Abstract
The mitochondrion is a dynamic membranous network whose morphology is conditioned by the equilibrium between ongoing fusion and fission of mitochondrial membranes. In the budding yeast, Saccharomyces cerevisiae, the transmembrane GTPase Fzo1p controls fusion of mitochondrial outer membranes. Deletion or overexpression of Fzo1p have both been shown to alter the mitochondrial fusion process indicating that maintenance of steady-state levels of Fzo1p are required for efficient mitochondrial fusion. Cellular levels of Fzo1p are regulated through degradation of Fzo1p by the F-box protein Mdm30p. How Mdm30p promotes degradation of Fzo1p is currently unknown. We have now determined that during vegetative growth Mdm30p mediates ubiquitylation of Fzo1p and that degradation of Fzo1p is an ubiquitin-proteasome–dependent process. In vivo, Mdm30p associates through its F-box motif with other core components of Skp1-Cullin-F-box (SCF) ubiquitin ligases. We show that the resulting SCFMdm30p ligase promotes ubiquitylation of Fzo1p at mitochondria and its subsequent degradation by the 26S proteasome. These results provide the first demonstration that a cytosolic ubiquitin ligase targets a critical regulatory molecule at the mitochondrial outer membrane. This study provides a framework for developing an understanding of the function of Mdm30p-mediated Fzo1p degradation in the multistep process of mitochondrial fusion.
INTRODUCTION
Mitochondria are dynamic in nature and collectively all mitochondria in a cell functionally constitute a single tubular network, the morphology of which is determined by an equilibrium between fusion and fission (Bleazard et al., 1999; Sesaki and Jensen, 1999; Fritz et al., 2003; Okamoto and Shaw, 2005; Hoppins et al., 2007). Disruption of fission in the budding yeast Saccharomyces cerevisiae drives the equilibrium toward fusion resulting in net-like mitochondrial structures that appear to collapse to one side of the cell (fused). Conversely, abrogation of fusion shifts the equilibrium toward fission, which is manifest as dot-like fragmented mitochondria (fizzed; see examples in Figure 1A). Such alterations are linked to apoptosis in mammals and compromise cell life span in fungus (Okamoto and Shaw, 2005; Chan, 2006; Heath-Engel and Shore, 2006; Martinou and Youle, 2006; Scheckhuber et al., 2007). Maintaining the capacity of mitochondria to fuse normally is essential to inheritance of mitochondrial DNA and in turn for respiration (Hermann et al., 1998).
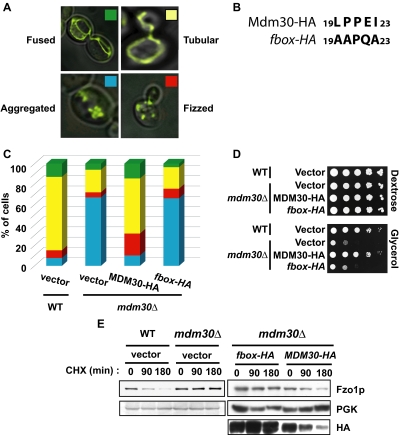
Figure 1.
The F-box of Mdm30p is required for mitochondrial fusion and respiration as well as degradation of Fzo1p. (A) Typical images of fused (green), tubular (yellow), fizzed (red), and aggregated (blue) mitochondria quantified in Figures 1C and Supplemental Data Table S2. (B) Amino acids 19, 20, 22, and 23 conserved in F-box motifs were mutated as indicated in Mdm30-HA to yield the fbox-HA mutant. (C) Mitochondrial morphology was assessed in wild-type (vector; W303 background), mdm30Δ (vector), or mdm30Δ cells expressing either Mdm30-HA or an F-box mutant of Mdm30-HA (fbox-HA) under control of the TEF promoter. (D) The same strains as described in C were grown at 37°C on selective media containing either dextrose or glycerol as the only carbon source. (E) Rate of Fzo1p degradation was analyzed in the indicated strains (W303 background) after shift to 37°C and treatment with CHX. Yeast extracts were prepared at the indicated times and remaining Fzo1p or Mdm30-HA evaluated by immunoblotting. Levels of a stable protein, phosphoglucokinase (PGK), are shown as a loading control.
Mitochondrial outer membrane fusion is controlled by mitofusins, a family of GTPases integral to the mitochondrial outer membrane. The yeast mitofusin, Fzo1p, is unstable during both vegetative and nonvegetative growth. Regulating levels of expression of Fzo1p is of critical importance as either deletion or overexpression of Fzo1p alters the mitochondrial fusion process resulting in fizzed or abnormal aggregated mitochondria, respectively (Hermann et al., 1998; Rapaport et al., 1998; Fritz et al., 2003; Escobar-Henriques et al., 2006).
The ubiquitin-proteasome system (UPS) constitutes the major mechanism by which cells acutely alter levels of cytosolic, nuclear, and endoplasmic reticulum (ER) proteins in a highly regulated manner. This occurs generally, but not exclusively, by conjugation with chains of ubiquitin linked through lysine 48 (K48) of ubiquitin, which targets modified proteins to the 26S proteasome for degradation (Glickman and Ciechanover, 2002). Ubiquitin ligases (E3s) mediate the transfer of ubiquitin from ubiquitin-conjugating enzymes (E2s) to specific substrates and are the primary determinants of specificity in ubiquitylation (Fang and Weissman, 2004). During nonvegetative growth, corresponding to mating conditions, a role for the 26S proteasome in degradation of Fzo1p has been reported. However, evidence for ubiquitylation has surprisingly been lacking, and no ubiquitin ligase has been implicated in this process (Neutzner and Youle, 2005; Escobar-Henriques et al., 2006).
During vegetative growth, the level of Fzo1p is regulated by Mdm30p. Mdm30p is a member of the F-box family of proteins. The F-box is a 50-amino acid protein interaction motif encoded in ∼15 genes in S. cerevisiae and ∼70 genes in mammals. F-box proteins are generally thought to serve as substrate recognition elements of ubiquitin ligases of the Skp1-Cullin-F-box (SCF) family (Cardozo and Pagano, 2004; Willems et al., 2004; Petroski and Deshaies, 2005). In this regard, Mdm30p has been shown to promote ubiquitylation and proteasomal degradation of the Gal4p transcription factor (Muratani et al., 2005). Nonetheless, to date not all F-box proteins have been positively linked to the SCF complex or even to a specific function in the UPS (Galan et al., 2001; Frescas et al., 2007).
Mdm30p associates with mitochondria (Fritz et al., 2003), appears to physically interact with Fzo1p (Escobar-Henriques et al., 2006), and has been shown to target Fzo1p for degradation (Fritz et al., 2003; Escobar-Henriques et al., 2006). The importance of Mdm30p in mitochondrial function is underscored by the finding that deletion of MDM30 abrogates mitochondrial fusion and leads to aggregated mitochondria (Dürr et al., 2006; see examples in Figure 1A) and consequently to defective mitochondrial DNA inheritance and a failure to respire (Fritz et al., 2003). However, the mechanism by which Mdm30p promotes Fzo1p degradation has not been elucidated.
In this study, we investigated the mechanism by which Fzo1p is targeted for degradation during vegetative growth. We establish that Fzo1p is ubiquitylated and targeted for proteasomal degradation. This ubiquitylation is mediated by an SCF ubiquitin ligase that includes Mdm30p as the substrate recognition factor (SCFMdm30p). Thus, we have identified a mechanism whereby a critical protein integral to the mitochondrial outer membrane is targeted for destruction by cytosolic components of the UPS.
MATERIALS AND METHODS
Yeast Strains and Media
The S. cerevisiae strains used in this study are listed in Supplemental Data, Table S1. Standard methods were used for growth, transformation and genetic manipulation of S. cerevisiae. Complete (YPDextrose) and minimal (SDextrose, SGlycerol) media supplemented with either 2% dextrose or 3% glycerol were prepared as described (Sherman et al., 1986). In the indicated strains (see Supplemental Data, Table S1), FZO1 was chromosomally tagged with three copies of an HA epitope sequence, as previously described (Longtine et al., 1998).
Plasmids
The plasmids used in this study are listed in Supplemental Data, Table S1. Ubiquitin wild type, K48R, or K63R, expressed under the control of the CUP1 promoter were overproduced by growing cells for 1 h in the presence of 0.1 mM copper sulfate. The MDM30-hemagglutinin (HA) construct was generated as follows. The 500 base pairs upstream of the ATG and downstream of the STOP codon of MDM30 were subcloned in the pRS316 vector, yielding the pRS316 MDM30prom/ter vector. MDM30 lacking its STOP codon or both its STOP codon and sequence encoding its F-box (amino acids 1-58) were cloned into pRS316 MDM30prom/ter followed by insertion of the HA tag downstream of the MDM30 coding sequence, resulting in MDM30-HA and Δfbox-HA, respectively. MDM30-HA was then subcloned in the p426TEF vector and previously described mutations of amino acids in its F-box motif (fbox-HA; Escobar-Henriques et al., 2006) that are critical for its association with Skp1p (Galan and Peter, 1999) were generated by site-directed mutagenesis.
Antibodies
Antiserum raised against Fzo1p was generously provided by J. Nunnari (University of California, Davis, CA). Monoclonal antibodies utilized were against HA and Myc epitopes (12CA5 and 9E10; Santa Cruz Biotechnology, Santa Cruz, CA) phosphoglucokinase (PGK; Monoclonal 22C5; Molecular Probes, Eugene, OR), Tom20p (kindly provided by A. Azem, Tel Aviv University), Hexokinase1 (Hxk1p; kindly provided by A. Azem), or Cdc53p (yC-17; Santa Cruz Biotechnology). Antiserum recognizing Cue1p was provided by Z. Kostova (National Cancer Institute).
Yeast Extracts and Cycloheximide Chase
Cells grown in YPD or SD were collected during the exponential growth phase. Total protein extracts were prepared by the NaOH-trichloroacetic acid (TCA) lysis method (Avaro et al., 2002). To monitor constitutive turnover of Fzo1, cycloheximide (CHX) was added to yeast cultures growing at 37°C to a final concentration of 100 μg/ml. Thermosensitive strains were incubated for 2 h at 37°C before adding CHX. Total protein extracts were prepared at the indicated time points after addition of CHX.
Pulse-Chase Metabolic Labeling
Cells grown in YPD were collected during the exponential growth phase, incubated for 50 min at 23°C in 1 ml labeling media (SD-Met) lacking methionine, and pulsed with 25 μCi of 35S methionine (Perkin-Elmer Cetus, Norwalk, CT) per OD of cells for 15 min at 30°C followed by 15 min at 37°C. After addition of 1 ml chase media (SD-Met supplemented with 6 mg/ml methionine and 2 mg/ml BSA), cells were incubated at 37°C. After addition of chase media ∼2 ODs of cells were collected immediately (time = 0) and at 30 and 60 min and treated as described previously (Moreau et al., 1997). Results were quantified by Storm Phosphorimager and ImageQuant software (GE Healthcare, Waukesha, WI). Error bars were determined by calculating the SD from three independent experiments.
Immunoprecipitation
For coimmunoprecipitation assays, cells were lysed at 4°C with glass beads in immunoprecipitation (IP) buffer (50 mM HEPES, pH 7.5, 50 mM sodium chloride, 0.6% Triton X-100, 10% glycerol, 20 mM iodoacetamide, and protease inhibitor; Complete mini, Roche, Indianapolis, IN). Insoluble material was removed by centrifugation for 30 min at 13,000 × g. An aliquot of the supernatant was precipitated with 50% TCA (pre-IP lysate). The remaining supernatant was incubated with Anti-HA Affinity Matrix (Roche) for 2 h at 4°C. Beads washed with IP buffer were heated in sample buffer before resolution by SDS-PAGE and analysis by immunoblotting. For immunoprecipitation of Fzo1-HA or Mdm30-HA, yeast extracts were prepared using the NaOH-TCA lysis method (Avaro et al., 2002). Extracts were then boiled for 10 min at 70°C in SDS loading buffer and insoluble material removed by centrifugation for 5 min at 13,000 ×g. Resulting supernatants were diluted 10-fold in IP buffer, and immunoprecipitations were performed as described above.
Subcellular Fractionation
Cell fractionation was performed as described (Meisinger et al., 2000). In brief, cell cultures were grown to midlog phase (OD6001-2), and spheroplasts were prepared by treating the cells with Zymolyase-20T (MP Biomedicals, Solon, OH). After gentle homogenization of spheroplasts and centrifugation at 1500 × g, the supernatant (total fraction) was further subject to centrifugation at 12,000 × g for 10 min, yielding a supernatant (S) and a mitochondrial enriched pellet fraction (P). Subcellular fractions were assayed for cytosolic and mitochondrial proteins; Tom20 and Hexokinase1 were used as mitochondrial and cytosolic markers, respectively.
Glycerol Growth Analysis
For glycerol growth assays, cultures grown overnight in SD medium were pelleted, resuspended at OD600 = 1, and diluted 1:10 five times in water. Three microliters of the dilutions were spotted on plates and grown for 2 d (SD) or 4 d (SG) at 30°C or 37°C.
Microscopy
For visualization of mitochondria, yeast strains were transformed with plasmid pYX232-mtGFP, encoding mitochondria-targeted GFP (mtGFP; Westermann and Neupert, 2000). Cultures in logarithmic growth were fixed with 3.7% formaldehyde (Sigma, St. Louis, MO) for 10 min, washed in KPi buffer (0.02 M KH2PO4, 0.08 M K2HPO4, 1 M sorbitol, pH 7.5) and mounted on Superfrost microscope slides (Esco Products, Oak Ridge, NJ) in phosphate-buffered saline. Cells were then analyzed by epifluorescence microscopy on an Axiovert 200M microscope (Carl Zeiss MicroImaging, Thornwood, NY) using a 100× oil-immersion objective. Images were recorded with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics, Hamamatsu City, Japan). For each field of cells, between 30 and 40 pictures were taken in the Z-coordinate, and cells were deconvoluted using Improvision Openlab 4.0.2 software (Improvision, Lexington, MA). The morphology of mitochondria was assessed by counting more than 300 cells per strain. Quantification was confirmed by independent counting by a second individual blinded to the identity of the strains. Results are displayed in Supplemental Data Table S2 and Figure 1B.
RESULTS
The F-Box Motif of Mdm30p Is Essential for Mitochondrial Fusion and Respiration as well as for Fzo1p Degradation
To assess the mechanism by which Mdm30p regulates mitochondrial morphology and particularly the importance of its F-box domain, we analyzed mitochondria from mdm30Δ cells expressing either wild-type Mdm30p or a form mutated in its F-box (Figure 1, B and C, and Supplemental Data, Table S1). This mutation has previously been shown to result in increased steady-state levels of Fzo1p (Escobar-Henriques et al., 2006). About 70% of wild-type cells displayed mitochondria characterized as being tubular. The remaining 30% were partitioned between those scored as having fused, fizzed, and aggregated mitochondria (see examples in Figure 1A). In agreement with previous reports (Fritz et al., 2003; Dürr et al., 2006; Escobar-Henriques et al., 2006), among mdm30Δ cells almost 70% displayed aggregated mitochondria (Figure 1C, vector transformation). Reintroducing MDM30 into mdm30Δ cells (MDM30-HA), under a constitutive TEF promoter, restored mitochondrial morphology to a distribution similar to wild-type cells and completely reversed the marked increase in aggregated mitochondria seen in mdm30Δ cells (Figure 1C). However, cells expressing the F-box mutant (fbox-HA) retained a mitochondrial distribution similar to that observed in the mdm30Δ mutant, pointing to an essential role for the F-box in maintaining a normal distribution of mitochondrial morphology.
To confirm the functional significance of these findings, we evaluated the impact on respiration of mutating the F-box of Mdm30p. Loss of mitochondrial fusion has been shown to manifest itself as a failure of mdm30Δ cells to respire properly and a decreased capacity to grow on media containing only a nonfermentable carbon source (Fritz et al., 2003; Dürr et al., 2006). Strains from Figure 1C were therefore tested for growth on selective media containing either dextrose (fermentable) or glycerol (nonfermentable) as the sole carbon source (Figure 1D). As shown previously (Fritz et al., 2003), mdm30Δ cells are defective for growth on glycerol media. Consistent with the morphological findings, this growth defect was rescued by expression of Mdm30-HA but not the F-box mutant Mdm30-HA (fbox-HA), once again pointing to an essential role for the F-box motif in mitochondrial function as well as morphology. Similar findings for both mitochondrial morphology and growth on glycerol were obtained when wild-type and truncated Mdm30p lacking its F-box (Δf-box-HA) were expressed from the endogenous MDM30 promoter (Supplemental Data, Figure S1).
Mitochondrial aggregation in mdm30Δ cells has been shown to correlate with accumulation of Fzo1p (Fritz et al., 2003). More recently, Mdm30p was shown to be essential for Fzo1p degradation during vegetative growth (Escobar-Henriques et al., 2006). In agreement with this latter study, we observed that endogenous Fzo1p, which is naturally turned over in cells, was completely stabilized upon genomic deletion of MDM30 (Figure 1E, left panels). Given our findings that the F-box of Mdm30p is required for normal mitochondrial morphology and respiration (Figure 1, C and D), we asked whether the F-box is also required for Fzo1p degradation. mdm30Δ strains expressing Mdm30-HA or its F-box mutant (fbox-HA) were used to monitor Fzo1p turnover (Figure 1E, right panels). Although Fzo1p was degraded upon expression of Mdm30-HA, it remained stable upon expression of the F-box mutant (fbox-HA), consistent with previous findings (Escobar-Henriques et al., 2006). This differential stability was also observed with wild-type and F-box–deleted Mdm30p expressed from its endogenous promoter (Supplemental Data, Figure S1, C and D). Together, results from Figure 1 indicate that the F-box of Mdm30p is essential for efficient mitochondrial fusion and respiration as well as for Fzo1p degradation.
Critical Components of the SCF Complex Participate in Fzo1p Degradation
Most F-box proteins are thought to act as substrate recognition subunits of SCF ubiquitin ligase complexes (Cardozo and Pagano, 2004; Willems et al., 2004; Petroski and Deshaies, 2005), thereby promoting substrate ubiquitylation followed by proteasomal degradation. In S. cerevisiae Skp1p serves as an adaptor between F-box proteins and the cullin, Cdc53p. The Skp1p-Cdc53p core together with a small RING finger protein serves as a molecular scaffold that also recruits a specific ubiquitin-conjugating enzyme (E2), Cdc34p.
Our finding that the F-box of Mdm30p is essential for Fzo1p degradation raises the possibility that Mdm30p functions as part of an SCF E3 ubiquitin ligase (SCFMdm30p), potentially providing a mechanism by which Mdm30p targets Fzo1p for degradation. To directly assess whether Mdm30p associates with Skp1p-Cdc53p in cells, HA-tagged Mdm30p was immunoprecipitated from whole cell extracts prepared from mdm30Δ cells expressing wild-type Mdm30p (Mdm30-HA) or Mdm30p mutated in its F-box (fbox-HA). Immunoprecipitates were resolved on SDS-PAGE and immunoblotted with Cdc53p antibody (Figure 2A, middle panel). Cdc53p coimmunoprecipitated with wild-type Mdm30-HA but not the F-box mutant. This result establishes that Mdm30p associates with core components of the SCF and that this interaction is dependent on an intact Mdm30p F-box.
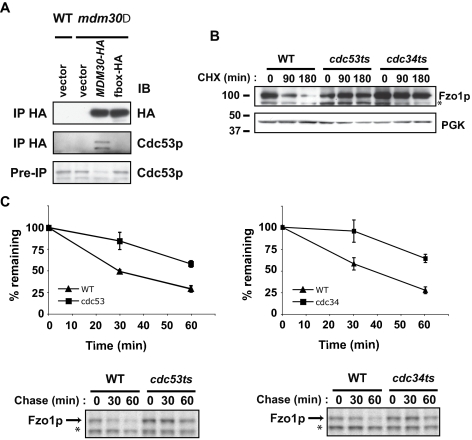
Figure 2.
Core SCF components interact with Mdm30p and are required for degradation of Fzo1p. (A) Epitope-tagged Mdm30p (Mdm30-HA) was immunoprecipitated (IP) from the indicated cell lysates (W303 background) with antibody recognizing HA. Whole cell lysates (pre-IP; bottom panel) and immunoprecipitates from equal amounts of lysates from each strain were analyzed by immunoblotting (IB) with antibody recognizing either Cdc53p or HA. Excess starting material (lysate) was utilized in IPs to maximize visualization of endogenous Cdc53. (B) Degradation of Fzo1p in wild-type, cdc53ts, or cdc34ts strains was evaluated by CHX chase as in Figure 1E. (C) Fzo1-HA turnover was analyzed by 35S pulse-chase metabolic labeling in wild-type (triangle), cdc53ts (square, left), and cdc34ts (square, right) strains. Graphs represent quantification of three independent experiments. Representative experiments are shown below. Data are plotted relative to the amount at the beginning of the chase. Asterisks indicate nonspecific band.
Having established that Mdm30p is a bona fide component of an SCF E3 (SCFMdm30p), we evaluated the requirements for Cdc34p and Cdc53p on Fzo1p degradation by CHX chase (Figure 2B). Compared with the wild-type strain, a marked stabilization of Fzo1p was observed in the conditional mutants, cdc53ts and cdc34ts, at the restrictive temperature (37°C). To assess effects on protein turnover without inhibiting protein synthesis, Fzo1p half-life in cdc53ts and cdc34ts strains was assessed by metabolic labeling using 35S methionine (Figure 2C). Although Fzo1p was degraded with a half-life of ∼30 min in wild-type cells, it was markedly stabilized in thermosensitive mutants of either CDC53 or CDC34 (cdc53ts and cdc34ts) with half-lives of greater than 60 min. These results establish that, in addition to Mdm30p, critical components of the SCF complex participate in Fzo1p degradation.
Fzo1p Is Modified with Ubiquitin at the Mitochondria
The observation that the ubiquitin ligase SCFMdm30p is required for Fzo1p degradation strongly suggests that Fzo1p is a substrate for ubiquitylation. Ubiquitylated intermediates are frequently difficult to detect as they are rapidly degraded by the proteasome. Using a polyclonal antibody directed against Fzo1p, we observed multiple immunoreactive species above the major Fzo1p band on long exposure of immunoblots (Figure 3A, right panel). These higher molecular weight bands were reproducibly and specifically observed in extracts prepared from wild-type yeast but not from fzo1Δ cells. The same pattern was observed whether monitoring endogenous Fzo1p or chromosomally tagged FZO1-HA using either anti-Fzo1p or anti-HA (Figure 3B). A slight retardation in the migration pattern of Fzo1p and the higher molecular weight bands was observed in the FZO1-HA strain when detected by Fzo1p antibody, consistent with increased mass conferred by the three copies of an HA tag. This indicates that these higher molecular weight species represent modified forms of Fzo1p.
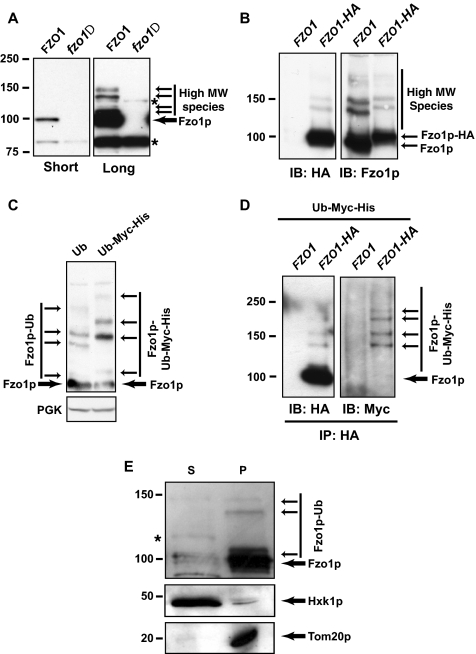
Figure 3.
Mitochondrial Fzo1p is modified with ubiquitin. (A) Short and long exposures of an identical anti-Fzo1p immunoblot probing equal amounts of wild-type (BY4742) and fzo1Δ whole cell extracts. Unmodified Fzo1p is indicated by a thick arrow; modified forms are marked with thin arrows. Nonspecific bands (also detected in fzo1Δ extracts) are indicated by asterisks. (B) Crude extracts from FZO1 (W303 background) and FZO1–3HA strains were processed for immunoblotting with antibodies that recognize either HA or Fzo1p. (C) Crude extracts from Ub (SUB280) and Myc-His6-Ub (SUB595) strains were processed for immunoblotting to detect Fzo1p. (D) Immunoprecipitates with HA antibody from Ub and Myc-His6-Ub strains having chromosomally HA-tagged (FZO1-HA) or untagged (FZO1) FZO1 were immunoblotted with either HA antibody or Myc antibody. (E) Distribution of Fzo1p between cytosolic (S) and mitochondrially enriched (P) fractions prepared from wild-type cells (W303 background). Hexokinase1p (Hxk1p) and Tom20p were used as cytosolic and mitochondrial markers, respectively. Asterisks correspond to nonspecific bands observed also in fzo1Δ (data not shown).
The ladder-like pattern of these higher molecular weight forms of Fzo1p is highly suggestive of ubiquitylation. To confirm this possibility, extracts from yeast strains expressing ubiquitin tagged with both Myc and His6 epitopes as the sole source of ubiquitin were processed for immunoblotting with anti-Fzo1p and the migration pattern compared with similar extracts prepared from a strain expressing untagged ubiquitin (Figure 3C). We observed that although migration of unmodified Fzo1p band was similar in both strains, all of the higher molecular weight species were shifted upward in the strain expressing Myc-His6-Ub. This upward shift is consistent with differential incorporation of tagged Myc-His6-Ub in one strain and untagged Ub in the other. To unequivocally and directly demonstrate ubiquitylation of Fzo1p, lysates from chromosomally tagged FZO1-HA cells expressing Myc-His6-Ub as the sole ubiquitin source were immunoprecipitated with HA antibody and immunoblotted with Myc antibody. This led to the specific detection of high-molecular-weight species that comigrated with those seen when replicate blots were probed with HA antibody (Figure 3D). These results conclusively establish for the first time that Fzo1p, expressed at endogenous levels, undergoes ubiquitylation.
As Fzo1p is an integral mitochondrial outer membrane protein, we wanted to determine the cellular location of ubiquitylated Fzo1p. Whole cell extract was fractionated and cytosolic (S) and mitochondria-enriched fractions (P) were tested for Fzo1p content by immunoblotting with Fzo1p antibody (Figure 3E). Both unmodified and ubiquitylated forms of Fzo1p were found almost exclusively in the mitochondria-enriched fraction. The mitochondrial localization of ubiquitylated Fzo1p was further confirmed by sucrose gradient analysis (Supplemental Data, Figure S2). These data indicate that ubiquitylated Fzo1p is localized to mitochondria and does not represent mis-targeted or mis-localized Fzo1p.
SCFMdm30p Mediates K48-linked Ubiquitylation of Fzo1p
Having established that the higher molecular weight species of Fzo1p correspond to Fzo1p-ubiquitin conjugates, we asked whether ubiquitylation of Fzo1p is dependent on Mdm30p and other SCF components. As is apparent, the ubiquitylated forms of Fzo1p that were detected in wild-type cells were undetectable in the mdm30Δ mutant (Figure 4A, cf. left two lanes). Moreover, although MDM30-HA restored Fzo1p ubiquitylation in mdm30Δ cells, the F-box mutant (fbox-HA) did not. Similarly, Fzo1p ubiquitylation was decreased in both conditional SCF mutants, cdc34ts and cdc53ts, when grown at the restrictive temperature compared with isogenic wild-type controls (Figure 4B). We conclude that Mdm30p, Cdc34p, and Cdc53p are essential for the Fzo1p ubiquitylation that is observed in wild-type cells.
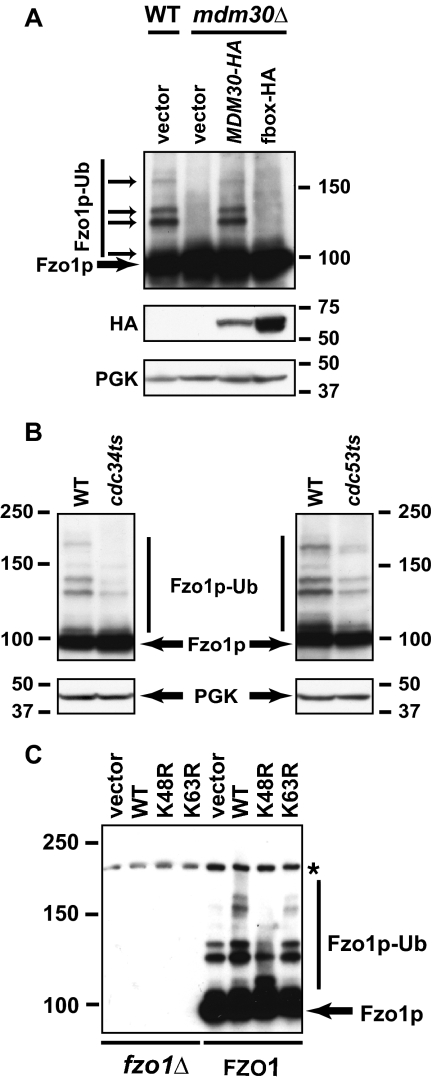
Figure 4.
Mdm30p, Cdc34p, and Cdc53p are required for K48-linked ubiquitylation of Fzo1p. (A) Cellular levels of Fzo1p, Mdm30p (Mdm30-HA), or a version of Mdm30p with a mutated F-box incapable of binding to Skp1p (fbox-HA) were analyzed in whole cell extracts prepared from the indicated strains (W303 background) grown at 30°C. Unmodified and ubiquitylated forms of Fzo1p are indicated by thick and thin arrows, respectively. PGK is shown as a loading control. (B) Ubiquitylated forms of Fzo1p were analyzed in whole cell extracts prepared from wild-type, cdc53ts, or cdc34ts cells grown at 23°C and shifted to 37°C. PGK serves as a loading control. (C) Anti-Fzo1p immunoblot of whole cell extracts prepared from fzo1Δ and wild-type cells (BY4742) transformed with vectors overexpressing either ubiquitin (WT), K48R ubiquitin, K63R ubiquitin, or empty vector as control.
The best characterized function of ubiquitylation, targeting of proteins to the 26S proteasome for degradation, occurs largely, although perhaps not exclusively, as a consequence of covalent modification of substrates with chains of ubiquitin that contain ubiquitin linked together through K48 (Fang and Weissman, 2004). Other cellular functions are known to involve monoubiquitylation or polyubiquitylation through other lysines of ubiquitin, especially K63 (Mukhopadhyay and Riezman, 2007). To gain insight into the linkages being generated on Fzo1p, wild-type and fzo1Δ cells were transformed with a high copy vector for overexpression of ubiquitin (wild type) or ubiquitin in which either K48 or K63 was mutated to arginine (K48R or K63R). Overexpression of wild-type ubiquitin is known to enhance the steady-state level of substrate ubiquitylation. Incorporation of overexpressed K48R or K63R ubiquitins into chains precludes elongation of polyubiquitin chains linked through these residues (Galan and Haguenauer-Tsapis, 1997). Strikingly, we observed that overexpression of K48R ubiquitin resulted in a downward shift in ubiquitylated forms of Fzo1p, reflecting either multiple mono-ubiquitylation events or short chains “capped” with unextendable K48R ubiquitin (Figure 4C). In contrast, overexpression of wild-type or K63R ubiquitin resulted in an upward shift in ubiquitylated species of Fzo1p, consistent with increased availability of ubiquitin competent for K48 chain elongation. Taken together, these data not only confirm that Fzo1p is ubiquitylated, but also provide strong evidence that the higher molecular weight forms include modification with K48-linked polyubiquitin chains.
Fzo1p Is Degraded by the 26S Proteasome during Vegetative Growth
The results presented thus far establish that Fzo1p is ubiquitylated at the mitochondria in a manner that is dependent on an intact SCFMdm30p ubiquitin ligase, that this ubiquitylation appears to be largely K48-linked in nature, and that this modification strongly correlates with degradation of Fzo1p, because deletion of MDM30 or mutation of its F-box abolishes both ubiquitylation and degradation of Fzo1p. Collectively these observations raise the possibility that Fzo1p turnover during vegetative growth is a consequence of proteasomal degradation.
To test the effect of altered proteasome function on Fzo1p degradation, Fzo1p turnover was assayed by CHX chase in two different proteasome mutant strains (cim3-1 and pre1-1 pre2-2), which we compared with their wild-type isogenic controls. The conditional proteasome mutations either slowed (pre1-1 pre2-2) or completely inhibited (cim3-1) degradation of Fzo1p (Figure 5A). These results strongly suggest that Fzo1p is a target for the ubiquitin-proteasome system.
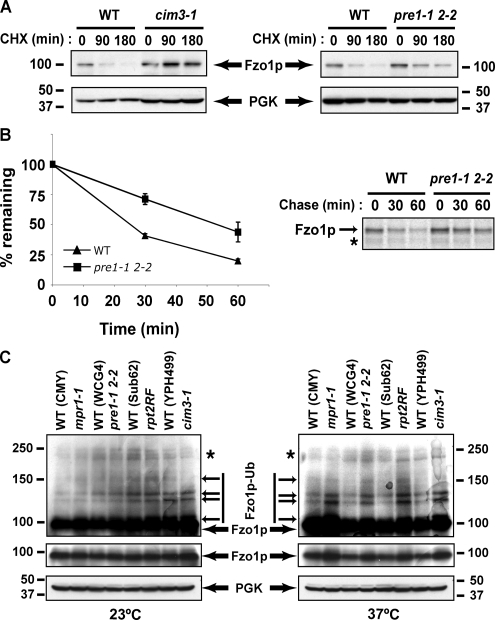
Figure 5.
Fzo1p is degraded by the 26S proteasome. (A) Fzo1p degradation was assessed in pre1-1 pre2-2 and cim3-1 proteasome thermosensitive strains and their corresponding isogenic wild-type strains (see Supplemental Data, Table S1) by treating with CHX. Yeast extracts were prepared at the indicated times and remaining Fzo1p evaluated by immunoblotting. (B) Fzo1-HA turnover was analyzed by 35S pulse-chase metabolic labeling in wild-type and pre1-1 pre2-2 strains. Graph on the left represents quantification of three experiments. A representative experiment is shown on the right. (C) Fzo1p levels were analyzed in four proteasome mutant strains and their isogenic wild-type controls grown at the permissive temperature (23°C) or after a 3-h shift to the restrictive temperature (37°C). Long (top panels) and short (middle panels) exposures of identical anti-Fzo1p blots are displayed. Note relative increase in levels in mutant strains at the restrictive temperature. PGK is utilized as a loading control.
To confirm the CHX chase results, pulse-chase metabolic labeling was performed (Figure 5B). We chose to use the pre1-1 pre2-2 strain, which carries thermosensitive mutations in two of the catalytic subunits of the 20S catalytic core of the proteasome, because it showed only a partial effect on Fzo1p degradation by CHX chase and therefore would be most important to confirm through a different approach. As is evident, mutation of these two core proteasome units doubled the half-life of Fzo1p (30 min in wild-type cells vs. 60 min in pre1-1 pre2-2 cells).
Finally, steady-state levels of unmodified and ubiquitylated Fzo1p were assayed in yeast strains bearing thermosensitive mutations in 26S proteasome subunits from the 19S lid (mpr1-1), 19S base (rpt2RF, cim3-1) or 20S core (pre1-1 pre2-2). In each of these four examples, steady-state levels of unmodified as well as ubiquitylated Fzo1p increased in proteasome mutants relative to the four different wild-type isogenic control strains at 37°C but not at 23°C (Figure 5C, cf. right and left panels). These results confirm the importance of the proteasome in Fzo1p degradation and provide further evidence that ubiquitylated forms of this protein are targeted for degradation. Together, the results presented in Figure 5 establish a clear role for proteasomes in regulating the constitutive turnover of Fzo1 during vegetative growth.
DISCUSSION
The major mechanism for acutely regulating levels of cellular proteins is the UPS. In addition to myriad cytoplasmic and nuclear proteins, the UPS is implicated in the degradation of proteins from other organelles, most notably the ER. ER-associated degradation (ERAD) consists of a complex set of processes that are responsible for ubiquitylating and degrading many transmembrane proteins as well as ER luminal proteins. In contrast to ERAD, a role for the UPS in degradation of outer mitochondrial membrane proteins remains surprisingly obscure.
A number of reports have provided indirect links between the UPS and mitochondria (Fisk and Yaffe, 1999; Sutovsky et al., 1999; Hitchcock et al., 2003; Peng et al., 2003; Thompson et al., 2003; Rinaldi et al., 2004; Altmann and Westermann, 2005; Dürr et al., 2006). The best evidence so far for involvement of the UPS in directly regulating mitochondrial outer membrane proteins is in mammals where a specific E3, MARCHV/ MITOL, is implicated in ubiquitylating two components of the mitochondrial fission apparatus, DRP1 and FIS1, which are the human orthologues of yeast Dnm1p and Fis1p, respectively. However, there is a lack of consensus as to whether this ubiquitylation serves to target these factors for proteasomal degradation or facilitates other nonproteolytic functions (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007).
The literature regarding the mitochondrial mitofusins is even more complicated. During nonvegetative (mating type) growth, degradation of the single yeast mitofusin, Fzo1p, has been suggested to be dependent on the proteasome. However, there is no direct evidence for ubiquitylation or involvement of a specific E3, and a role for Mdm30p has been excluded (Neutzner and Youle, 2005).
The groups of Westermann and Langer have demonstrated, and we confirm herein, that during vegetative growth, degradation and maintenance of normal levels of Fzo1p is dependent on the F-box protein Mdm30p (Fritz et al., 2003), with a specific requirement for an intact F-box (Escobar-Henriques et al., 2006). However, Escobar-Henriques et al. concluded that this degradation of Fzo1p is independent of ubiquitin, the SCF and proteasomes (Escobar-Henriques et al., 2006). This led the authors to conclude that Fzo1p is degraded during vegetative growth by a novel UPS-independent proteolytic pathway that was still dependent on Mdm30p having an intact F-box. The findings presented in the current study lead to a very different conclusion. We provide direct evidence of ubiquitylation of endogenously expressed Fzo1p that is highly suggestive of K48-linked ubiquitin chains. This ubiquitylation is dependent on Mdm30p capable of assembling with other SCF components through its F-box motif. Moreover, both CHX and 35S pulse-chase metabolic labeling experiments implicate the UPS and particularly SCFMdm30p in Fzo1p degradation. Our internally consistent positive findings unequivocally establish an important role for the UPS in determining the fate of Fzo1p. The discrepancy between the conclusions reached in Escobar-Henriques et al. and our findings are difficult to reconcile. However, we certainly cannot discount the possibility that in addition to the UPS other means of degrading Fzo1p could exist including through mitochondrial and other cytosolic proteases as well as by autophagy. Regardless, the clear involvement of the UPS in Fzo1p degradation, established herein, should lay the groundwork for further investigation of the signals that target Fzo1p for degradation and the significance of this degradation in mitochondrial function.
The findings presented in this study are in accord with a paradigm based on cells lacking Mdm30p (Fritz et al., 2003): increased levels of Fzo1p due to failure to regulate the level of this protein results in mitochondrial aggregation and a failure to respire. Despite the requirement for the Mdm30p F-box in Fzo1p degradation found in Escobar-Henriques et al., the same group found that Mdm30 lacking its F-box overexpressed from the CUP promoter surprisingly restored normal mitochondrial morphology in mdm30Δ cells (Dürr et al., 2006). In contrast, we find that, in these cells, expression of F-box mutants of Mdm30p from either the TEF or MDM30 promoter results in a failure to reverse the abnormal aggregated mitochondrial morphology seen in mdm30Δ cells, a finding confirmed by the failure to restore growth on a nonfermentable carbon source (glycerol). As suggested (Dürr et al., 2006), the restoration of mitochondrial morphology obtained with the CUP promoter in Dürr et al. could be a consequence of overexpression of truncated Mdm30 that binds to and inactivates excess Fzo1p. If this is the case, it is unlikely of physiological significance.
The determination that an integral membrane protein of the mitochondrial outer membrane is ubiquitylated while still mitochondria-associated and unambiguously degraded by the proteasome leads us to posit a general UPS-dependent process of mitochondrial-associated degradation (MAD). Analogous to ERAD, ubiquitin ligases intrinsic to the mitochondrial outer membrane, such as MARCHV/ MITOL (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007) and the newly described MULAN (Li et al., 2008), as well as E3s that are recruited to the mitochondrial outer membrane, such as the SCF, will be involved in this process. As with ERAD there is likely to be a high degree of complexity, and we predict that a number of mitochondrial and cytosolic proteins will be implicated in playing roles either in protecting proteins or facilitating their targeting to the UPS and in retro-translocation from mitochondrial membranes through as yet to be established mechanisms. With definitive proof for involvement of the UPS at the mitochondria now established, a number of exciting questions arise including how individual substrates are recognized; the extent to which the UPS might be involved in the fate of proteins in the intramembranous space, the inner mitochondrial membrane, and the mitochondrial matrix; and the degree to which MAD targets misfolded proteins as well as highly regulated normal proteins involved in critical mitochondrial functions such as fusion and fission.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Drs. Abdussalam Azem (Tel Aviv University), Jodi Nunnari (University of California, Davis), Maurits Kleijnen (Harvard Medical School), and Benedikt Westermann (University of Bayreuth, Germany) for their generous gifts of antibodies, expression plasmids, and yeast strains. We thank Dr. Zlatka Kostova, other members of the LPDS, and Dr. Catherine Dargemont for invaluable discussions and comments. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and a grant from the USA-Israel Binational Science Foundation (BSF) to M.H.G. and A.M.W.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0227) on March 19, 2008.
REFERENCES
- Altmann K., Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avaro S., Belgareh-Touze N., Sibella-Arguelles C., Volland C., Haguenauer-Tsapis R. Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast. 2002;19:351–371. doi: 10.1002/yea.838. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J. M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chan D. C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Dürr M., Escobar-Henriques M., Merz S., Geimer S., Langer T., Westermann B. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol. Biol. Cell. 2006;17:3745–3755. doi: 10.1091/mbc.E06-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Weissman A. M. A field guide to ubiquitylation. Cell Mol. Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk H. A., Yaffe M. P. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- Fritz S., Weinbach N., Westermann B. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell. 2003;14:2303–2313. doi: 10.1091/mbc.E02-12-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Peter M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J. M., Wiederkehr A., Seol J. H., Haguenauer-Tsapis R., Deshaies R. J., Riezman H., Peter M. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 2001;21:3105–3117. doi: 10.1128/MCB.21.9.3105-3117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M. H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Heath-Engel H. M., Shore G. C. Mitochondrial membrane dynamics, cristae remodelling and apoptosis. Biochim. Biophys. Acta. 2006;1763:549–560. doi: 10.1016/j.bbamcr.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock A. L., Auld K., Gygi S. P., Silver P. A. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R. J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Martinou J. C., Youle R. J. Which came first, the cytochrome c release or the mitochondrial fission? Cell Death Differ. 2006;13:1291–1295. doi: 10.1038/sj.cdd.4401985. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Sommer T., Pfanner N. Purification of Saccharomyces cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- Moreau V., Galan J. M., Devilliers G., Haguenauer-Tsapis R., Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol. Biol. Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Muratani M., Kung C., Shokat K. M., Tansey W. P. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A., Youle R. J. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Rinaldi T., Pick E., Gambadoro A., Zilli S., Maytal-Kivity V., Frontali L., Glickman M. H. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem. J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckhuber C. Q., Erjavec N., Tinazli A., Hamann A., Nystrom T., Osiewacz H. D. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B. In: Methods in Yeast Genetics. C.S.H., editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. 1986. [Google Scholar]
- Sutovsky P., Moreno R. D., Ramalho-Santos J., Dominko T., Simerly C., Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Thompson W. E., Ramalho-Santos J., Sutovsky P. Ubiquitination of prohibitin in mammalian sperm mitochondria: possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol. Reprod. 2003;69:254–260. doi: 10.1095/biolreprod.102.010975. [DOI] [PubMed] [Google Scholar]
- Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Willems A. R., Schwab M., Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Yonashiro R., et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.