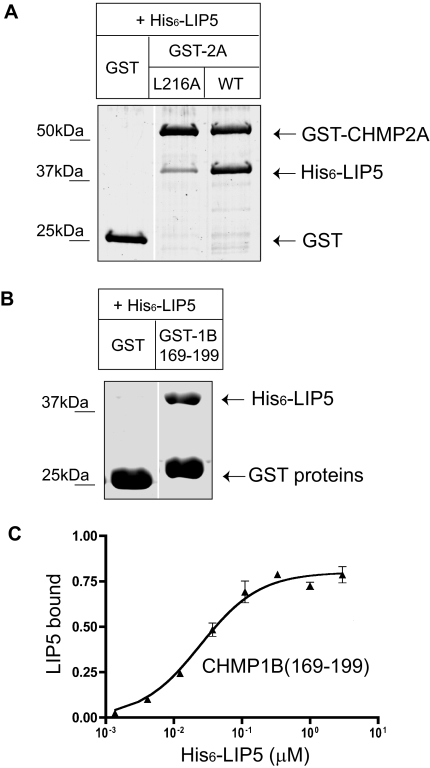
Because deleting sequences from ESCRT-III proteins significantly changes their conformation (Shim et al., 2007), there remained the possibility of deletions indirectly impairing binding to a site or sites located elsewhere in the protein. To rule this out, we carried out additional experiments. We started by changing a single conserved residue within α6 of CHMP2A from leucine to alanine (L216A). This mutation significantly decreased binding to LIP5 (Figure 3A). A comparable leucine residue in other ESCRT-III proteins has previously been shown to be important for binding of VPS4B to CHMP1B (Stuchell-Brereton et al., 2007) and binding of Alix to CHMP4 (von Schwedler et al., 2003), pointing to a likely common role for the surface of α6 in binding between ESCRT-III proteins and other factors.
Figure 3.
CHMP2A and CHMP1B α6 region is responsible for LIP5 binding. (A) Effect of CHMP2A L216A mutation on interaction with LIP5. GST and GST-CHMP2A proteins immobilized on beads were incubated with E. coli lysate containing His6-LIP5. Bound material was analyzed by staining with Coomassie blue. Where necessary, gel lanes were rearranged as shown by a white line. (B) Binding of His6-LIP5 to GST-CHMP1B(169-199). This CHMP1B fragment contains α6 and surrounding sequences but does not include α5. (C) Solid phase assay of His6-LIP5 binding to GST-CHMP1B(169-199) carried out as described in Figure 1. The EC50 of 25 nM is similar to that of His6-LIP5 for full-length CHMP1B. Absorbance data were normalized to the Bmax for full-length CHMP1B measured in parallel.
To ask whether the C-terminal region is by itself sufficient for interaction of these ESCRT-III proteins with LIP5, we expressed α6 and surrounding linker sequences from CHMP1B (169-199) as a GST fusion protein and asked if it could bind to LIP5. Indeed, LIP5 bound to this 31-amino acid fragment (Figure 3B) with an EC50 of 25 nM (Figure 3C), similar to what we observed above for full-length CHMP1B. These results argue that all of the determinants needed for LIP5 binding are encoded within the C-terminal regions of these ESCRT-III proteins.
Studies in Mammalian Cells Suggest That ESCRT-III Interaction with LIP5 May Be Regulated by ESCRT-III Assembly Status
As mentioned above, the high-affinity binding of LIP5 to the core ESCRT-III protein CHMP2A was entirely unexpected. Indeed, this finding is at first glance inconsistent with a published report in which endogenous LIP5 was not immunoisolated with overexpressed CHMP2A from transfected mammalian cells (Ward et al., 2005). We therefore examined interaction of LIP5 with CHMP2A in HEK293T cells transiently transfected with tagged versions of each protein. When we immunoprecipitated FLAG-CHMP2A from the solubilized lysate of doubly transfected cells, we also did not recover significant amounts of LIP5 (data not shown). However, we noticed that overexpressed FLAG-CHMP2A had a strong tendency to form large complexes or aggregates that were insoluble in Triton X-100 and therefore pelleted during preparation of the solubilized lysate. Although LIP5-GFP expressed alone is soluble, we found that when coexpressed with CHMP2A it associated with this insoluble material (Figure 4A). LIP5 remained soluble when coexpressed with CHMP2A fragments lacking their α6 region despite the fact that the CHMP2A proteins still sedimented. In addition, LIP5 only associated with pelleted CHMP2A when its N-terminus—previously shown in yeast to mediate interaction with the ESCRT-III–related protein Vps60—was intact.
Figure 4.
LIP5 associates preferentially with polymerized CHMP2A and CHMP1B in transfected mammalian cells. (A) Cosedimentation of LIP5 with CHMP2A. HEK293T cells cotransfected with GFP-LIP5 or GFP-LIP5ΔN, and the indicated FLAG-CHMP2A constructs were solubilized in 1% Triton X-100 and centrifuged. The distribution of CHMP2A and LIP5 in the resulting supernatant (S) and pellet (P) was visualized by immunoblotting. LIP5ΔN is equivalent to a deletion in Vta1 that impairs binding to Vps60 (Azmi et al., 2006). Experiments with LIP5ΔN were performed separately from those with full-length LIP5 and are therefore shown in a separate box. (B) Cosedimentation of LIP5 with CHMP1B. The same experiments performed with FLAG-CHMP1B constructs.
Parallel studies with cells transfected with CHMP1B and LIP5-GFP demonstrated that CHMP1B similarly recruited LIP5 to sedimentable complexes only when its α6 region was intact (Figure 4B). These results are consistent with our analysis of recombinant proteins above and confirm that LIP5 binds to extreme C-terminal sequences in CHMP2A and CHMP1B. Based on our earlier study of ESCRT-III homopolymers (Shim et al., 2007), the preferential association of LIP5 with pelleted CHMP2A and CHMP1B suggests that LIP5 may bind to these proteins in their “open” conformation and tend to stabilize this state.
LIP5 and VPS4 MIT Binding Sites Overlap in CHMP2A and CHMP1B
Interestingly, the extreme C-terminal regions of CHMP2A and CHMP1B were recently shown to contain a short α-helix that binds to VPS4 via a twelve residue sequence referred to as the MIT interaction motif (MIM; Obita et al., 2007; Stuchell-Brereton et al., 2007). The MIM helix largely coincides with the sequence we defined as α6 by secondary structure prediction (Shim et al., 2007). We found above (Figure 3A) that mutating a conserved hydrophobic residue in this helix reduced binding of LIP5 to CHMP2A. This impairment is in accordance with the reported 10-fold decrease in VPS4 binding when the equivalent change was made in CHMP1B (Stuchell-Brereton et al., 2007). VPS4 and LIP5 may therefore share elements of a common binding site in these ESCRT-III proteins.
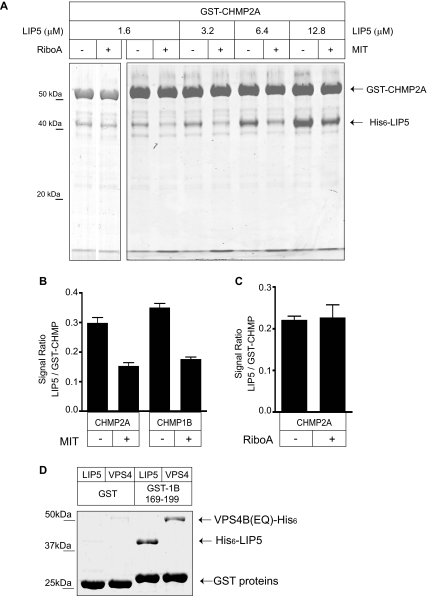
To determine if this is the case, we asked whether the VPS4 MIT domain competes with LIP5 for binding to CHMP2A and CHMP1B. We expressed and purified the His6-tagged MIT domain of VPS4A from E. coli and added it to GST pulldown experiments (Figure 5A). We found that high concentrations of the MIT domain reduced but did not abolish binding between LIP5 and both CHMP2A and CHMP1B (Figure 5B). Parallel control experiments demonstrated that adding 300 μM ribonuclease A had no effect on LIP5 binding (Figure 5, A and C).
Figure 5.
Binding sites for VPS4 and LIP5 in CHMP2A and CHMP1B overlap. (A) VPS4A MIT domain reduces LIP5 binding to GST-CHMP2A. GST-CHMP2A was incubated with His6-LIP5 alone or together with 300 μM His6-VPS4A MIT domain or ribonuclease A as indicated. Bound LIP5 was visualized by staining with colloidal Coomassie blue and quantified by infrared fluorescence scanning. The bound MIT domain can be seen as an increased intensity in the dye front. (B) Quantitation of the effect of MIT domain on binding of 3.2 μM LIP5 to CHMP2A or (in parallel experiments) CHMP1B. (C) Quantitation of lack of effect of the same concentration (300 μM) of ribonuclease A on binding of 1.6 μM LIP5 to GST-CHMP2A. (D) LIP5 and VPS4B(E235Q) bind similarly to GST-CHMP1B(169-199). Material retained on GST or GST-CHMP1B(169-199) after incubation with the indicated 5 μM protein is shown on a gel stained with Coomassie Blue.
Given this apparent overlap in binding sites, the question of how the affinity of these ESCRT-III proteins for LIP5 compares with that for VPS4 becomes important. We were unable to quantitate VPS4B (full-length or MIT domain) binding to immobilized GST-ESCRT-III proteins because of high background in the microtiter plates. In recent studies of VPS4 MIT domain binding to ESCRT-III MIM fragments the observed EC50 values were significantly higher (i.e., lower affinity) than those we measured between LIP5 and full-length ESCRT-III proteins (Obita et al., 2007; Stuchell-Brereton et al., 2007). For a first assessment of the relative ability of VPS4 and LIP5 to bind to their shared binding site, we compared binding of full-length proteins to GST-CHMP1B(169–199) (Figure 5D). After incubating this α6 fragment with 5 μM His6-VPS4B(E235Q) or His6-LIP5, we recovered similar amounts of bound protein, suggesting that full-length VPS4B and LIP5 may have similar affinity for the MIM-containing ESCRT-III proteins.
A Second Binding Site for VPS4 in ESCRT-III Proteins
If VPS4 and LIP5 have overlapping binding sites in this subset of ESCRT-III proteins, how do they function together? One possibility is that the C-terminal α6 sequences, preferentially exposed when the proteins assemble into ESCRT-III complex, cooperate to bring VPS4 and LIP5 together. Another, not mutually exclusive, possibility is that the interaction between α6 sequences and these proteins is only one step in the reaction leading to ESCRT-III disassembly, with additional steps and interactions required. On the basis of what is known about other AAA+ proteins, we wondered whether there might be a second, yet unidentified, binding site for VPS4 within ESCRT-III proteins. If so, this might also enable simultaneous interaction of ESCRT-III with VPS4 and LIP5.
To search for such a binding site, we took advantage of the fact that the detergent insoluble polymers formed when CHMP proteins are overexpressed in mammalian cells (see Figure 4) create a high avidity matrix for their binding partners. We carried out sedimentation assays using HEK293T cells coexpressing CHMP2A deletion mutants and VPS4B(E235Q). Similar to what we saw with LIP5, coexpressed VPS4B (E235Q) sedimented with full-length CHMP2A (Figure 6A). Interestingly, however, small C-terminal deletions (including the MIMs) that eliminated interaction between CHMP2A and LIP5 did not affect association of VPS4B(E235Q) with CHMP2A. On the other hand, further deleting α5 and surrounding sequences abolished the CHMP2A dependent recruitment of VPS4B(E235Q). Note that the basal association of VPS4B(E235Q) with the insoluble fraction is somewhat higher than that of LIP5, presumably because VPS4B(E235Q) traps and binds to polymerized endogenous ESCRT proteins (Lin et al., 2005). Similar results were obtained with CHMP1B and VPS4B(E235Q) (Figure 6B), suggesting that there might be a secondary binding site for VPS4 around or within the predicted α5 helix of both proteins. The orthogonal and exposed position of α5 in the currently available crystal structure of CHMP3 (Muziol et al., 2006) suggests that this helix may move as a function of ESCRT-III conformation, making it an attractive candidate for engaging VPS4. Our initial attempts to define this potential binding site more precisely using purified proteins in GST pulldown experiments failed, both because the affinity of this interaction appears to be low and because the nonspecific binding of VPS4B to truncated ESCRT-III proteins was variable and relatively high.
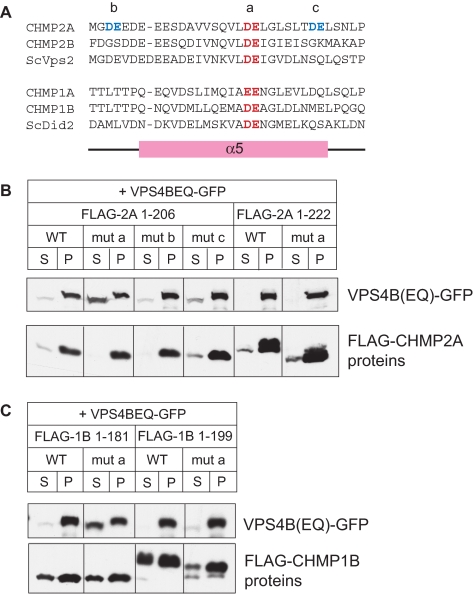
To gain additional insight into the nature of this binding site, we instead turned to site-directed mutagenesis in our cell-based sedimentation assay. We noted that the region within and around α5 is highly acidic in all ESCRT-III proteins and contains a glutamic acid that is the only residue conserved among all ESCRT-III proteins (Muziol et al., 2006; Figure 7A). To determine if this region is involved in the secondary association of VPS4B with ESCRT-III proteins, we replaced pairs of acidic residues within and around α5 in CHMP2A with alanines. One pair included the conserved glutamic acid (mut a), and the others (mut b and mut c) were nearby but less conserved pairs. We also replaced the conserved pair of acidic residues (mut a) in CHMP1B. We made these mutations in both full-length and α6-deleted proteins, with the prediction that association of VPS4B with the full-length proteins would be mediated largely by their MIM and would therefore be independent of a secondary binding site, whereas association of VPS4B with the truncated (α6-deleted) proteins would instead be fully dependent on the secondary binding site. Strikingly, we found that mut a eliminated recruitment of VPS4B(E235Q) to α6-deleted but not full-length CHMP2A (Figure 7B). Mut b and mut c had no effect. The fact that mut a did not affect recruitment of VPS4B to polymers of full-length CHMP2A confirms that the alanine replacements did not induce significant protein misfolding. In support of these results, we found the same effect of mut a replacements on the recruitment of VPS4B (E235Q) to CHMP1B (Figure 7C). We conclude that conserved acidic residues at the center of α5 are an important component of the secondary VPS4 binding site. Because these experiments were carried out in cells that highly overexpress VPS4B and the ESCRT-III protein in question, we consider it unlikely but cannot exclude that an intermediate protein such as Ist1 (Dimaano et al., 2008) mediates this secondary interaction between VPS4 and the acidic α5 residues in ESCRT-III proteins.
Figure 7.
Conserved acidic residues in α5 are part of secondary VPS4-binding site. (A) Sequences of predicted α5 and surrounding sequences in CHMP2 and CHMP1 proteins. Highly conserved acidic residues near the center of the helix are colored red; less conserved pairs of acidic residues in CHMP2A are colored blue. Pairs of alanine replacements in CHMP2A studied below are designated mut a, mut b, and mut c as indicated. The conserved central pair of acidic residues was also mutated in CHMP1B and designated as mut a. (B) Effect of double alanine mutants on cosedimentation of VPS4B(E235Q) with full-length (1-222) or α6-deleted (1-206) CHMP2A in cotransfected HEK293 cells. Cells were solubilized in 1% Triton X-100 and centrifuged. The resulting distribution of VPS4B(E235Q) and CHMP2A between supernatant (S) and pellet (P) was visualized by immunoblotting. (C) Effect of mut a on cosedimentation of VPS4B(E235Q) with full-length (1-199) or α6-deleted α1–α5 (1-181) CHMP1B.
LIP5 Complex with CHMP5 Is Unique
Finally, we wondered how the previously described interaction between LIP5 and CHMP5 (Ward et al., 2005) compares to its binding to the MIM-containing ESCRT-III proteins studied above. The fact that LIP5 and CHMP5 have been reciprocally identified as binding partners in unbiased pulldowns from cultured mammalian cell cytosol (Ward et al., 2005; Ma et al., 2007), whereas none of the other complexes have been detected, suggests that there could be important differences. CHMP5 does not have a MIM, and in fact its predicted secondary structure does not include strong indication of a C-terminal helix comparable to α6 in the other ESCRT-III proteins (Figure 8A). To characterize the interaction between LIP5 and CHMP5, we began by performing in vitro binding experiments. Our initial attempts to use full-length GST-CHMP5 purified from E. coli were unsuccessful because the protein was not well behaved, forming aggregates that did not consistently bind to LIP5 (data not shown). The C-terminal half of the protein (GST-CHMP5 121-219), however, was readily soluble and reproducibly bound to LIP5 (Figure 8, B and C). In solid phase-binding assays, we found that LIP5 bound to this C-terminal fragment with an EC50 of 1–2 nM (Figure 8D), confirming an even tighter interaction between CHMP5 and LIP5 than between LIP5 and the other ESCRT-III proteins.
To define the structural requirements for interaction of CHMP5 with LIP5, we made a series of GST-CHMP5 fragments and tested their ability to bind to GFP-LIP5 present in a transfected cell extract (Figure 8B). Deleting sequences C-terminal to the predicted α5 helix (GST-CHMP5 121-175) had little effect on binding, whereas removing the predicted α5 region (GST CHMP5 121-158 and 121-149) abolished LIP5 binding, suggesting an important role for α5. Indeed, a 27-residue fragment containing only linker sequences and α5 (GST CHMP5 149–175) bound to LIP5 as efficiently as the longer fragments. Sequences within and around α5 are thus both necessary and sufficient for binding of LIP5. To confirm this result with purified proteins, we examined binding of His6-LIP5 expressed in E. coli to the CHMP5 α4 + α5 or α5 fragments. As was the case with GFP-LIP5 from mammalian cell extracts, both fragments were able to bind efficiently to LIP5, confirming that sequences within and around α5 are responsible for high-affinity binding between these two proteins (Figure 8C).
To further compare the interaction of LIP5 with CHMP5 to that with CHMP1B or 2A, we again carried out a sedimentation assay in cotransfected HEK293T cells. As seen previously for CHMPs 1B and 2A, a substantial portion of overexpressed CHMP5 formed complexes or aggregates and ended up in the pellet. Interestingly, however, this insoluble material did not recruit LIP5, which was exclusively found in the soluble fraction (Figure 8E). To confirm that soluble CHMP5 actually interacts with LIP5 in these cells, we immunoprecipitated GFP-LIP5 and found that, as expected, FLAG-CHMP5 was efficiently recovered (Figure 8F). Deleting the N-terminal 75 residues from LIP5 (LIP5ΔN) abolished this binding as it has been reported to do with the comparable proteins in yeast (Azmi et al., 2006). Together, our data suggest that LIP5 interacts with CHMP5 in a distinct manner that may or may not be compatible with ESCRT-III polymer assembly.
DISCUSSION
LIP5 (Vta1 in yeast) emerged in recent years as a protein involved in late stages of MVB formation and viral budding. It participates in these events at least in part by binding via its C-terminal “VSL domain” to the AAA ATPase VPS4 to enhance oligomerization and ATPase activity (Scott et al., 2005a; Azmi et al., 2006; Lottridge et al., 2006; Vajjhala et al., 2006). At the same time, LIP5 also binds via its N-terminus to the ESCRT-III–related protein CHMP5 (Vps60 in yeast; Shiflett et al., 2004; Ward et al., 2005; Rue et al., 2007), and in yeast Vta1 has been shown to bind to Vps46, another ESCRT-III–related protein (Lottridge et al., 2006). Whether, and if so how, these interactions affect VPS4 activity toward ESCRT-III complexes has been unclear. Here, we define new and unexpected relationships among these proteins, including a high-affinity connection between LIP5 and the C-termini of a subset of ESCRT-III proteins and a second binding site for VPS4 further inside these proteins. In addition, comparison of LIP5's interaction with CHMP5 and the other ESCRT-III proteins revealed important differences in where and how the proteins bind to each other, suggesting the possibility of a unique role for CHMP5. These findings lead us to propose that there are at least two ways in which LIP5 is involved in ESCRT-III disassembly.
LIP5 has been clearly shown to be a positive modulator of the MVB sorting pathway. Reducing its expression by RNAi decreases degradation of the EGF receptor and blocks HIV viral particle release, whereas overexpressing it has no effect (Ward et al., 2005). In yeast, mutations in VTA1 impair membrane protein degradation and create a weak class E phenotype, the severity of which may depend on the flux of cargo through the endosomal pathway (Shiflett et al., 2004; Azmi et al., 2006; Rue et al., 2007). Although LIP5's known role in VPS4 oligomerization might explain these effects, our results reveal that LIP5 also directly and efficiently engages a number of ESCRT-III proteins, including in particular those that contain the C-terminal MIM known to bind VPS4 (Figures 1 and 2; Obita et al., 2007; Stuchell-Brereton et al., 2007).
How might LIP5 bound to ESCRT-III proteins modulate progress through the MVB pathway? Because LIP5 and ESCRT-III are already thought to be cofactor and substrate of VPS4, respectively, it is logical to think that their interaction will affect VPS4 function. This idea is supported by the fact that the same ESCRT-III proteins that bind well to VPS4 (CHMP1, CHMP2, and CHMP3) bind well to LIP5 (Howard et al., 2001; Scott et al., 2005b; Nickerson et al., 2006; Obita et al., 2007; Stuchell-Brereton et al., 2007). One possibility is that the extra link between LIP5 and ESCRT-III complex ensures that VPS4 oligomerizes only where it is needed. In the simplest scenario, this would predict that interactions between these three proteins would reinforce each other. Indeed, VPS4 interacts with ESCRT-III and LIP5/Vta1 via separate domains (Azmi et al., 2006; Vajjhala et al., 2006). Similarly, LIP5/Vta1 binds to VPS4 and the ESCRT-III like protein CHMP5/Vps60 via its C-terminus and N-terminus, respectively (Azmi et al., 2006). However, we found that the VPS4 MIT domain reduces LIP5 binding to both CHMP2A and CHMP1B, indicating that everything cannot happen simultaneously (Figure 5).
At the same time, we found evidence for a second, more internal, binding site for VPS4 in these ESCRT-III proteins (Figures 6 and 7), leading us to suggest that the interaction of VPS4 with the MIM motifs in α6 may represent only one step in ESCRT-III disassembly. Although we were unable to precisely define the second VPS4 binding motif in vitro, deletion and alanine scanning studies in both CHMP2A and CHMP1B indicated that this interaction depends on conserved sequences within these proteins' α5 helix and in particular on two acidic residues that are conserved across ESCRT-III proteins. Interestingly, α5 occupies an exposed position in the crystal structure of CHMP3 (Muziol et al., 2006), which probably represents the “open” form of these proteins (Saksena et al., 2007). We propose that interaction of VPS4–using its MIT domain, elements within its AAA+ domain such as the “pore loops” known to be important for its function (Scott et al., 2005a), or possibly an associated cofactor such as the recently described Ist1 (Dimaano et al., 2008)—with α5 in all ESCRT-III proteins is likely to be an important additional step in ESCRT-III complex disassembly.
Although LIP5 is clearly established as a positive modulator of the MVB sorting pathway, the role played by CHMP5 (Vps60 in yeast) is less clear. CHMP5 binds with high affinity to LIP5 (Figure 8), and deleting these two proteins in yeast has overlapping rather than additive effects (Rue et al., 2007). However, reducing or eliminating CHMP5 expression in mammalian tissues or cells does not prevent formation of MVBs (nor incorporation of TGF-β receptors into the internal vesicles; Shim et al., 2006) and in fact enhances HIV budding from cells (Ward et al., 2005). These effects, together with the fact that LIP5's interaction with CHMP5 is fundamentally different from its interaction with the other ESCRT-III proteins, lead us to suggest that CHMP5 bound to LIP5 might negatively regulate LIP5 for engagement with other ESCRT-III proteins and VPS4. This possibility remains to be further explored.
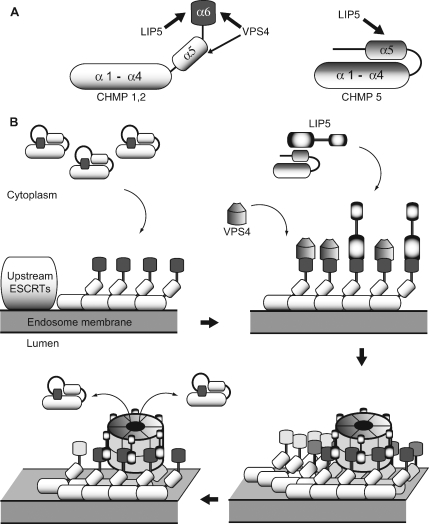
A model that summarizes our results and how they impact thinking about the cooperation between LIP5 and VPS4 in regulating ESCRT-III is shown in Figure 9. Binding sites for LIP5 (α5 in CHMP5 and α6 in CHMP1B and CHMP2A) and for VPS4B (previously described primary site in α6, secondary binding site in α5) are shown in Figure 9A. The relationship between ESCRT-III subunits, ESCRT-III complex, and LIP5 and VPS4 is depicted in Figure 9B. ESCRT-III proteins are closed monomers in the cytosol. In this state, our results suggest that only CHMP5 binds to LIP5. When ESCRT-III proteins polymerize into complexes on the endosomal membrane (presumably nucleated by upstream factors that are connected to cargo) the subunits open, exposing sequences at their C-termini for binding to LIP5 and/or VPS4. How these two proteins share their overlapping binding sites remains to be determined, but their separate ability to bind each other (via domains that are not engaged with the ESCRT-III proteins, the β domain in VPS4 (Scott et al., 2005a; Vajjhala et al., 2006) and the VSL domain in LIP5 (Azmi et al., 2006) is likely to reinforce their association. Once some threshold is reached (perhaps full assembly of a VPS4 oligomer; Hartmann et al., 2008; Yu et al., 2008), we hypothesize that VPS4 engages its secondary contact site. This in turn may allow VPS4 to unfold individual ESCRT-III subunits and release them into the cytoplasm, where they revert to their closed and monomeric states. Although aspects of this model remain to be confirmed, and importantly any ESCRT-III disassembly reaction has yet to be reconstituted, the intricacies of this important step in MVB biogenesis are finally starting to come into focus.
Figure 9.
Model showing proposed engagement of LIP5 with ESCRT-III proteins and VPS4. (A) Binding sites for LIP5 and Vps4 in individual ESCRT-III subunits. Sites defined in this study in CHMP2A and CHMP1B are shown at left and in CHMP5 at right. The schematic structure of the ESCRT-III subunits is based on the crystal structure of CHMP3 (Muziol et al., 2006). (B) Proposed model of ESCRT-III assembly, disassembly, and interaction with LIP5 and VPS4. Most ESCRT-III proteins are closed monomers in the cytoplasm and do not bind to LIP5, although CHMP5 interacts differently with LIP5 and can bind in the cytoplasm. As ESCRT-III complexes assemble on the endosomal membrane, individual subunits open and expose sequences at their C-termini for binding to LIP5 and/or VPS4. How these two proteins share their overlapping binding sites remains to be determined, but their separate ability to bind each other (via domains that are not engaged with the ESCRT-III proteins, the β domain in VPS4 (Scott et al., 2005a; Vajjhala et al., 2006) and the VSL domain in LIP5 (Azmi et al., 2006) is likely to reinforce their association. Once some threshold is reached (perhaps assembly of VPS4 rings), we propose that VPS4 engages its secondary contact site in α5 of the ESCRT-III proteins. Based on analogy to other AAA+ proteins, this may allow VPS4 to unfold individual ESCRT-III subunits and release them into the cytoplasm where they revert to their closed and monomeric state.
While this article was being reviewed and revised, two articles examining the structure and interactions of Vta1 (the yeast equivalent of LIP5) were published (Azmi et al., 2008; Xiao et al., 2008). In one, the high-resolution crystal structure of the Vta1 N-terminus revealed two MIT-like domains, each consisting of three α-helices (Xiao et al., 2008). This structure strongly supports our finding of a high-affinity interaction between LIP5 and the MIM-containing ESCRT-III proteins CHMP1B and CHMP2A and suggests that one or both of LIP5's MIT domains binds to these proteins. In the second article, the interaction between Vta1 and Vps60 (yeast CHMP5) was explored in more detail with results that largely agree with what we report here for mammalian proteins (Azmi et al., 2008). Significant differences are the failure to see a high-affinity interaction between Vta1 and Vps2 in the yeast system and mapping of the Vta1 binding site in Vps60 to α4 instead of α5 as found here for CHMP5 and LIP5. Whether these differences reflect differences in the protein interactions or in the conditions used to study them remains to be established.
ACKNOWLEDGMENTS
We thank Lisa Kimpler for an early visual screen of LIP5 binding to ESCRT-III proteins, Teresa Naismith for help with site-directed mutagenesis, and members of the Hanson Lab for helpful discussions. We also thank Diane Ward and Jerry Kaplan (University of Utah) for a LIP5 expression plasmid. This work was supported by a grant from the American Heart Association (0550148Z) and a faculty fellowship from Andrew and Virginia Craig (P.I.H.). S.S. is a student in the Markey Pathway for Pathobiology and S.A.M. a student in the Medical Scientist Training Program.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1263) on April 2, 2008.
REFERENCES
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D. J. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 2006;172:705–717. doi: 10.1083/jcb.200508166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. doi: 10.1016/j.devcel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. Escrt–III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T. K., Banta L. M., Emr S. D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P. D. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Bishop N., Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. G., Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Demirov D. G., Freed E. O. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Dimaano C., Jones C. B., Hanono A., Curtiss M., Babst M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 2008;19:465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Yamanaka M., Imamura K., Tanaka Y., Nara A., Yoshimori T., Yokota S., Himeno M. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci. 2003;116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- Fujita H., Umezuki Y., Imamura K., Ishikawa D., Uchimura S., Nara A., Yoshimori T., Hayashizaki Y., Kawai J., Ishidoh K., Tanaka Y., Himeno M. Mammalian class E proteins, SBP1 and mVps2/CHMP2A, interact with and regulate the function of an AAA-ATPase SKD1/Vps4B. J. Cell Sci. 2004;117:2997–3009. doi: 10.1242/jcs.01170. [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Hanson P. I., Roth R., Lin Y., Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C., Chami M., Zachariae U., de Groot B. L., Engel A., Grutter M. G. Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J. Mol. Biol. 2008;377:352–363. doi: 10.1016/j.jmb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Howard T. L., Stauffer D. R., Degnin C. R., Hollenberg S. M. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Emr S. D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Odorizzi G., Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kranz A., Kinner A., Kolling R. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell. 2001;12:711–723. doi: 10.1091/mbc.12.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Kimpler L. A., Naismith T. V., Lauer J. M., Hanson P. I. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7–1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- Lottridge J. M., Flannery A. R., Vincelli J. L., Stevens T. H. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA. 2006;103:6202–6207. doi: 10.1073/pnas.0601712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. M., Boucrot E., Villen J., el Affar B., Gygi S. P., Gottlinger H. G., Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sundquist W. I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Muziol T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Gottlinger H., Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson D. P., West M., Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- Piper R. C., Katzmann D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue S. M., Mattei S., Saksena S., Emr S. D. Novel Ist1-Did2 Complex Functions at a Late Step in MVB Sorting. Mol. Biol. Cell. 2007;19:475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S., Sun J., Chu T., Emr S. D. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Scott A., et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005a;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., Sundquist W. I. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA. 2005b;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett S. L., Ward D. M., Huynh D., Vaughn M. B., Simmons J. C., Kaplan J. Characterization of Vta1p, a class E Vps protein in S. cerevisiae. J. Biol. Chem. 2004;279:10982–10990. doi: 10.1074/jbc.M312669200. [DOI] [PubMed] [Google Scholar]
- Shim J. H., et al. CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J. Cell Biol. 2006;172:1045–1056. doi: 10.1083/jcb.200509041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Kimpler L. A., Hanson P. I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Vajjhala P. R., Wong J. S., To H. Y., Munn A. L. The beta domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS J. 2006;273:2357–2373. doi: 10.1111/j.1742-4658.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- von Schwedler U. K., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Ward D. M., Vaughn M. B., Shiflett S. L., White P. L., Pollock A. L., Hill J., Schnegelberger R., Sundquist W. I., Kaplan J. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 2005;280:10548–10555. doi: 10.1074/jbc.M413734200. [DOI] [PubMed] [Google Scholar]
- Williams R. L., Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- Xiao J., Xia H., Yoshino-Koh K., Zhou J., Xu Z. Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 2007;374:655–670. doi: 10.1016/j.jmb.2007.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Xia H., Zhou J., Azmi I. F., Davies B. A., Katzmann D. J., Xu Z. Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev. Cell. 2008;14:37–49. doi: 10.1016/j.devcel.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo S. C., et al. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 2003;116:3957–3970. doi: 10.1242/jcs.00751. [DOI] [PubMed] [Google Scholar]
- Yu Z., Gonciarz M. D., Sundquist W. I., Hill C. P., Jensen G. J. Cryo-EM structure of dodecameric Vps4p and Its21 complex with Vta1p. J. Mol. Biol. 2008;377:364–377. doi: 10.1016/j.jmb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]