Abstract
GRASP55 is a Golgi-associated protein, but its function at the Golgi remains unclear. Addition of full-length GRASP55, GRASP55-specific peptides, or an anti-GRASP55 antibody inhibited Golgi fragmentation by mitotic extracts in vitro, and entry of cells into mitosis. Phospho-peptide mapping of full-length GRASP55 revealed that threonine 225 and 249 were mitotically phosphorylated. Wild-type peptides containing T225 and T249 inhibited Golgi fragmentation and entry of cells into mitosis. Mutant peptides containing T225E and T249E, in contrast, did not affect Golgi fragmentation and entry into mitosis. These findings reveal a role of GRASP55 in events leading to Golgi fragmentation and the subsequent entry of cell into mitosis. Surprisingly, however, under our experimental conditions, >85% knockdown of GRASP55 did not affect the overall organization of Golgi organization in terms of cisternal stacking and lateral connections between stacks. Based on our findings we suggest that phosphorylation of GRASP55 at T225/T249 releases a bound component, which is phosphorylated and necessary for Golgi fragmentation. Thus, GRASP55 has no role in the organization of Golgi membranes per se, but it controls their fragmentation by regulating the release of a partner, which requires a G2-specific phosphorylation at T225/T249.
INTRODUCTION
In mammalian cells, the Golgi apparatus is composed of 40–100 individual Golgi stacks, which are connected by membrane tubules and localize to the perinuclear region. How and why do Golgi membranes retain their organization during protein transport and completely fall apart in mitosis? What is the significance of Golgi fragmentation during mitosis? We found that inhibiting Golgi fragmentation prevented entry of cell into mitosis (Sütterlin et al., 2002). Golgi fragmentation, therefore, begins in G2 (before entry into mitosis), and the cells have a mechanism to sense this change. Blocking this change activates a signal [checkpoint] and entry into mitosis. This checkpoint seems to be independent of ataxia-telangiectasia mutated/ATM/Rad3-related- dependent inactivation of CDc2 kinase (Sütterlin et al., 2004).
Golgi stacks in plants, Drosophila, and unstacked Golgi cisternae in Saccharomyces cerevisiae, are dispersed throughout the cytosol (Preuss et al., 1992; Ripoche et al., 1994; Nebenfuhr et al., 2000). Interestingly, in Drosophila S2 cells, Golgi stacks are severed by an actin-dependent process in G2, and this event is necessary for entry into mitosis (Kondylis et al., 2007). Several other reagents that block Golgi fragmentation affect entry into mitosis (Sütterlin et al., 2002; Hidalgo Carcedo et al., 2004; Preisinger et al., 2005; Yoshimura et al., 2005; Colanzi et al., 2007; Feinstein and Linstedt, 2007). The cells to regulate mitotic entry, therefore, sense a feature of the overall Golgi organization. What is this feature and what is significance compared with other cellular features that regulate mitotic entry and progression? An understanding of the mechanism of Golgi fragmentation could provide insight into this surprising new role of Golgi membranes.
Our in vitro assay that reconstitutes mitosis-specific Golgi fragmentation revealed the requirement of polo-like kinase (Plk) and the Raf1-mitogen-activated protein kinase kinase (MEK)1 pathway (Acharya et al., 1998; Colanzi et al., 2000, 2003; Sütterlin et al., 2001). The role of MEK1 in mitosis-specific events was a surprise. Even more intriguing is our finding that the well-known substrates of MEK1, extracellular signal-regulated kinase (ERK)1/ERK2 are not involved in this process (Acharya et al., 1998). Our results indicate that MEK1 is differentially modified in mitosis, and we suggest that MEK1 must have new Golgi-associated targets (Colanzi et al., 2003). It was therefore encouraging to find that a novel isoform of ERK, ERK1b/1c (1b, mouse; 1c, humans) has been identified, which is recruited to the Golgi membranes, activated by MEK1, and required for Golgi fragmentation and for entry into mitosis (Abershold et al., 2004; Shaul and Seger, 2006). GRASP55 was identified as a mitogen-activated protein kinase (MAPK) substrate by functional proteomics, and subsequently it was found to be phosphorylated by ERK2 both in vitro and in vivo (Lewis et al., 2000; Jesch et al., 2001). A connection therefore exists between Raf-MEK1 and GRASP55, but whether this is through the conventional ERK2 or its spliced version ERK1b/1c remains unclear.
The Golgi-associated protein GM130 is phosphorylated by Cdc2 kinase, but a direct role of GM130 in mitosis-specific Golgi fragmentation in vivo is not clear (Lowe et al., 1998; Kano et al., 2000; Wang et al., 2005; Yoshimura et al., 2005). Additionally, loss of GM130 in a temperature-sensitive conditional-lethal mutant IdlG cells has no obvious affect on the overall organization of Golgi apparatus (Vasile et al., 2003). It is therefore unclear whether Cdc2 kinase has a role in Golgi fragmentation through GM130. Cdc2 kinase and Polo-like kinase also phosphorylate the Golgi membrane-associated protein GRASP65 (Preisinger et al., 2005; Wang et al., 2003, 2005). Interestingly, GM130 and GRASP65 are required for normal spindle dynamics during mitosis and for the separation of Golgi stacks latterly (Sütterlin et al., 2005; Puthenveedu et al., 2006; Kodani and Sütterlin, 2007; Marra et al., 2007). Thus, the connection between Cdc2 kinase and Plk in Golgi fragmentation, directly, remains unclear.
What changes in the Golgi organization control entry of cells into mitosis? We found that disrupting microtubules that disconnect stacks of Golgi cisternae or by fusing Golgi membranes with the endoplasmic reticulum (ER) alleviated the block in entry imposed by inhibiting Golgi fragmentation (Sütterlin et al., 2004). MEK-dependent phosphorylation of GRASP55, and BARS 50, are reportedly involved in disconnecting lateral connections between Golgi stacks and to permit entry into mitosis (Hidalgo Carcedo et al., 2004; Colanzi et al., 2007; Feinstein and Linstedt, 2007). For a scholarly description of these new developments see a recent review by Catherine Rabouille (Rabouille and Kondylis, 2007).
Here, we report on the role of GRASP55 in the overall Golgi organization and their fragmentation during mitosis. Our findings suggests that although it has no direct role in Golgi organization, its phosphorylation specifically at T225 and T249 is required for Golgi fragmentation and subsequent entry of cells into mitosis. The description of our findings follows.
MATERIALS AND METHODS
Expression and Purification of Recombinant Proteins
GRASP55 constructs were generated by polymerase chain reaction (PCR) using a C6-glioma cDNA library (generated by Yusuke Maeda, Research Institute for Microbial Diseases, Osaka University, Japan). The PCR fragments, full-length GRASP55 (upstream primer 1: 5′-ggctggatccggctcctcgcagagcgtcgag-3′, downstream primer 1: 5′-ccacaagcttttaagaagccccagaagcatttgcatcc-3′), C254 (amino acids 201-454) (upstream primer 2: 5′-ggctggatcccgaatacctacgcgtccctttg-3′, downstream primer 1: 5′-ccacaagcttttaagaagccccagaagcatttgcatcc-3′); C204 (amino acids 251-454), (upstream primer 3: 5′-cctcggatccggagttgagcagagtctgtctgga-3′, downstream primer 1: 5′-ccacaagcttttaagaagccccagaagcatttgcatcc-3′); C150 (amino acids 301-454), (upstream primer 4: 5′-ttatggatccctgatgcctttgtcagcagggc-3′, downstream primer 1: 5′-ccacaagcttttaagaagccccagaagcatttgcatcc-3′); C100 (351-454 amino acids), (upstream primer 5: 5′-ttatggatccaacttacctggcattgcacctctc-3′, downstream primer 1: 5′-ccacaagcttttaagaagccccagaagcatttgcatcc-3′); [201-304] (amino acids 201-304), (upstream primer 2: 5′-ggctggatcccgaatacctacgcgtccctttg-3′, downstream primer 2: 5′-tattaagcttttaactggtaatgtggcagc-3′) were subcloned into the BamHI and HindIII restriction sites of the pQE30 bacterial expression vector (QIAGEN, Valencia, CA). Recombinant proteins were induced to express with 0.5 mM isopropyl β-d-thiogalactoside in either 71-18 (gift from Mike Yaffe, University of California, San Diego) or XL-1-Blue (Stratagene, La Jolla, CA) competent bacterial cells, lysed in lysis buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl, and 10% glycerol) and purified with nickel-agarose beads. The elutions were dialyzed in a low-salt buffer (50 mM sodium phosphate buffer, pH 8.0, 50 mM NaCl, and 10% glycerol) using a Slide-A-Lyzer slide (Pierce Chemical, Rockford, IL). The dialyzed protein was then passed over a DEAE-Sephacel column (BD Biosciences PharMingen, San Diego, CA), and the GRASP55-containing flow-through was collected. Purified proteins were dialyzed against KHM (25 mM HEPES-KOH, pH 7.4, 125 mM potassium acetate, and 2.5 mM magnesium acetate) plus 10% glycerol and concentrated with Microcon spin columns (Millipore, Billerica, MA). It is important to note that all recombinant proteins were purified in the complete absence of detergent.
Generation of Anti-GRASP55 Antibody and Antibody Affinity Purification
After the purification of 6His-tagged GRASP55 using nickel-agarose (QIAGEN), the protein fractions were loaded onto a large preparative gel for additional purification from copurifying contaminants. The GRASP55 band was then cut out of the SDS-polyacrylamide gel and eluted from the gel slices by electroelution and used to inject rabbits. Each bleed was tested for reactivity with recombinant protein and total normal rat kidney (NRK) lysate by Western blot analysis and by immunofluorescence microscopy. The anti-GRASP55 antibody was affinity purified. In brief, an antigen column was generated by coupling approximately 4 mg of recombinant full-length GRASP55 to activated cyanogen bromide beads (Sigma-Aldrich, St. Louis, MO); the column was washed with PBS and incubated with the crude antiserum. Immediately after elution of the antibody with 100 mM glycine, pH 2.5, the eluted sample was brought to neutral pH and dialyzed against PBS containing 10% glycerol in a Slide-A-Lyzer slide (Pierce Chemical). The affinity-purified antibody was then concentrated using Microcon spin columns (Millipore) to a final concentration of 10 mg/ml. Preimmune immunoglobulin G (IgG) was purified by binding the crude preimmune serum to a protein A-Sepharose (GE Healthcare, Chalfont St. Giles, United Kingdom) column. IgG was eluted with 100 mM glycine, pH 2.5, dialyzed, and concentrated as described for the anti-GRASP55 antibody.
Additional Antibodies and Reagents
Antibodies used in this study were obtained from the following sources: GRASP65 and GRASP55 (Malhotra laboratory), GM130 and p230 (BD Biosciences, San Jose, CA), phospo-histone-H3 (Upstate Biotechnology, Charlottesville, VA), actin (AC-15 clone; Sigma-Aldrich), mannosidase II (ManII) (Covance Research Products, Princeton, NJ). The small interfering RNA (siRNA) oligonucleotides (oligos) used in this study were purchased from Dharmacon RNA Technologies (Lafayette, CO) and are as follows: control siRNA (nontargeting siRNA #1; Dharmacon RNA Technologies) and GR55 (target sequence: nncuauucagccuuaucgaaa). To create the signal squence-horseradish peroxidase (SS-HRP)-Flag plasmid (pSS-HRP-Flag), the SS-HRP cDNA sequence was amplified by PCR from a previously described plasmid encoding SS-HRP-KDEL (Connolly et al., 1994) and cloned into pEGFP-N1 (BD Biosciences) by using SmaI–AgeI targets. The enhanced green fluorescent protein (EGFP) cDNA in the vector was removed by AgeI–NotI digestion, and the resulting digested plasmid was ligated with preannealed complementary primers encoding both the Flag epitope and the AgeI–NotI restriction sites. The plasmid pManII-green fluorescent protein (GFP) was created by amplification by PCR of the first 100 amino acids of rat mannosidase II enzyme. This fragment was inserted in frame with GFP between EcoRI and BamHI sites in pEGFP-N1 plasmid (Clontech, Mountain View, CA).
Cell Culture and Transfections
HeLa cells were grown in complete medium consisting of DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (FCS) and l-glutamine at 37°C in 6% CO2 incubator. Knockdown transfections were performed on ∼50–70% confluent HeLa cells by using Lipofectamine 2000 reagent (Invitrogen), according to manufacturer's instructions. Transfection reagents were washed away 5 h after transfection to prevent cell death. One day after the transfection, cells were passaged to either new plates or onto coverslips at the desired confluence, depending on the needs of each experiment. Unless otherwise stated, experiments were performed 3 d after siRNA transfection, when GRASP55 knockdown was found to be optimal.
Immunofluorescence Microscopy
For immunofluorescence microscopy studies, cells were fixed with 4% formaldehyde in PBS, blocked with blocking buffer (2.5% FCS, 0.1% Triton-X-100, and 0.05% sodium azide in PBS). Cells were incubated with primary antibody (as mentioned above) followed by a PBS wash and 30-min incubation in secondary antibody (rabbit or mouse anti-goat Alexa Fluor-488 or-594; Invitrogen). Cells were mounted using Fluor Save Reagent (Calbiochem, San Diego, CA) and visualized by immunofluorescence microscopy. Pictures were taken with a Leica SPE confocal laser-scanning microscope. All pictures were opened in Adobe Photoshop and Illustrator (Adobe Systems, Mountain View, CA).
Fluorescence Recovery after Photobleaching
Forty-eight hours after transfection with siRNA, the cells were plated in 35-mm dishes (MatTek, Ashland, MA). Twenty-four hours later, cells were transfected with plasmid pMannII-GFP by using FuGENE 6 as a transfection reagent and following manufacturer's instructions. Seventy-two hours after the initial siRNA transfection, cells were imaged using a Leica SP5 confocal laser-scanning microscope equipped with an environmental control system set to 37°C and 5% CO2 atmosphere. Cells were imaged on Opti-MEM (Invitrogen) without phenol red containing 10% FCS by using the argon laser line at 488 nm and using a 63× 1.4 numerical aperture PlanAPO oil immersion objective. The sample was observed every 5 s for 50 s, and then the central area of the Golgi complex, labeled with ManII-GFP, was bleached using two passages of the 488-nm argon laser line at full power. Recovery of the fluorescence was followed every 5 s for 500 s. After background correction, the recovery of fluorescence was calculated as the ratio of the average intensity in the bleached area to the average intensity of the total Golgi complex. Normalization was set between the values before the bleach and the first time point after the bleach.
Electron Microscopy
Control and GRASP55-depleted cells were fixed with 1% glutaraldehyde in HEPES buffer, embedded in Epon 812, and processed for electron microscopy. Analysis of thin sections was performed with a Tecnai-12 electron microscope (FEI, Philips, Einhoven, The Netherlands). Images were taken using a ULTRA VIEW charge-coupled device digital camera.
Preparation of Mitotic, Nonsynchronous Cytosol, and the Golgi Fragmentation Assay
NRK cells were grown in complete medium consisting of α-MEM (Invitrogen) containing 10% FCS, l-glutamine, 10 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2 incubator. Cytosol from NRK cells arrested in mitosis or from a nonsynchronous population, at a concentration of 12–14 mg/ml, was prepared as described previously (Acharya et al., 1998). Permeabilization of cells grown on coverslips and the mitotic Golgi fragmentation assay was performed as described previously (Acharya et al., 1998). In brief, cells were treated with 2 mM thymidine for 8–14 h and permeabilized on ice with 30 μg/ml digitonin in KHM buffer (25 mM HEPES-KOH, pH 7.4, 125 mM potassium acetate, and 2.5 mM magnesium acetate). Permeabilized cells were washed with 1 M KCl-containing KHM and incubated with nonsynchronous or mitotic cytosol in the presence of 200 μg/ml anti-GRASP55 antibody, preimmune IgG, recombinant rat GRASP55, or its C-terminal fragments, and an ATP-regenerating system. Incubations were carried out in a volume of 50 μl. After 60 min of incubation at 32°C, cells were fixed and processed for immunofluorescence microscopy by using anti-mannosidase II antibody to visualize the structure of the Golgi. For each experiment, 200–300 cells per coverslip were analyzed for their Golgi morphology.
Microinjections of GRASP55-specific Reagents
Affinity-purified anti-GRASP55 antibody or preimmune-IgG, all at a concentration of 5 mg/ml, was injected into ∼400 aphidicolin-arrested NRK cells (aphidicolin was used at 2.5 μg/ml) roughly 45 min after removal of the S phase block. C100 and the [201-304] peptide microinjections were performed in 200–300 cells at a concentration of 3–5 mg/ml. In both cases, cells were then incubated in complete medium for ∼7.5 h before fixation. For brefeldin A (BFA) treatment, BFA was added 3 h before fixation as determined previously. In all cases, cells were stained with anti-phospho-histone H3 and Hoechst 33342 to visualize the mitotic state and organization of the DNA.
SS-HRP Secretion Assay
Forty-eight hours after transfection with siRNA, the cells were transfected with the SS-HRP-Flag plasmid, by using Lipofectamine 2000 (Invitrogen). Thirty microliters of extracellular media was harvested 72 h after the initial siRNA transfection. HRP activity was measured using enhanced chemiluminescence (ECL) as described previously (Bard et al., 2006).
Vesicular Stomatitis Virus-G Protein (VSV-G) Transport Assay
Forty-eight hours after transfection with siRNA, the cells were transfected with ts045VSV-G-GFP construct and cultured at 40°C for 20 h. Cycloheximide (100 μg/ml) was then added before 2-h incubation at 20°C. After incubation at 32°C for 40 min, the cells were harvested with a cell dissociation buffer (Invitrogen) and fixed with 4% paraformaldehyde. After blocking with PBS containing 1.5% serum and 0.1% sodium azide, the labeling of surface VSV-G-GFP was performed for 30 min of incubation at 4°C with the anti-VSV-G mAb 8G5F11, which is specific for the extracellular domain of VSV-G. After washings with the blocking buffer, the cells were incubated with the secondary allophycocyanin (APC)-labeled anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 min at 4°C. After washing, the cells were analyzed on a FACScalibur flow cytometer (BD Biosciences). The amount of VSV-G present at the cell surface (APC positive) of cells transfected with both siRNA (Cy3 positive) and VSV-G (GFP positive) after subtracting the background was normalized by the GFP intensity.
Phosphopeptide Mapping
Recombinant GRASP55 was incubated for 1 h at 32°C with either interphase or mitotic cytosol. Recombinant GRASP55 was immunoprecipitated with nickel-agarose beads, washed in buffer, and eluted with 100 mM imidazole containing phosphate buffer. Protein in the eluate was precipitated using MeOH/CHCl3 (Wessel and Flugge, 1984) and resuspended in 30 μl of 8.0 M urea/100 mM Tris, pH 8.5. The sample was reduced with dithiothreitol (final concentration 5 mM) for 30 min at 50°C. The sample was cooled and alkylated with iodoacetamide (final concentration 15 mM) for 30 min in the dark at room temperature. The resulting reduced and alkylated sample was divided into three equal fractions and digested with three different proteases as described previously (MacCoss et al., 2002). Briefly, fraction 1 was diluted threefold with 100 mM Tris, pH 8.5/1 mM CaCl2 to bring the urea concentration to ∼2 M. Modified trypsin (Promega, Madison, WI) was added at an estimated enzyme: substrate ratio of 1:50, and the reaction was incubated for 12 h at 37°C. Formic acid was added to a final concentration of 5%, and the sample was stored at −20°C until mass spectrometric analysis. Fraction 2 was diluted threefold with 100 mM Tris, pH 8.5, and subtilisin (Sigma-Aldrich) was added at an estimated enzyme: substrate ratio of 1:50. The reaction was incubated for 3 h at 37°C. Formic acid was added to a final concentration of 5% and the sample was stored at −20°C until mass spectrometric analysis. Fraction 3 was diluted threefold with 100 mM Tris, pH 8.5, and elastase (Roche Diagnostics, Indianapolis, IN) was added at an estimated enzyme: substrate ratio of 1:50. The reaction was incubated for 12 h at 37°C. Formic acid was added to a final concentration of 5% and the sample was stored at −20°C until mass spectrometric analysis. All three digests (fractions 1–3) were pooled and analyzed using multi-dimensional protein identification technology (MudPIT) as described previously (Washburn et al., 2001; MacCoss et al., 2002). Briefly, protein digests were pressure-loaded onto a fused silica triphasic microcapillary column constructed from 100-μm-i.d. fused silica capillary tubing pulled to a 5-μm-i.d. tip (P-2000 laser puller; Sutter Instruments, Novato, CA) and packed with 7-cm 5-μm Aqua C18 material (Phenomenex, Torrance, CA), 3-cm 5-μm Partisphere strong cation exchanger (Whatman, Trenton, NJ), and 3-cm 5-μm hydrophilic interaction chromatography material (PolyLC, Columbia, MD). The column was place in-line with a Surveyor quaternary HPLC pump (Thermo Fisher Scientific, Waltham, MA) and analyzed using a six-step separation as described previously (MacCoss et al., 2002). Peptides were eluted directly into an LCQ-deca mass spectrometer (ThermoFisher) with the application of a 2-kV spray voltage applied distally to the waste. A cycle of one full-scan mass spectrum (400–1400 m/z) was followed by three data-dependent tandem mass spectrometric (MS/MS) spectra at 35% normalized collision energy. MS/MS spectra were analyzed sequentially using the protocol described previously (MacCoss et al., 2002). Briefly, 2TO3 (Sadygov et al., 2002) was used to determine the appropriate charge state of multiply charged peptide mass spectra, delete spectra of poor quality, and identify spectra containing a prominent 98-Da (-H3PO4) neutral loss from the precursor. The filtered MS/MS data were searched using the database searching algorithm Sequest running on a 31-node Beowulf computer cluster against the RefSeq human database (downloaded on 27 February 2004). The resulting Sequest output files were filtered using the program DTASelect (Tabb et al., 2002). The MS/MS data were then researched to consider modifications of +80 on serine, threonine, and tyrosine.
RESULTS
Cloning of GRASP55 and the Production of a GRASP55-specific Antibody
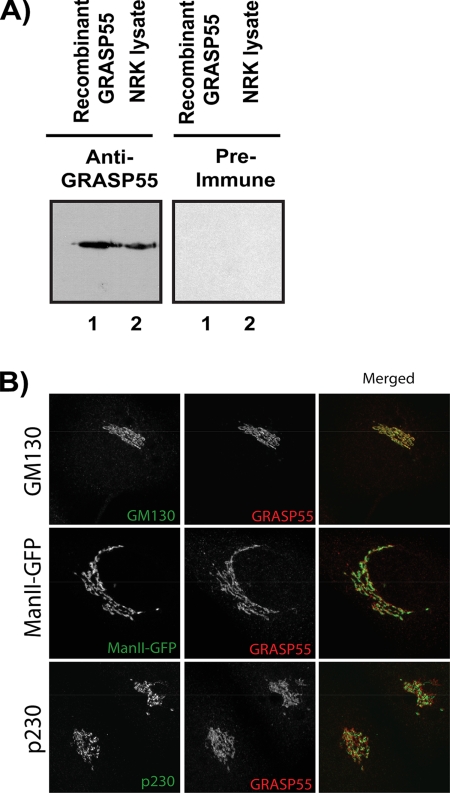
The GRASP55 gene was cloned from a rat C6-glioma cDNA library and transformed into bacterial cells for recombinant protein expression. The full-length epitope-tagged protein was used to generate polyclonal antiserum. The crude anti-GRASP55 serum was affinity-purified on a GRASP55 antigen column and tested by Western blotting recombinant GRASP55 and total NRK cell lysate. The GRASP55 antiserum recognized the recombinant protein and a single polypeptide of 55 kDa in the total NRK cell lysate (Figure 1A). Immunofluorescence microscopy of HeLa cells with the affinity-purified antibody revealed colocalization with GM130 of the cis-Golgi cisternae (Figure 1B). The pattern of GRASP55 also overlaps partially with the cis/medial-Golgi enzyme mannosidase II. However, there was no colocalization with the late Golgi marker protein p230. We conclude that the anti-GRASP55 antibody specifically recognizes GRASP55, which is concentrated on the cis-side of the Golgi stack.
Figure 1.
The GRASP55 antibody recognizes a single-polypeptide in NRK cell extracts. (A) Affinity-purified antiserum and preimmune serum were tested by Western blotting of either purified recombinant protein (lane 1) or total NRK lysate (lane 2). (B) Preimmune or affinity-purified anti-GRASP55 antibodies were tested by immunofluorescence microscopy of NRK cells. The anti-GRASP55 antibody recognizes the Golgi apparatus as revealed by colocalization with bona fide Golgi proteins. (C) HeLa cells were stained with antibodies for Golgi markers (green) GM130 and p230 or transfected with ManII-GFP (cis/medial-Golgi marker). Colocalization with GRASP55 (red) was assessed using confocal laser scanning microscopy. Each picture is the Z-projection of a stack made up of six planes.
GRASP55 Is Not Required for Stacking of Golgi Cisternae
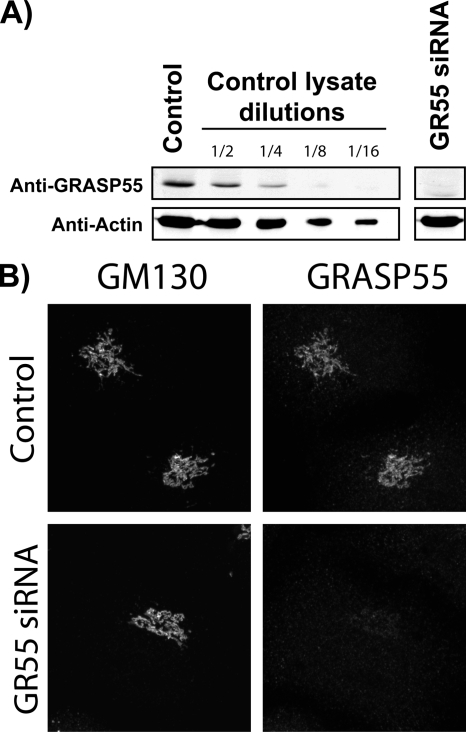
HeLa cells were transfected with two individual GRASP55-specific siRNA oligos (Figure 2A). The reduction in GRASP55 levels by siRNA was quantitated by Western blotting cell lysates and immunofluorescence microscopy with an anti-GRASP55 antibody (Figure 2, A and B). In addition, serial dilution of control cell lysate was Western blotted with anti-GRASP55 to estimate the percentage of residual GRASP55 in siRNA-depleted cells. Our findings revealed that endogenous GRASP55 levels were lowered by >85% compared with control cells. Immunofluorescence microscopy of GRASP55-depleted cells revealed that Golgi membranes retained their localization and organization.
Figure 2.
Depletion of GRASP55 by siRNA. (A) HeLa cells were transfected with either control siRNA (Dharmacon RNA Technologies standard control) or GR55 siRNA. After 72 h, the cells were lysed, and the lysate was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by Western blot with antibodies against GRASP55. Actin was used as a loading control. Efficiency of the knockdown was determined by comparing GRASP55 bands in diluted control lysates to depleted lysates. (C) HeLa cells were transfected with either control siRNA (Dharmacon RNA Technologies standard control) or GR55 siRNA. After 72 h, cells were processed for immunofluorescence microscopy and stained with anti-GRASP55 and anti-GM130 antibodies.
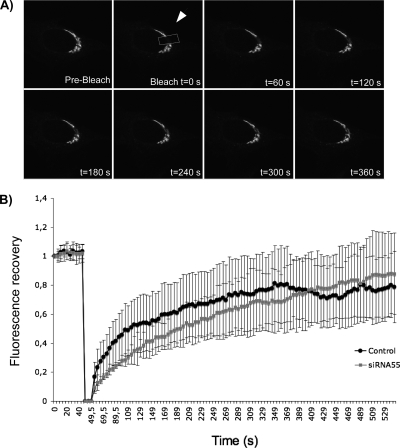
Linstedt and colleagues have recently reported that MEK1-dependent unlinking of Golgi stacks is through GRASP55 and required for entry of cell into mitosis (Feinstein and Linstedt, 2007). To test whether GRASP55 depletion affected lateral Golgi connections, we used the approach of Linstedt and colleagues (Puthenveedu et al., 2006). In brief, HeLa cells were treated with control or GRASP55 siRNA as described above and transfected with ManII-GFP. Because GRASP55 is localized preferentially on the cis face of the Golgi stacks, we tested the distribution of ManII of the cis/medial Golgi cisternae in these experiments. An area of the Golgi (Figure 3A, arrow) was bleached by two pulses of the argon laser followed by visualizing the recovery of fluorescence in the bleached area. 12 control cells were compared with 12 cells depleted of GRASP55 by siRNA and the kinetics of recovery of fluorescence revealed no obvious difference (Figure 3B).
Figure 3.
Lateral mobility of Golgi enzymes in GRASP55 depleted cells. HeLa cells expressing ManII-GFP were treated with control (A) or GR55 siRNA (data not shown). After 72 h, the area of the Golgi indicated by the arrow was bleached. Recovery of fluorescence was followed for 500 s. Representative images of the indicated times are shown. (B) The ratio of fluorescence of the bleached area to the unbleached area of the Golgi was calculated and normalized. The rate of recovery in control siRNA-treated cells (black circles) and GR55-treated cells (gray squares) was plotted.
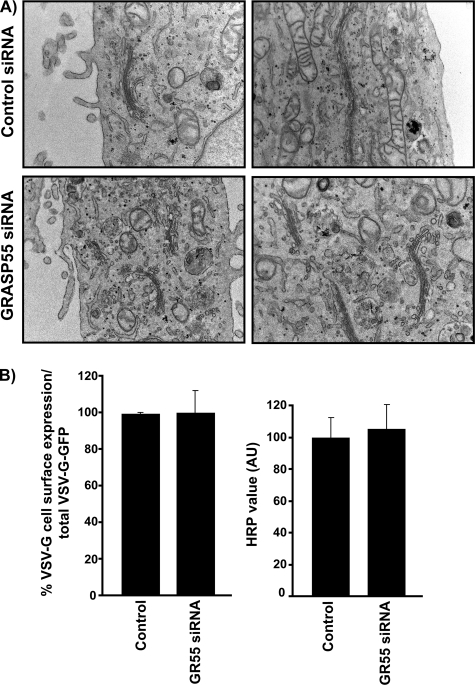
Cells depleted of GRASP55 were also processed for electron microscopy, which revealed no obvious change in the overall organization of Golgi membranes vis-à-vis the number, and the size of cisternae/stack, compared with control cells (Figure 4A).
Figure 4.
GRASP55 is not required for Golgi stacking or protein secretion. (A) HeLa cells were transfected with either control or GR55 siRNA oligo. After 72 h, cells were fixed in 1% glutaraldehyde and processed for electron microscopy. The stacking of Golgi cisternae is not affected by GRASP55 depletion. Cisternae numbers range from three to five cisternae per Golgi stack both in control and GRASP55-depleted cells. (B) Cell surface VSV-G was determined using the anti-VSV-G mAb 8G5F11, which is specific for the extracellular domain of VSV-G. FACS sorting revealed that VSV-G surface expression was not affected in both control and siRNA-transfected cells. Likewise, the secretion of ss-HRP was monitored as described previously (Bard et al., 2006). Secreted HRP was assayed using ECL and found to be at comparable levels at all time points tested in control and GRASP55-depleted cells.
We conclude that under our experimental conditions, >85% knockdown of GRASP55 does not affect the overall organization of Golgi apparatus in terms of cisternal stacking and lateral connections between stacks.
GRASP55 Is Not Required for Protein Secretion
We tested the requirement of GRASP55 in the transport of a soluble protein, SS-HRP (SS-HRP oxidase), and the integral membrane, G protein of the VSV. For SS-HRP secretion, a mammalian expression plasmid encoding the SS of human growth hormone and the enzyme HRP, containing a Flag tag, was used (Bard et al., 2006). On synthesis, SS-HRP-Flag is translocated into the ER where after cleavage of the signal sequence, the soluble HRP-Flag is transported along the secretory pathway. The release of HRP into the medium is quantitated by its enzymatic activity as described previously (Bard et al., 2006). Twenty-four hours after transfection of HeLa cells with the SS-HRP-Flag plasmid, the HRP activity in the extracellular medium was found to be ∼20-fold higher compared with nontransfected cells (data not shown) (Bard et al., 2006). When this secretion assay was carried out in cells previously transfected with GRASP55 siRNA, there was no change in the quantity or kinetics of secreted HRP activity compared with cells transfected with control siRNA (Figure 4B).
We next tested the transport of the temperature-sensitive mutant (ts04) VSV-G protein in GRASP55-depleted cells. Both control and GRASP55-depleted cells were transfected with VSV-G, blocked at the nonpermissive temperature, and then shifted to the permissive temperature to monitor its trafficking along the secretory pathway. Transport of VSV-G was monitored with an antibody that binds the extracellular domain of the protein, and measured against total VSVG-GFP expression by FACSCaliber flow cytometer. Trafficking of VSV-G protein to the cell surface was unaffected in GRASP55-depleted cells (Figure 4B).
GRASP55 Is Involved in Mitosis-specific Golgi Fragmentation
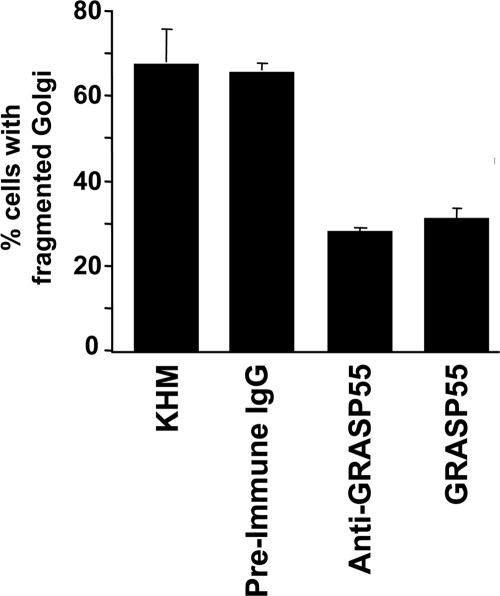
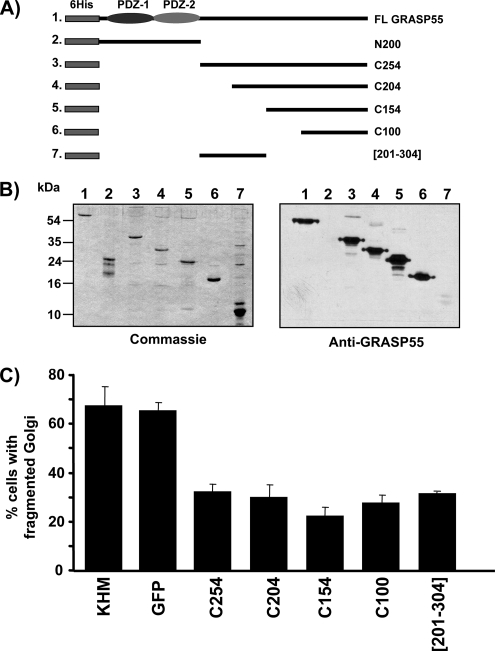
As reported previously, incubation of permeabilized NRK cells with mitotic cytosol and an ATP-regenerating system at 32°C caused fragmentation and dispersal of the Golgi membranes (Acharya et al., 1998). Addition of either the purified His-tagged full-length GRASP55 or the anti-GRASP55 antibody inhibited Golgi fragmentation, compared with purified preimmune IgG (Figure 5). The inhibition was ∼50%, comparable with the strongest inhibitory effects that have been seen with this permeabilized assay. To further ascertain the involvement of GRASP55 in this process, multiple regions of the protein were cloned, expressed, purified, and tested by Western blotting with the anti-GRASP55 antibody (Figure 6A). Western blot of the recombinant purified GRASP55 peptides revealed that the terminal 100 amino acids (aa) (termed C100, aa354-454) contained the anti-GRASP55 antibody-binding site (Figure 6B).
Figure 5.
Anti-GRASP55 antibody and full-length GRASP55 inhibits mitosis-specific Golgi fragmentation in permeabilized cells. An in vitro assay reconstituting Golgi fragmentation by mitotic cytosol described previously (Acharya et al., 1998), was incubated with affinity purified preimmune IgG, anti-GRASP55 antibody, or purified recombinant GRASP55. Cells were fixed and processed for immunofluorescence microscopy by using an anti-giantin or anti-mannosidase II antibody to determine the percentage of cells with fragmented Golgi membranes. The data represents the average of individual experiments (buffer, n = 12; preimmune, n = 4; anti-GRASP55, n = 4; recombinant GRASP55, n = 4).
Figure 6.
The C-terminal domain of GRASP55 is required for Golgi fragmentation. (A) Schematic drawing of the recombinant full-length GRASP55 protein and the peptide fragments N200, C254, C204, C150, C100, and [201-304] used in this study. All constructs contain an N-terminal 6 histidine tag (6His). The N-terminal PDZ domains are shown. (B) Recombinant full-length GRASP55, the N-terminal fragment (N200 represents amino acids 1-200), C254 (amino acids 201-454), C204 (amino acids 251-354), C150 (amino acids 301-354), C100 (amino acids 351-454), and the internal peptide [201-304] (amino acids 201-304) were expressed in bacteria and purified on a nickel-agarose column. The recombinant proteins were analyzed by SDS-PAGE, and they were either Western blotted using the affinity-purified anti-GRASP55 antibody, or with Coomassie Blue to determine total protein levels. Anti-GRASP55 antibody recognizes only the C-terminal region of the protein. Note the low affinity of the antibody against the internal peptide [201-304] representing amino acids 201-304. (C) The recombinant C-terminal fragments C254 (amino acids 201-454), C204 (amino acids 251-354), C150 (amino acids 301-354), C100 (amino acids 351-454), [201-304] (amino acids 201-304), KHM buffer alone, or His-tagged GFP as a control were tested in the in vitro assay reconstituting Golgi fragmentation in NRK cells by mitotic cytosol. The percentage of cells with fragmented Golgi membranes was determined by immunofluorescence microscopy using an anti-giantin or anti-mannosidase II antibody. The data shows an average of four independent experiments per sample.
Because GRASP55-specific antibody inhibited Golgi fragmentation, we tested the C-terminal peptides for their effects on Golgi fragmentation. Surprisingly, we found that each recombinant C-terminal fragment of GRASP55, when added to the semi-intact assay, inhibited fragmentation similar to the full-length protein. Recombinant purified His-tagged GFP had no effect on Golgi fragmentation (Figure 6C). To test whether GRASP55-C100 is necessary and sufficient, an internal peptide not recognized by the GRASP55 antibody, termed [201-304], was included in the in vitro Golgi fragmentation assay. This region, [201-304], is reported to contain a threonine residue, T222 or T225, that is phosphorylated by ERK2 (Jesch et al., 2001). The [201-304] peptide inhibited Golgi fragmentation to the same extent as C100 (Figure 6C). These findings prompt us to conclude that multiple regions within the C terminus of GRASP55 (amino acids 201-454) are involved in Golgi fragmentation. Interestingly, multiple C-terminal peptides of GRASP65 also inhibit Golgi fragmentation and at least two kinases, Plk and Cdc2, are reported to regulate its activity during mitosis (Preisinger et al., 2005; Sütterlin et al., 2001; Wang et al., 2005; Yoshimura et al., 2005).
GRASP55 Contains Two Mitosis Specific Phosphorylation Sites
Linstedt and colleagues have reported that GRASP55 is phosphorylated at the predicted MAP kinase (ERK) consensus site T222 and/or T225 (Jesch et al., 2001; Feinstein and Linstedt, 2007). Overexpression of GRASP55-T222A/T225A delayed entry into mitosis, suggesting the involvement of MAP kinase-dependent phosphorylation of GRASP55 at the G2/M transition (Feinstein and Linstedt, 2007). Our findings reveal that in addition to the peptide [201-304] containing T222/T225, the C100 peptide also inhibited Golgi fragmentation. This suggests that multiple sites within GRASP55 are required for Golgi fragmentation.
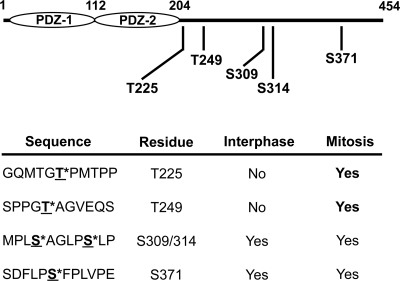
To identify mitosis-specific phosphorylation sites, an equal amount of recombinant His-tagged GRASP55 was incubated with either interphase or mitotic cytosol. Interphase cytosol was prepared from cells arrested in S phase by aphidicolin treatment. Mitotic cytosol was prepared from cells arrested in pre-metaphase by nocodazole treatment. The mitotic cytosol, therefore, contains all kinases that are active from G2 up until pre-metaphase stage of the cell cycle. After 1 h at 32°C, GRASP55 was immunoprecipitated with nickel-agarose beads, washed, and eluted. The samples were digested with protease and analyzed using MudPIT. This procedure revealed only two mitosis specific phosphorylation sites, T225 and T249 in GRASP55 (Figure 7). Surprisingly, C100 peptide did not contain any mitosis-specific phosphorylation sites. We suggest that the C100 peptide inhibits Golgi fragmentation by sequestering kinases/effectors independent of [201-304], and that event might be necessary for the phosphorylation of T225 and T249.
Figure 7.
Phosphopeptide mapping reveals two mitotic specific phosphorylation sites in GRASP55. Full-length recombinant GRASP55 was incubated with either interphase or mitotic cytosol. GRASP55 was isolated by affinity chromatography, digested by proteolysis, and analyzed using MudPIT followed by MS/MS to identify phosphorylated amino acids.
Phosphorylated GRASP55 Regulates Entry of Cells into Mitosis
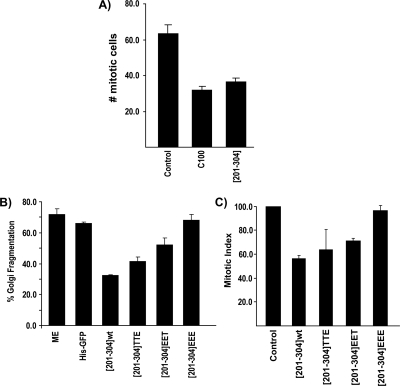
NRK cells were treated with aphidicolin to synchronize cells in S phase. The cells were released from S phase and incubated at 37°C for 5.5–9.5 h. At each time point, cells were fixed and incubated with an anti-phosphohistone H3 antibody and Hoechst 33342 to determine mitotic index as described previously (Sütterlin et al., 2002). The highest mitotic index was observed at 7.5 h post-S phase release, and this time point was used for subsequent experiments. The same procedure was followed with cells microinjected with either the GRASP55 fragment [201-304] or C100. Cells microinjected with either peptide revealed a 50% decrease in mitotic index compared with noninjected cells or cells injected with bovine serum albumin (BSA) as control (Figure 8A). The GRASP55-specific reduction in mitotic index was abrogated in cells treated with BFA to artificially induce Golgi fragmentation (data not shown). This is consistent with our previous findings that fragmentation of the Golgi membranes is required for entry into mitosis (Sütterlin et al., 2002).
Figure 8.
GRASP55 is required for Golgi fragmentation and mitotic entry in a phosphorylation-dependent manner. (A) As reported previously (Sütterlin et al., 2002) NRK cells were arrested in S phase with thymidine. The cells were washed to remove thymidine, and then they were injected with either C100, [201-304], or BSA as a control. Between 5.5 and 9.5 h post S-phase release, cells were fixed and stained with phospho-histone H3 and Hoechst 33342 to determine the number of mitotic cells. For each time point 200 cells were counted. The data shows an average of four independent experiments. The maximum mitotic index was observed at 7.5 h postthymidine release. (B) Wild-type [201-304], [201-304]EET, [201-304]TTE, [201-304]EEE, KHM buffer, or His-tagged GFP were tested in an in vitro assay reconstituting Golgi fragmentation in NRK cells by mitotic extract. The percentage of cells with fragmented Golgi membranes was determined by immunofluorescence microscopy using anti-giantin antibody. (C) Wild-type [201-304], [201-304]EET, [201-304]TTE or [201-304]EEE were injected into post-S phase-arrested NRK cells as described previously (Sütterlin et al., 2002). Cells were fixed at 7.5 h and processed to determine mitotic index compared with control or noninjected cells. The data represents and average of the total experiments (control, n = 14; C100 injected, n = 4; each [201-304] peptide, n = 4).
To determine whether phosphorylation of GRASP55 at T225 and T249 are necessary for Golgi fragmentation and mitotic entry, these sites were mutated within the [201-304] peptide. The mutant constructs (T/A) or (T/E) were tested for affects on Golgi fragmentation in vitro and mitotic entry in vivo. The [201-304] peptides T222E/T225E or T249E when incubated individually in the assay reconstituting Golgi fragmentation by mitotic cytosol inhibited this process. However, the [201-304] peptide T222E/T225E/T249E did not inhibit Golgi fragmentation by mitotic cytosol (Figure 8B). The [201-304] peptides T222A/T225A and/or T249A inhibited Golgi fragmentation (data not shown).
These peptides were also tested for their ability to block entry of cells into mitosis. NRK cells were released from an S phase block and incubated at 37°C for 7.5 h after microinjection with either wild-type peptide or the mutated peptides. Injection of the peptide T222E/T225E/T249E did not inhibit entry of NRK cells into mitosis. The peptides T222E/T225E or T249E, in contrast, were inhibitory (Figure 8C). Similarly, injection of the [201-304] T222A/T225A and/or T249A peptides dramatically reduced the mitotic index (data not shown). Therefore, phosphorylation of GRASP55 at both T225 and T249 is necessary for Golgi fragmentation and entry of cells into mitosis.
DISCUSSION
The pericentriolar Golgi apparatus in mammalian cells is fragmented in mitosis. But when does the process of fragmentation begin? Inhibition of fragmentation and dispersal of the Golgi apparatus from its pericentriolar localization arrests cells in G2. This block can be alleviated by artificially dismantling Golgi membranes with drugs such as nocodazole and brefeldin A (Sütterlin et al., 2002). Therefore, the signal that triggers Golgi fragmentation is activated in early G2, although visibly fragmented Golgi membrane are not evident until late prophase. What is the molecular identity of this signal?
Kinases Involved in GRASP55 Phosphorylation
Linstedt and colleagues found that residues T222/T225, the consensus ERK phosphorylation site, in GRASP55 is recognized by the CDK related phospho-antibody, MPM2. These residues were phosphorylated by ERK2. Furthermore, over expressing GRASP55-T222A/T225A lowered the mitotic index, suggesting the significance of these residues in Golgi fragmentation and mitotic entry (Feinstein and Linstedt, 2007; Jesch et al., 2001). These results are interesting in light of our previous findings that MEK1 (the activator of ERK1 and ERK2) is required for Golgi fragmentation in vitro (Acharya et al., 1998). Surprisingly, however, MEK1-dependent Golgi fragmentation was independent of ERK1 and ERK2 activity (Acharya et al., 1998). A possible resolution to this conundrum comes from the recent findings of Seger and colleagues that a spliced variant of ERK1, called ERK1c, which is localized to the Golgi apparatus by monoubiquitination, is required for Golgi fragmentation (Shaul and Seger, 2006). The substrate specificity of ERK1c is not fully understood; it can, however, phosphorylate the known ERK1 and ERK2 substrates in vitro (Shaul and Seger, 2006).
Our findings reveal that in addition to T225, T249 of GRASP55 is also phosphorylated in a mitosis-specific manner, and both phosphorylations are required for Golgi fragmentation and mitotic progression. Thus, at least two different kinases phosphorylate GRASP55, one of which might be a MAPK (ERK1c).
GRASP55 Is a Scaffold for Effectors That Regulates Different Cellular Functions
It has been proposed that unlinking of Golgi stacks is necessary for mitotic entry (Colanzi et al., 2007; Feinstein et al., 2007). Inhibiting phospholipase A2, and blocking protein transport from ER to the Golgi also disconnect Golgi stacks, but it is not known whether these processes are required for mitotic entry (Chan et al., 2004; Marra et al., 2007). Stacks of Golgi cisternae in Drosophila S2 cells that are unlinked (dispersed) undergo severing by an actin-dependent process, and surprisingly, this event is necessary for entry into mitosis (Kondylis et al., 2007). Surprisingly, depletion of cargo receptor Surf4 together with ERGIC-53 or depleting p25 also causes unlinking of Golgi stacks. GM130 and GRASP65 remain attached to the dissociated Golgi stacks under these conditions (Mitrovic et al., 2008). It is not known whether depletion of cargo receptors arrests cells in G2. Clearly, a large number of diverse proteins are [somehow] required for linking Golgi stacks. What is the common connection between these proteins, unlinking of Golgi stacks and entry into mitosis?
We do not see unlinking of Golgi stacks in cells depleted of GRASP55 or GRASP65 (as reported previously, Sütterlin et al., 2005). If GRASP55 depletion by siRNA does not unlink Golgi stacks, how is it involved in regulating Golgi fragmentation and mitotic entry? We suggest that GRASP55 sequesters a specific effector, which is released upon phosphorylation of GRASP55. The liberated effector is phosphorylated and used for the reactions leading to Golgi fragmentation. Addition of GRASP55 C-terminal peptides inhibits phosphorylation of endogenous GRASP55 thereby preventing the release of its effector and subsequent activation. The net result is a failure in Golgi fragmentation and mitotic entry. In cells depleted of GRASP55, its effector is free for phosphorylation in G2 to initiate Golgi fragmentation. The role of GRASP55 at the Golgi, therefore, is to withhold a protein from participating in Golgi fragmentation until the cells are in G2. Simply put, GRASP55 (and GRASP65) provide a safeguard mechanism to prevent inadvertent Golgi fragmentation, which is lost upon phosphorylation but only in G2.
It has been reported recently that the single GRASP gene in Dictyostelium and Drosophila is required for protein secretion that does not involve the conventional secretory pathway (Kinseth et al., 2007; Schotman et al., 2008). The Drosophila GRASP relocates to the cell surface to mediate trafficking independently of the Golgi membranes (Schotman et al., 2008). We suggest that GRASP proteins mediate different functions through their effectors, which are recruited in a cell cycle- and development-specific manner. The N-terminal postsynaptic density 95/disc-large/zona occludens (PDZ) domains of GRASPs bind different effectors, and the c-terminal region regulates their binding by phosphorylation.
ACKNOWLEDGMENTS
We thank the current and past members of the Malhotra laboratory. We thank Carolan Buckmaster and Dr. James Feramisco (University of California, San Diego [UCSD]) for performing the antibody microinjection experiments. National Institutes of Health grants and a senior investigator award from the Sandler's program for Asthma Research supported the Malhotra laboratory at UCSD.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-0998) on April 2, 2008.
REFERENCES
- Acharya U., Mallabiabarrena A., Acharya J. K., Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 1998;92:183–192. doi: 10.1016/s0092-8674(00)80913-7. [DOI] [PubMed] [Google Scholar]
- Aebersold D. M., Shaul Y. D., Yung Y., Yarom N., Yao Z., Hanoch T., Seger R. Extracellular signal-regulated kinase 1c (ERK1c), a novel 42-kilodalton ERK, demonstrates unique modes of regulation, localization, and function. Mol. Cell. Biol. 2004;24:10000–10015. doi: 10.1128/MCB.24.22.10000-10015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F., et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- Chan D., Strang M., Judson B., Brown W. J. Inhibition of membrane tubule formation and trafficking by isotetrandrine, an antagonist of G-protein-regulated phospholipase A2 enzymes. Mol. Biol. Cell. 2004;15:1871–1880. doi: 10.1091/mbc.E03-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Carcedo C. H., Persico A., Cericola C., Turacchio G., Bonazzi M., Luini A., Corda D. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26:2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Deerinck T. J., Ellisman M. H., Malhotra V. A specific activation of the mitogen-activated protein kinase kinase 1 (MEK1) is required for Golgi fragmentation during mitosis. J. Cell Biol. 2000;149:331–339. doi: 10.1083/jcb.149.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colanzi A., Sütterlin C., Malhotra V. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. J. Cell Biol. 2003;161:27–32. doi: 10.1083/jcb.200208099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly C. N., Futter C. E., Gibson A., Hopkins C. R., Cutler D. F. Transport into and out of the Golgi complex studied by transfecting cells with cDNAs encoding horseradish peroxidase. J. Cell Biol. 1994;127:641–652. doi: 10.1083/jcb.127.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T. N., Linstedt A. D. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol. Biol. Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo Carcedo C., Bonazzi M., Spanò S., Turacchio G., Colanzi A., Luini A., Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Jesch S. A., Lewis T. S., Ahn N. G., Linstedt A. D. Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol. Biol. Cell. 2001;12:1811–1817. doi: 10.1091/mbc.12.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano F., Takenaka K., Yamamoto A., Nagayama K., Nishida E., Murata M. MEK and Cdc2 kinase are sequentially required for Golgi disassembly in MDCK cells by the mitotic Xenopus extracts. J. Cell Biol. 2000;149:357–368. doi: 10.1083/jcb.149.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinseth M. A., Anjard C., Fuller D., Guizzunti G., Loomis W. F., Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Kodani A., Sütterlin C. The golgi protein GM130 regulates centrosome morphology and function. Mol. Biol. Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., van Nispen tot Pannerden H. E., Herpers B., Friggi-Grelin F., Rabouille C. The Golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev. Cell. 2007;12:901–915. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lowe M., Rabouille C., Nakamura N., Watson R., Jackman M., Jamsa E., Rahman D., Pappin D. J., Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- MacCoss M. J., et al. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr, Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M. A. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol. Biol. Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr A., Frohlick J. A., Staehelin L. A. Redistribution of Golgi stacks and other organelles during mitosis and cytokinesis in plant cells. Plant Physiol. 2000;124:135–151. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovic S., Ben-Tekaya H., Koegler E., Gruenberg J., Hauri H. P. The cargo receptors Surf4, ERGIC-53 and p25 are required to maintain the architecture of ERGIC and Golgi. Mol. Biol. Cell. 2008;19:1976–1990. doi: 10.1091/mbc.E07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C., Korner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Franzusoff A., Segev N., Botstein D. Characterization of the Saccharomyces Golgi complex through the cell cycle by immunoelectron microscopy. Mol. Biol. Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- Ripoche J., Link B., Yucel J. K., Tokuyasu K., Malhotra V. Location of Golgi membranes with reference to dividing nuclei in syncytial Drosophila embryos. Proc. Natl. Acad. Sci. USA. 1994;91:1878–1882. doi: 10.1073/pnas.91.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov R. G., Eng J., Durr E., Saraf A., McDonald H., MacCoss M. J., Yates J. R., 3rd Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- Schotman H., Karhinen L., Rabouille C. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell. 2008;14:171–182. doi: 10.1016/j.devcel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Shaul Y. D., Seger R. ERK1c regulates Golgi fragmentation during mitosis. J. Cell Biol. 2006;172:885–897. doi: 10.1083/jcb.200509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C., Hsu P., Mallabiabarrena A., Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Sütterlin C., Lin C. Y., Feng Y., Ferris D. K., Erikson R. L., Malhotra V. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proc. Natl. Acad. Sci. USA. 2001;98:9128–9132. doi: 10.1073/pnas.161283998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L., McDonald W. H., Yates J. R., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile E., Perez T., Nakamura N., Krieger M. Structural integrity of the Golgi is temperature sensitive in conditional-lethal mutants with no detectable GM130. Traffic. 2003;4:254–272. doi: 10.1034/j.1600-0854.2003.00080.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Satoh A., Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P., Wolters D., Yates J. R., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wessel D., Flugge U. I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Yoshioka K., Barr F. A., Lowe M., Nakayama K., Ohkuma S., Nakamura N. Convergence of cell cycle regulation and growth factor signals on GRASP65. J. Biol. Chem. 2005;280:23048–23056. doi: 10.1074/jbc.M502442200. [DOI] [PubMed] [Google Scholar]