Abstract
Few components of the mitochondrial fission machinery are known, even though mitochondrial fission is a complex process of vital importance for cell growth and survival. Here, we describe a novel protein that controls mitochondrial fission. This protein was identified in a small interfering RNA (siRNA) screen using Drosophila cells. The human homologue of this protein was named Mitochondrial fission factor (Mff). Mitochondria of cells transfected with Mff siRNA form a closed network similar to the mitochondrial networks formed when cells are transfected with siRNA for two established fission proteins, Drp1 and Fis1. Like Drp1 and Fis1 siRNA, Mff siRNA also inhibits fission induced by loss of mitochondrial membrane potential, it delays cytochrome c release from mitochondria and further progression of apoptosis, and it inhibits peroxisomal fission. Mff and Fis1 are both tail anchored in the mitochondrial outer membrane, but other parts of these proteins are very different and they exist in separate 200-kDa complexes, suggesting that they play different roles in the fission process. We conclude that Mff is a novel component of a conserved membrane fission pathway used for constitutive and induced fission of mitochondria and peroxisomes.
INTRODUCTION
Mitochondria are dynamic organelles that continually divide and fuse. Mitochondrial fission facilitates the redistribution of mitochondria in response to local changes in the demand for ATP, whereas mitochondrial fusion is needed to exchange mitochondrial DNA and other components that may become damaged over time (Okamoto and Shaw, 2005; Chan, 2006). The rates of fission and fusion are usually balanced, but they do vary between different cell types, and they may respond to environmental changes. There are circumstances in which the rates of mitochondrial fission are greatly accelerated. This occurs when cytochrome c is released from mitochondria during apoptosis (Desagher and Martinou, 2000) and also when mitochondria are depolarized, for example, with the protonophore carbonylcyanide 3-chloro phenyl hydrazone (CCCP) (Ishihara et al., 2003). Induced fission can lead to fragmented mitochondria, whereas mutations in fission proteins lead to a closed network of mitochondria as a result of continuing fusion without fission.
Mitochondrial fission and fusion are controlled by the opposing actions of different dynamin family members on or in mitochondria. The dynamins are large proteins with an amino-terminal guanosine triphosphatase (GTPase) domain followed by a middle domain and a GTPase effector domain (GED) (Praefcke and McMahon, 2004). The GTPase domain is thought to provide mechanical force, but it may also act as a molecular switch, similar to roles of other GTP-binding proteins (Song and Schmid, 2003). The middle domain and the GED of dynamin family members mediate assembly of these proteins into multimeric complexes. The archetypal family member, dynamin, which is required for endocytosis (van der Bliek and Meyerowitz, 1991; van der Bliek et al., 1993), wraps around constrictions at the necks of vesicles that are budding from the plasma membrane (Hinshaw, 2000; Praefcke and McMahon, 2004).
Mitochondrial dynamin proteins are called Drp1 in mammals and Dnm1 in yeast (van der Bliek, 1999). These proteins are largely cytosolic, but cycle on and off of mitochondria as needed for fission (Bleazard et al., 1999; Labrousse et al., 1999; Smirnova et al., 2001). After translocating to mitochondria, they wrap around constricted parts of the mitochondria where they induce a late stage of mitochondrial outer membrane fission (Bleazard et al., 1999; Labrousse et al., 1999). Consistent with this role, yeast Dnm1 was shown to form rings or spirals with a diameter that matches the minimal diameter of a double membrane constriction (Ingerman et al., 2005). Mutations in Dnm1 and Drp1 give rise to a highly interconnected mesh of mitochondrial tubules. This mesh forms because fusion still occurs while fission is blocked (Bleazard et al., 1999; Labrousse et al., 1999; Smirnova et al., 2001). The rates of mitochondrial fission are dramatically increased during apoptosis (Desagher and Martinou, 2000). Fission occurs just before or at the same time as cytochrome c release (Munoz-Pinedo et al., 2006). Mutations in Drp1 delay cytochrome c release, suggesting that Drp1 is proapoptotic (Frank et al., 2001).
Other members of the dynamin family are involved in fusion between mitochondria. Mitochondrial outer membranes are fused by proteins called Fzo1 in yeast (Hermann et al., 1998) and Mitofusins in mammals (Mfn1 and Mfn2) (Chen et al., 2003). These proteins have two transmembrane segments that anchor them in the mitochondrial outer membrane. Mutations in Fzo1 and the Mitofusins give rise to fragmented mitochondria, but this can be reversed by mutations in yeast Dnm1 and mammalian Drp1 (Hales and Fuller, 1997; Hermann et al., 1998; Santel and Fuller, 2001). Mitochondrial inner membranes are fused by dynamin family members called Opa1 in mammals (Alexander et al., 2000; Delettre et al., 2000), EAT-3 in Caenorhabditis elegans (Kanazawa et al., 2008), and Mgm1 in yeast (Shepard and Yaffe, 1999; Wong et al., 2000). Mutations in these proteins also fragment mitochondria and this fragmentation is reversed by mutations in mitochondrial fission proteins, similar to the reversal of outer membrane fusion defects by mutations in mitochondrial fission proteins (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Sesaki et al., 2003; Kanazawa et al., 2008).
Two more components of the mitochondrial fission machinery were identified with genetic screens aimed at reversal of the mitochondrial fusion defects in yeast Mgm1 and Fzo1 mutants (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001). The proteins identified in these screens were called Fis1 and Mdv1. Mdv1 and its close relative Caf4 are cytosolic WD proteins that bind to Fis1 and Dnm1 in yeast (Tieu et al., 2002; Griffin et al., 2005; Bhar et al., 2006; Naylor et al., 2006; Wells et al., 2007; Zhang and Chan, 2007). There are no obvious homologues of Mdv1 in metazoans, but there are metazoan homologues of Fis1. Transfection of mammalian cells with Fis1 small interfering RNA (siRNA) increases the numbers of connections between mitochondria, similar to the connections observed with Drp1 siRNA, whereas overexpression of Fis1 causes mitochondria to fragment (James et al., 2003; Yoon et al., 2003; Stojanovski et al., 2004). Fis1 proteins have two tetratricopeptide (TPR) repeats, which are common protein–protein interaction domains, and a carboxy-terminal membrane-spanning segment that anchors Fis1 in the mitochondrial outer membrane. Mammalian Fis1 and Drp1 bind to each other in vitro, suggesting transient interactions during the fission process (Yoon et al., 2003). A new twist to this story came with the discovery that Fis1 and Drp1 proteins affect peroxisomal fission (Koch et al., 2003; Li and Gould, 2003; Koch et al., 2005; Kobayashi et al., 2007). Peroxisomes are semiautonomous single-membrane organelles that can bud from the endoplasmic reticulum, but they also undergo further growth and fission after their release into the cytosol. Utilization of Fis1 and Drp1 suggests that the mechanisms of peroxisome and mitochondrial fission are very similar if not identical (Schrader and Yoon, 2007).
Mitochondrial fission and fusion are controlled by several regulatory mechanisms. Drp1 is activated by cyclin-dependent kinase 1/cyclin B-mediated phosphorylation during mitosis (Taguchi et al., 2007) and inactivated by cAMP-dependent protein kinase (PKA) in quiescent cells (Chang and Blackstone, 2007; Cribbs and Strack, 2007). Reversal of PKA phosphorylation by the calcium-dependent phosphatase calcineurin triggers mitochondrial fission (Cribbs and Strack, 2007). These newly discovered connections among cAMP, calcium, and the actions of Drp1 are gratifying, because it has long been known that these molecules influence the rates of mitochondrial fission (Bereiter-Hahn and Voth, 1994). Mitochondrial fission and fusion are also regulated by ubiquination as shown with F-box proteins that determine the amounts of Fzo1 in yeast (Fritz et al., 2003; Kondo-Okamoto et al., 2006) and an E3 ubiquitin ligase that controls mitochondrial fission in mammalian cells (Karbowski et al., 2007). In addition, mitochondrial fission might also be regulated by sumoylation in mammalian cells (Zunino et al., 2007), but the targets of sumoylation and ubiquitination have not yet been unequivocally determined.
It is clear from the above-mentioned description that mitochondrial fission is a complex process that follows a sequence of well-defined stages. These stages include the activation of cytosolic Drp1 by calcineurin or other cytosolic signal transduction proteins and recruitment of Drp1 to spots on mitochondria, most likely through the actions of Fis1. However, the typical diameters of mitochondria (500–1000 nm) are too large to accommodate Drp1 rings (100 nm, as determined with yeast Dnm1) (Ingerman et al., 2005), suggesting that a separate constriction process is needed to reduce the mitochondrial diameter before Drp1 can form rings. Once this constriction occurs and Drp1 rings are assembled, those rings will sever the mitochondrial outer membrane through GTP hydrolysis. Drp1 then remains attached to one of the newly formed mitochondrial tips and slowly disassembles before returning to the cytoplasm (Labrousse et al., 1999). Considering that other membrane-trafficking processes, such as endo- and exocytosis, make use of many more proteins (Pfeffer, 2007), it seems likely that all of these steps require additional, as yet unknown proteins. We sought new mitochondrial fission and fusion proteins using a high-throughput screen of Drosophila siRNAs. With this approach, we identified a novel tail-anchored protein that controls mitochondrial fission. This protein is conserved in metazoans. It affects apoptosis and peroxisome fission, but it is in a complex that is separate from other known fission proteins, suggesting that it acts independently.
MATERIALS AND METHODS
Nucleic Acids
Human Mitochondrial fission factor (Mff) cDNA was cloned with reverse transcription-polymerase chain reaction (RT-PCR) by using RNA isolated from HeLa cells. The PCR product was cloned into pCRII-TOPO (Invitrogen, Carlsbad, CA) and sequenced to rule out mutations introduced by PCR. Our cDNA is derived from isoform 8, which lacks exons 5, 6, and 8 (Figure 1). The amino-terminal green fluorescent protein (GFP) fusion was made by recloning this Mff cDNA behind the GFP sequences of pEGFP-C1 (Clontech, Mountain View, CA) with PCR. A fusion construct lacking the carboxy-terminal transmembrane domain was made with a 3′ PCR primer that removes the last 20 amino acids of Mff. A 5XMYC epitope-tagged construct was made by cloning the Mff PCR fragment after the first five myc tags of pCS2+MT by using NcoI and XhoI sites. All constructs were resequenced. pcDNA3.1 myc::Mfn2(K109T) and pEGFP-N1::Mfn1(K88T) were kindly provided by Ansgar Santel (Atugen, Berlin, Germany).
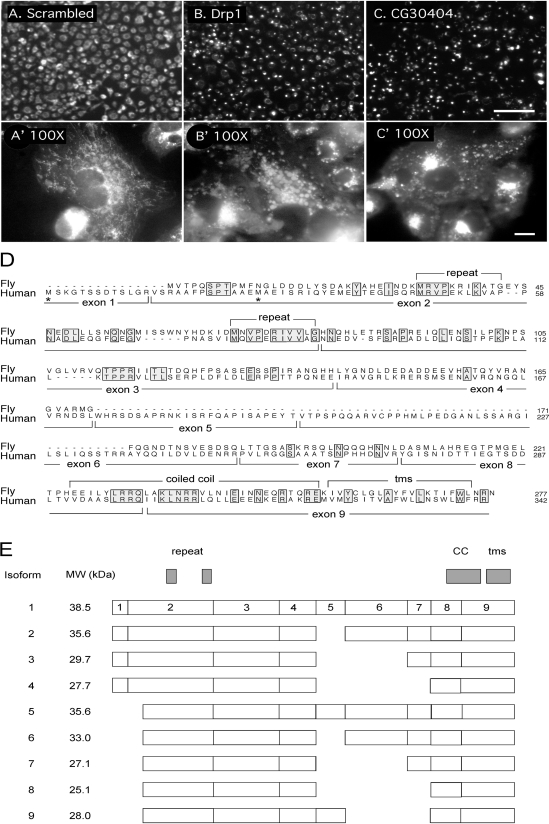
Figure 1.
Identification of an Mff homologue in a Drosophila siRNA screen. (A and A′) Untransfected Drosophila S2 cells stained with MitoTracker. (B and B′) Cells transfected with Drosophila Drp1 siRNA. (C and C′) Cells transfected with CG30404 siRNA. A, B, and C are images taken with a 20× lens in the primary screen (bar, 100 μm), and A′, B′, and C′ are images taken with a 100× lens in the secondary screen (bar, 10 μm). (D) Alignment of the Drosophila CG30404 protein with Mff, which is encoded by C2ORF33. Positions of exon boundaries of the human gene, as deduced from alignment with genomic sequences, are shown below the sequence and key features of the proteins are shown above. Alternative translation start sites identified in different cDNAs are marked with asterisks. (E) Schematic drawing of domains and splice variants identified with different cDNAs encoding Mff. The numbering refers to the exons shown in the alignment in D. The National Center for Biotechnology Information shows two additional isoforms, but those are most likely incomplete or aberrant clones, because they are exceedingly short and represented by single ESTs.
Oligonucleotides for siRNA were made by Sigma-Proligo (The Woodlands, TX). Two pairs of Mff siRNA oligonucleotides were tested: 5′-CGCUGACCUGGAACAAGGAdTdT-3′ for exon 2 and 5′-CUAAAUAGACGUCUACAACdTdT-3′ for exon 8. The results that are shown were obtained with the first pair, but the second pair was equally effective. A scrambled sequence was used as negative control (5′-AGGAACAAGGUCCAGUCGC-3′). Pairs of Drp1 and Fis1 siRNA oligonucleotides were based on the sequences 5′-GCAGAAGAAUGGGGUAAAU-3′ and 5′-AGGCCUUAAAGUACGUCCG-3′, respectively.
For expression analysis, a blot with RNA from a range of human tissues (OriGene, Rockville, MD) was hybridized with 32P labeled Mff cDNA prepared with a Random Primed StripAble DNA Probe Synthesis kit (Ambion, Austin, TX). Hybridization conditions were as specified in the manufacturer's protocol. Hybridizing bands were detected with a Typhoon phosphorimager (GE Healthcare, Chalfont St. Giles, United Kingdom).
Antibodies and Other Reagents
Rabbit polyclonal antibodies against Mff were produced by cloning human Mff cDNA into pET21d, which introduces a carboxy-terminal 6xHis tag. Bacterially expressed protein was purified with nickel-nitrilotriacetic acid agarose using 8 M urea. The protein was further purified with preparative SDS-polyacrylamide gel electrophoresis (PAGE) gels and then used to immunize rabbits. Anti-Fis1 antibodies were purchased from Alexis Laboratories (San Diego, CA), α-tubulin antibodies were from Sigma-Aldrich (St. Louis, MO), Tom20 and c-myc/9E10 were from Santa Cruz Biotechnology (Santa Cruz, CA), cytochrome c antibodies were from BD Biosciences PharMingen, prohibitin antibodies were from RDI (Flanders, NJ), and DLP1 antibodies were from BD Biosciences Transduction Laboratories (Lexington, KY). Secondary antibodies and horseradish peroxidase conjugates were from Pierce Chemical (Rockford, IL). Fluorescent-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA).
Mito-Tracker Red CMX-Ros (Invitrogen) was used at a final concentration of 20 nM. z-VAD-fmk (BIOMOL Research Laboratories, Plymouth Meeting, PA) was used at a final concentration of 25 μM. Staurosporine (Tocris Cookson, Ellisville, MO) was used at a final concentration of 1 μM. Actinomycin D (Sigma-Aldrich) was used at a final concentration of 10 μM. CCCP (Sigma-Aldrich) was used at a final concentration of 2 μg/ml.
Cell Culture and Transfections
HeLa cells and human embryonic kidney (HEK)293T cells were grown with DMEM and 10% fetal calf serum. HEK293 cells were transfected with Ca-phosphate precipitation. HeLa cells were transfected using Lipofectamine 2000 and Opti-MEM (Invitrogen). Cells were transfected with siRNA duplexes at a concentration of 100 pM, and then they were allowed to grow for an additional 3 d before analysis. For cotransfections with overexpression constructs, the siRNA-transfected cells were split after 3 d and retransfected with 50 pM siRNA oligonucleotides and the expression plasmids. These cells were analyzed by immunofluorescence 18 h later. The effectiveness of siRNA was also monitored by RT-PCR and Western blotting.
Drosophila DS2R+ cells were kindly provided by Buzz Baum (University College London) and Shin-ichi Yanagawa (University of Kyoto) (Yanagawa et al., 1998). These cells were grown with Schneider's medium (Invitrogen) and 10% fetal calf serum at 25°C. The primary screen was done in 384-well plates with clear bottoms and black walls. Two sets of 62 plates containing 0.25 μg of double-stranded RNA (dsRNA) per well were provided by the Drosophila RNAi Screening Center (DRSC; http://flyrnai.org). Each well was seeded with 104 DS2R+ cells in 10 μl of Schneider's medium. Plates were then incubated for 30 min at 25°C, followed by addition of 30 μl of Schneider's medium with 10% fetal bovine serum, penicillin, and streptomycin. These plates were sealed and incubated for 3 d at 25°C. The culture medium was then replaced with 50 μl of fresh medium with 20 nM MitoTracker followed by a 30-min incubation at 25°C, washes with phosphate-buffered saline (PBS), and fixation with 50 μl of chilled methanol:acetone (1:1) for 5 min at −20°C. The fixed cells were washed with PBS, and mounting medium (FluorSave; Calbiochem, San Diego, CA) with 0.3 μM 4,6-diamidino-2-phenylindole and sodium azide was added to each well. Mitochondria were observed with a 20× objective on an Axiovert 200M microscope (Carl Zeiss, Thornwood, NY). Abnormal mitochondrial distributions were classified clumped, punctate, and fragmented. Genes that gave rise to abnormal mitochondrial distributions in two independent wells, and they had no obvious nonmitochondrial functions (e.g., transcription, translation) based on their annotations were retested in a secondary screen. Double-stranded RNA was remade from PCR templates provided by the DRSC. The effects on mitochondrial morphology were tested with the same protocol as in the primary screen except that cells were plated in larger petri dishes with glass bottoms or on glass coverslips. The mitochondria could then be examined with a 100× oil immersion objective.
Immunofluorescence
Cells were grown on glass coverslips, and then they were fixed for 10 min at room temperature with prewarmed 3.7% formaldehyde. Where indicated, 20 nM MitoTracker Red was added 30 min before fixation. Cells were permeabilized for 5 min with 0.2% Triton X-100 in phosphate-buffered saline, blocked for 15–30 min with 1% bovine serum albumin in PBS, followed by an overnight incubation with primary antibodies in blocking buffer, three washes with PBS, and 1 h at room temperature with secondary antibodies. Coverslips were mounted on glass slides with FluorSave (Calbiochem). To quantify apoptosis with Hoechst, cells were grown on coverslips, washed three times with PBS, fixed with 3.7% paraformaldehyde, and stained with Hoechst, before counting apoptotic pycnotic nuclei with fluorescence microscopy. Images were acquired with a 100× oil immersion objective on a Zeiss Axiovert 200M microscope with an ORCA ER charge-coupled device camera (Hamamatsu, Bridgewater, NJ).
Biochemical Procedures
Fresh bovine brain (50 g) was minced in 250 mM sucrose, 10 mM Tris-HCl, pH 7.5, 1 mM EGTA, and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) and homogenized with a Dounce homogenizer in a final volume of 250 ml. This crude extract was centrifuged for 15 min at 1000 × g. The low-speed supernatant (S1) was recentrifuged for 15 min at 10,000 × g to yield medium-speed supernatant (S2) and pellet (P2) fractions. The P2 fraction, which contains mitochondria and lysosomes, was washed twice by resuspending and repelleting in homogenization buffer. Volume equivalents of S2 and P2 fractions were analyzed with 15% SDS-PAGE and Western blotting. Protease protection experiments were done with P2 fractions resuspended in homogenization buffer without protease inhibitors (50 μg of protein in 100 μl). Trypsin was added at the specified concentrations, and the samples were incubated for 30 min on ice. Proteolysis was then stopped with 30-fold excess soybean trypsin inhibitor, and the samples were analyzed with SDS-PAGE and Western blotting. Blots were incubated with primary antibodies and developed with a horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence reagents (GE Healthcare). For Blue Native Gel electrophoresis (BNGE), the HeLa cells transfected with siRNA were lysed with 0.1% N-dodecyl-β-d-maltoside HeLa and fractionated with a 6–16% Blue Native Gel (Schagger and von Jagow, 1991). The gels were blotted and probed with the antibodies as described above. Where indicated, expression levels were determined with densitometry using a Personal Densitometer SI and ImageQuant software (GE Healthcare).
RESULTS
RNA Interference (RNAi) Screen for Mitochondrial Morphology Defects in Drosophila DS2R+ Cells
We screened the Drosophila siRNA collection at Harvard (Echeverri and Perrimon, 2006) for novel proteins that affect mitochondrial morphology. Drosophila DS2R+ cells were transfected with siRNAs representing 22,000 transcripts in Drosophila. Abnormal morphologies were detected by staining the transfected cells with MitoTracker. Details of the screening protocol and the controls are given in Materials and Methods. The collection of Drosophila siRNAs was screened in duplo, yielding ∼500 genes that affect mitochondrial morphology. Genes with annotations suggesting an indirect effect were not further considered, nor were genes without human homologues or with potential off-target dsRNAs (Echeverri and Perrimon, 2006). The remaining 15 genes were retested by transfecting Drosophila DS2R+ cells with siRNA in individual culture dishes. Their names and Gene Ontology annotations are given in Supplemental Table 1. Three of these proteins are tubulins, and three others are associated with microtubules (lis-1/PAFAH1B1, nudE/NDE1, and par-1/MARK3), consistent with the effects that perturbations of microtubule dynamics have on mitochondrial morphology. We also found a PIP kinase (sktl/PIP5K1A), a Rab GTPase activation protein (CG9339/TBC1D24), a nucleocytoplasmic transporter (sbr/NXF1), a subunit of respiratory complex I (CG12203/NDUFS4), and some proteins of unknown function (CG3625/AIG1 and mys/ITGB1). Some of these proteins might directly affect mitochondrial morphology, but their siRNA phenotypes were variable and not as strong as Drp1 siRNA.
There was, however, one gene that showed a particularly severe and reproducible phenotype with perinuclear clustering of mitochondria, distinct from the even distribution of mitochondria in untransfected cells, but similar to the clustering of mitochondria in Drp1 siRNA transfected cells (Figure 1, A–C). This gene, which was first called CG30404, but recently renamed Tango11 (Bard et al., 2006), was chosen for further analysis. The predicted amino acid sequence of CG30404/Tango11 is conserved in other metazoans; it contains a domain of unknown function named DUF800. The remainder of our analysis was done with the human homologue of CG30404/Tango11, because more reagents are available for cell biological analyses of mammalian cells.
Mff, Encoded by C2orf33, Is the Human Homologue of Drosophila CG30404/Tango11
A BLAST search shows that humans have two proteins with substantial homology to CG30404/Tango11. The first protein was named FATE, because it is specifically expressed in fetal and adult testis, but the homology to this protein is limited to its carboxy-terminal half. Moreover, pilot experiments with FATE siRNA showed no effect on mitochondria in HeLa cells or other human cell lines commonly used in our laboratory (data not shown), as was expected because FATE is testis specific (Olesen et al., 2001). The second protein, encoded by C2ORF33 and renamed Mff, shows homology over the entire length of the protein (Figure 1D). The expression pattern of Mff was determined with an RNA blot representing a range of human tissues (Supplemental Figure 1). This blot shows that Mff is highly expressed in heart, kidney, liver, brain, muscle, and stomach and at low levels in other tissues. Because this pattern is compatible with a general mitochondrial function, we focused our analysis on this protein.
The main features of human Mff and Drosophila CG30404/Tango11 sequences are a short amino-terminal repeat, a middle segment with strong coiled coil forming propensity, and a carboxy-terminal hydrophobic segment that most likely serves as membrane anchor. The Mff gene encodes at least nine different isoforms represented by multiple expressed sequence tags (ESTs) in GenBank (Figure 1E). These isoforms are generated by the presence or absence of exon 1 and various combinations of exons 5, 6, and 7, which encode the middle part of the protein. The presence or absence of exon 1 most likely results from the use of alternative promoters. Isoforms lacking exon 1 have an alternative translation initiation codon embedded in exon 2. Alternatively spliced exons 5, 6, and 7 encode parts of the protein that are not conserved in Drosophila. However, other mammalian species have largely identical patterns of alternative splicing, suggesting that at least for mammalian cells sequence variations caused by the presence or absence of exons 5, 6, and 7 are functionally important.
Localization of Mff to Mitochondria
To determine the subcellular localization of Mff, we generated an antibody by immunizing rabbits with bacterially expressed Mff protein. Immunofluorescence analysis with this antibody shows colocalization with MitoTracker (Figure 2, A–C). Transfection with an amino-terminal myc-tagged version of Mff also shows mitochondrial localization (Figure 2, D–F). The mitochondrial staining detected with Mff antibody increases when cells are transfected with an Mff expression construct and decreases when they are transfected with Mff siRNA, confirming that staining with our Mff antibody reflects Mff localization (data not shown). The Mff antibody is also specific on Western blots, because the bands on blots are uniformly diminished in siRNA-transfected cells (Figure 3E). Transfection with a construct that encodes Mff with deletion of the carboxy-terminal hydrophobic segment gives rise to diffuse cytosolic staining, showing that the hydrophobic sequence is required for localization to mitochondria (Supplemental Figure 2). We conclude that Mff is localized to mitochondria and that this localization requires the carboxy-terminal transmembrane segment.
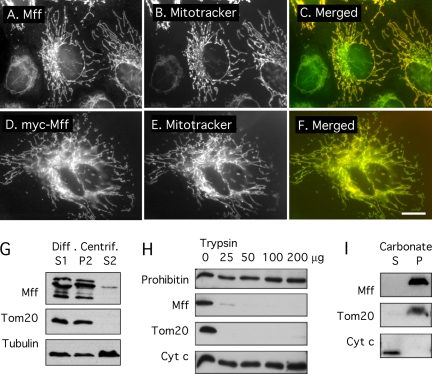
Figure 2.
Fluorescence and biochemical analysis of Mff localization. (A–C) Immunofluorescence of endogenous protein in HeLa cells. (D–F) Overexpression of myc-tagged Mff. The construct used for this experiment encodes isoform 8, which lacks exons 5, 6, and 7 (Figure 1). A and D show Mff or myc antibody staining, B and E show MitoTracker staining, and C and F show merged images with Mff or myc in green and MitoTracker in red. Bar, 10 μm. (G) Western blots of differential centrifugation fractions prepared from HeLa cells. Mff is present in the low speed supernatant (S1) and the medium speed pellet (P2), which contain mitochondria as shown with Tom20 antibody. Tubulin, which was used to track soluble proteins, is also present as a contaminant in the P2 fraction, but very little Mff is present in the S2 fraction, consistent with mitochondrial localization. The Mff antibody detects bands ranging in size from 25 to 39 kDa, most likely corresponding to different splice variants. (H) Protease protection experiment using the mitochondrial (P2) fraction from bovine brain to determine the topology of Mff. Our Mff antibody detects only a single band of 38-kDa in brain extracts, similar in size to the top band in HeLa cells (see G), suggesting that the repertoire of Mff isoforms is more limited in brain than it is in HeLa cells. The P2 fraction was subjected to increasing amounts of trypsin. Proteins that are exposed to the cytosol, like Tom20, are digested at the lowest concentrations, whereas proteins that are protected by membrane, such as the mitochondrial intermembrane space proteins prohibitin and cytochrome c are still protected at the highest concentration. Solubilization with detergents was used to verify that trypsin is able to digest prohibitin and cytochrome c were it not for protection by membrane (data not shown). Together, these data show that Mff is exposed to the cytosol. (I) Alkaline extraction shows that Mff is anchored in membrane. The P2 fraction from bovine brain extracts was resuspended in carbonate buffer, pH 11.5. Membranes were pelleted by centrifugation at 100,000 × g. Tom20 serves as marker for the membrane fraction, and cytochrome c as marker for the cytosolic fraction. These experiments show that Mff cofractionates with mitochondria, that it is exposed to the cytosol, and that it is a membrane-anchored protein.
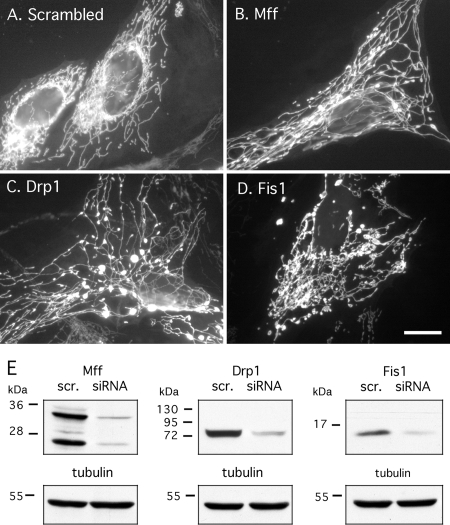
Figure 3.
Effects of Mff siRNA on mitochondrial morphology. (A) HeLa cell transfected with scrambled Mff siRNA oligonucleotides and stained with MitoTracker. These cells show wild-type mitochondrial morphologies. (B) HeLa cell transfected with Mff, (C) with Drp1 siRNA and (D) with Fis1 siRNA oligonucleotides. These transfected cells all show highly connected mitochondria, consistent with defects in mitochondrial fission. Bar, 10 μm. (E) Western blots showing protein levels in siRNA-transfected cells. The blots show reduced levels in HeLa cells transfected with Mff, Drp1 or Fis1 siRNA, in comparison with scrambled (scr.) controlled. The Mff antibody shows two prominent bands (25 and 35 kDa) and several fainter bands that may correspond to different isoforms produced by alternative splicing. Tubulin serves as loading control. The levels of Mff are reduced by 88%, of Drp1 by 93%, and of Fis1 by 82% as determined by densitometry.
To further analyze Mff localization, we fractionated bovine brain extracts with differential centrifugation. Mitochondria and other heavy organelles were enriched in the heavy membrane fraction (P2) as shown with Tom20, which serves as a mitochondrial marker (Figure 2G). This fraction contained almost all Mff protein. The topology of Mff on the membrane was determined with a protease protection experiment using increasing amounts of Trypsin. Mff was digested by trypsin at concentrations that affect the mitochondrial outer membrane protein Tom20, but not the intermembrane space protein cytochrome c or the inner membrane protein prohibitin (Figure 2H). These results show that a substantial part of Mff is exposed to the cytosol, similar to the topology of other tail-anchored proteins, such as Fis1 and Tom20. To verify that Mff truly is membrane anchored, HeLa cell homogenates were subjected to alkaline extraction (pH 11.5). Mff was found almost exclusively in the pellet fraction, as was a known integral membrane protein (Tom20), but not cytochrome c (Figure 2I), from which we conclude that Mff is firmly anchored in membrane. Mff thus belongs to the growing number of tail-anchored membrane proteins.
Mff siRNA Produces Interconnected Mitochondria and Tubular Peroxisomes
To determine the effects of Mff loss of function, HeLa cells were transfected with siRNA oligonucleotides. We tested two independent pairs of oligonucleotides, targeting sequences that are present in all isoforms (exon 2 and exon 8). The two pairs of oligonucleotides were similarly effective. Results are shown with the first pair (exon 2), which reduces the protein levels of Mff by ∼88% (Figure 3E). MitoTracker staining of cells transfected with Mff siRNA reveals highly interconnected mitochondrial networks. The networks are strikingly similar to the mitochondrial networks in cells transfected with Drp1 siRNA (Figure 3, B and C). The mitochondria of Mff siRNA-transfected cells had fewer free ends than those of Fis1 siRNA (Figure 3D), even though expression levels of both proteins were reduced to similar extents (Figure 3E). These phenotypes were observed in 94 ± 5% of cells (mean percentages and SD determined with 350–400 cells in each of four independent transfection experiments). We conclude that Mff siRNA affects mitochondrial morphology in ways that are indistinguishable from Drp1 siRNA and stronger than the effects of Fis1 siRNA.
Because Fis1 overexpression induces mitochondrial fission (James et al., 2003; Yoon et al., 2003), we tested whether Mff overexpression also induces fission by transfecting HeLa cells with an Mff expression construct. The percentage of untransfected cells with fragmented mitochondria is 7.7 ± 1.7%, whereas the percentage of transfected cells with fragmented mitochondria is 6.0 ± 3.8%, suggesting that Mff overexpression does not induce fission (mean percentages and SD were determined with 100 200 cells in each of 3 independent transfection experiments). These results are different from those obtained previously with Fis1 overexpression, which does cause fission (James et al., 2003; Yoon et al., 2003), but similar to results obtained with Drp1 overexpression, which does not cause fission (Smirnova et al., 2001). It seems likely that Mff and Drp1 are not rate limiting for fission or are more tightly controlled than Fis1.
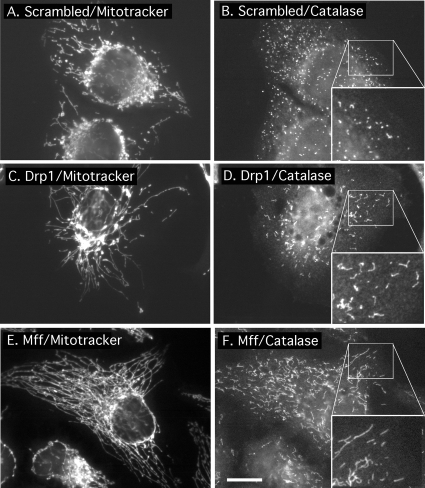
Because Fis1 and Drp1 were previously shown to affect peroxisomal morphologies as well, we investigated whether Mff siRNA also affects peroxisomal morphology. The effect on peroxisomes was determined by immunofluorescence with an antibody raised against the peroxisomal protein catalase and counterstaining with MitoTracker to detect mitochondria. Peroxisomal staining in untransfected cells is typified by diffuse punctae (Figure 4B). As shown previously, these punctae are converted to short tubules by transfection with Drp1 siRNA (Figure 4F) (Koch et al., 2003; Li and Gould, 2003). We observe similarly tubular peroxisomes upon transfection with Mff siRNA (Figure 4D). These phenotypes were observed in 84 ± 6% of cells (mean percentages and SD were determined with 350 cells in 3 independent transfection experiments). Counterstaining with MitoTracker Red shows that cells with affected peroxisomes invariably also have affected mitochondria (Figure 4, C and E). Because small amounts of peroxisomal staining might have been obscured by mitochondrial staining, we changed the mitochondrial distribution with Drp1 siRNA to make parts of the cell devoid of mitochondria. These areas show colocalization of myc-tagged Mff with a peroxisomal marker (Supplemental Figure 3). We conclude that there is a small but significant amount of Mff on peroxisomes, similar to Fis1, which is also localized to peroxisomes and mitochondria (Koch et al., 2005; Kobayashi et al., 2007). The dual roles and localizations of Drp1, Fis1, and Mff on peroxisomes and mitochondria suggest that these proteins are part of the same fission apparatus, acting on two different organelles.
Figure 4.
Effects of Mff siRNA on peroxisomal morphology. (A) HeLa cells transfected with scrambled oligonucleotides showing wild-type mitochondrial morphologies by staining with MitoTracker. (B) The same cells stained with catalase antibody, showing the punctate distribution of peroxisomes. The inset shows an enlargement of the peroxisomal staining. (C and D) HeLa cells transfected with Drp1 siRNA oligonucleotides, showing highly connected mitochondria and elongated peroxisomes. The enlargement shows the peroxisomal defect more clearly. (E and F) Similar patterns were obtained with Mff siRNA oligonucleotides. The boxed areas, enlarged in the bottom right-hand corners of D–F, show close-ups of peroxisomes. Bar, 10 μm.
The Drosophila homologue of Mff (CG30404/Tango11) was also identified in a screen for defects in constitutive secretion, suggesting that Mff might affect the secretory pathway (Bard et al., 2006). We tested whether Mff siRNA also affects secretion in mammalian cells, by using a luciferase assay. We were unable to detect significant differences in the rate of secretion or in Golgi morphology, even when there were large differences in mitochondrial and peroxisomal morphologies (data not shown). The secretion defects that were observed in Drosophila cells may have been due to respiratory defects, because these defects can result from mitochondrial fission defects (Estaquier and Arnoult, 2007). We conclude that Mff affects peroxisomal and mitochondrial morphologies in ways that are indistinguishable from the effects of Drp1 siRNA. It seems likely that Mff, Fis1, and Drp1 all affect different aspects of the pathways that mediate mitochondrial and peroxisomal fission.
Fusion Defects Caused by Dominant-Negative Mitofusin Mutants Are Partially Suppressed by Mff siRNA
It was previously shown that dominant-negative mutations in transfected Drp1 suppress mitochondrial fragmentation in mouse Mfn1 and Mfn2 knockout cell lines (Chen et al., 2003). Similar effects were observed with yeast where it was shown that mutations in Dnm1 are epistatic to mutations in Fzo1 (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000). To determine whether Mff contributes to fission, rather than negatively regulating fusion, we tested the ability of Mff siRNA to suppress a mitochondrial fusion defect. The mitochondrial fusion defect was introduced by transfecting HeLa cells with constructs that express dominant interfering Mitofusin mutants. We used Mfn1(K88T) and Mfn2(K109T), which have mutations in the critical lysines of the G1 consensus motifs in their GTP binding domains (Eura et al., 2003; Santel et al., 2003). Dominant interference most likely occurs through coassembly with endogenous protein complexes. Mitofusin complexes on the mitochondrial outer membrane can be heteromeric or homomeric (Chen et al., 2003) and act in trans with similar complexes on opposing mitochondrial membranes during fusion (Koshiba et al., 2004; Meeusen et al., 2004). Because of these heteromeric complexes, dominant interfering mutants are more effective than siRNA, which is hampered by the partial redundancy of Mfn1 and Mfn2 (Chen et al., 2003).
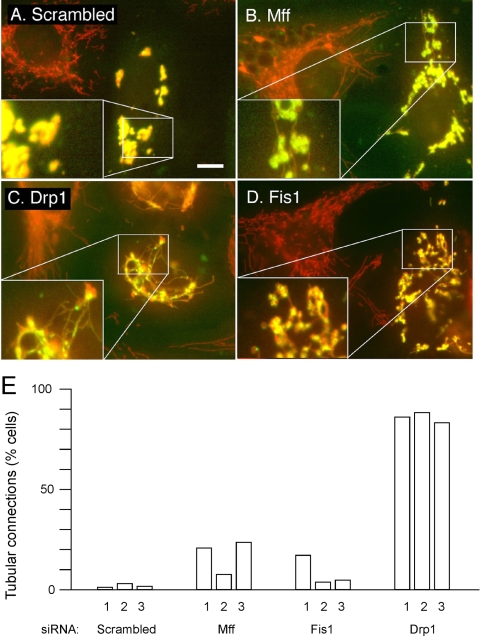
In this experiment, HeLa cells were first transfected with scrambled controls, Mff, Fis1, or Drp1 siRNA followed by a second transfection 3 d later with the dominant-negative Mitofusin constructs. These constructs also have epitope tags to identify the cells that express dominant-negative Mitofusins. All cells transfected with dominant negative Mitofusin constructs show mitochondrial fragmentation, consistent with a complete block of mitochondrial fusion. These mitochondrial fragments are also clustered near the nucleus, which is most likely due to nonspecific perturbations of mitochondrial outer membranes as seen previously with other overexpressed mitochondrial proteins (Smirnova et al., 2001). Perinuclear clustering was not averted by Mff, Drp1, or Fis1 siRNA (Figure 5, B–D), but there was a notable increase in mitochondrial connectivity in almost all Drp1 siRNA cells and in a small fraction of Mff and Fis1 siRNA-transfected cells (Figure 5E). Cotransfection of Mff and Fis1 siRNA oligonucleotides did not enhance the fission defect, but in our hands cotransfected siRNA oligonucleotides are often not as effective as individual transfections (data not shown). It therefore remains possible that Mff and Fis1 are partially redundant. Alternatively, we may not have achieved low enough levels of Mff and Fis1 to fully block their function or Mff and Fis1 are not absolutely required for fission. We can, nevertheless, conclude that Mff and Fis1 siRNA reverse some of the fragmentation caused by dominant-negative Mitofusins, whereas Drp1 siRNA fully reverses their effects.
Figure 5.
Reversal of mitochondrial fragmentation caused by dominant-negative Mitofusin expression constructs. (A) HeLa cells were transfected with scrambled Mff siRNA oligonucleotides as a control and with a myc-Mfn2(K109T) expression construct. Mitochondria were detected with MitoTracker (red). Cells expressing mutant Mfn2 were detected with myc antibody (green). These cells invariably had fragmented and often clumped mitochondria, most likely due to overexpression of aberrant mitochondrial outer membrane protein. Similar transfections were done with Mff (B), Drp1 (C), and Fis1 (D) siRNA oligonucleotides along with the myc-Mfn2(K109T) expression construct. The cotransfected cells show clumped mitochondria, but these mitochondria often have thin tubular connections, which were never detected when the myc-Mfn2(K109T) construct was transfected alone. The enlargements in the bottom left corners show these connections more clearly. Bar, 10 μm. The percentages of cells with mitochondrial connections are shown in E. The experiments numbered 1 were done with GFP tagged Mfn1(K88T), and the experiments numbered 2 and 3 were done with myc-Mfn2(K109T). For each point, 250–400 cells were counted.
Mff siRNA Partially Inhibits CCCP-induced Mitochondrial Fragmentation
It was previously shown that loss of mitochondrial membrane potential caused by treatment with the protonophore CCCP induces rapid mitochondrial fission. Within minutes, the mitochondria of cells treated with CCCP are converted to small, dispersed, fragments (Griparic et al., 2007). We used this method to investigate the effects of Drp1, Mff, and Fis1 siRNA on inducible mitochondrial fission (Figure 6, A–H). Cells were transfected with siRNA oligonucleotides and stained with MitoTracker 3 d later, followed by a 60-min incubation with CCCP to induce fragmentation and analysis by fluorescence microscopy.
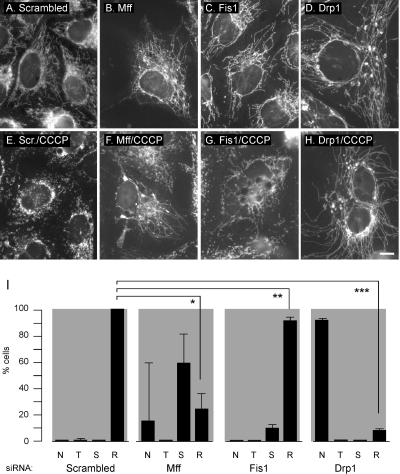
Figure 6.
Reversal of fission induced by loss of mitochondrial membrane potential. HeLa cells were transfected with scrambled, Mff, Fis1, or Drp1 siRNA oligonucleotides, respectively (A–D), stained with MitoTracker, and then incubated for 60 min with the membrane-depolarizing drug CCCP at 2 μg/ml to induce mitochondrial fission (E–H). Transfection with Drp1 siRNA inhibits almost all of the induced fragmentation. Transfection with Mff siRNA inhibits some, but not all, of the induced fragmentation, whereas transfection with Fis1 siRNA had only a limited effect. Bar, 10 μm. I shows quantification of these effects. N, highly connected tubular networks; T, mixture of tubules as observed in untreated cells; S, short tubules; and R, round fragments. The percentages are means of three independent experiments with 300–400 cells per data point. The error bars show SD for the three experiments. The bars show values obtained after CCCP induction. Although Drp1 siRNA is the only one that strongly inhibits CCCP-induced fission, the effects of Mff and Fis1 siRNA are significant when the percentages of cells with round fragments are compared with those percentages in cells transfected with scrambled oligonucleotides. An unpaired Student's t test shows p < 0.0005 for Mff (*), p < 0.005 for Fis1 (**), and p < 0.0001 for Drp1 (***). The amounts of Mff protein were reduced by 91%, Fis1 by 93%, and Drp1 by 99% (average values as determined by densitometry of Western blots).
A histogram with the distributions of different mitochondrial morphologies is shown in Figure 6I. The results show that Drp1 siRNA renders mitochondria resistant to CCCP-induced fragmentation (Figure 6, C and D). Mff siRNA renders some mitochondria resistant to induced fragmentation, but a range of transition forms (a mixture of tubule lengths or short tubules) are observed as well (Figure 6, E and F). Much weaker effects are observed with Fis1 siRNA (Figure 6, E and F). Quantitation of these results is shown in Figure 6I. We conclude that Mff siRNA has modest but reproducible inhibitory effects on CCCP-induced fragmentation of mitochondria.
Mff siRNA Inhibits Apoptosis
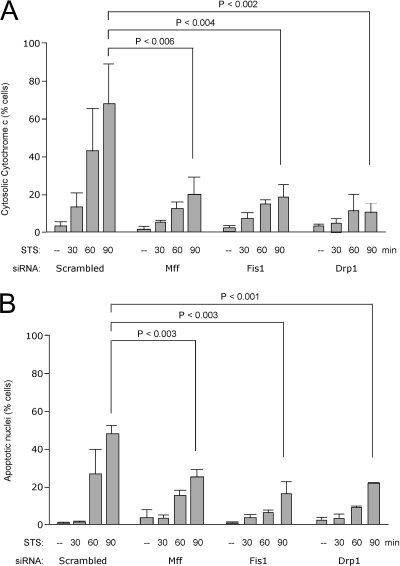
Mutations in Drp1 and Fis1 siRNA were previously shown to inhibit apoptosis by preventing cytochrome c release from mitochondria (Frank et al., 2001; James et al., 2003; Lee et al., 2004). To test whether Mff siRNA also inhibits these processes, HeLa cells were transfected with scrambled control, Mff, Drp1, and Fis1 siRNA oligonucleotides, and apoptosis was induced with staurosporine 3 d later. The pan caspase inhibitor ZVAD-fmk was included to prevent rounding up and detachment of the cells. Samples of cells were taken at fixed intervals and processed for immunostaining with anti-cytochrome c antibodies. The histogram in Figure 7A shows the percentages of cells with cytochrome c released from mitochondria into the cytosol. Mff siRNA strongly inhibits cytochrome c release in the majority of cells, similar to Drp1 and Fis1 siRNA. These experiments were done with the pan-caspase inhibitor z-VAD-fmk to prevent rounding up of cells and thus underestimating the number of cells that have cytochrome c release. The inhibition of cytochrome c release from mitochondria is not absolute, because at 5 h after induction all cells, whether they are transfected with Mff, Drp1, and Fis1 siRNA or with scrambled oligonucleotides, have cytosolic cytochrome c (data not shown). Comparable results were obtained with actinomycin D, as an alternative apoptosis-inducing agent (Supplemental Figure 4). In a separate experiment, z-VAD-fmk was omitted to determine the effects on further progression of apoptosis. The percentages of cells undergoing apoptosis were determined by counting the numbers of pycnotic nuclei in cells stained with Hoechst. At time points up to 3 h, these percentages are significantly lower in transfections with Mff, Fis1, or Drp1 siRNA oligonucleotides than with scrambled oligonucleotides (Figure 7B). This inhibition is also not absolute, because in due time all cells have pycnotic nuclei (Mff, Fis1, or Drp1 siRNA transfected cells are indistinguishable from cells transfected with scrambled oligonucleotides after 5 h; data not shown). We conclude that Mff siRNA inhibits apoptotic release of cytochrome c and further progression of apoptosis similar to the inhibitory effects of Drp1 and Fis1 siRNA, but these effects are not absolute.
Figure 7.
Inhibition of apoptosis by Mff siRNA. HeLa cells were transfected with scrambled, Mff, Fis1, and Drp1 siRNA oligonucleotides as indicated, and apoptosis was induced by treating the cells with staurosporine or as control with solvent (dimethyl sulfoxide). Cells were fixed and stained at fixed intervals after the addition of staurosporine as indicated. (A) Effects on cytochrome c release from mitochondria as detected by staining the cells with cytochrome c antibody. Along with Staurosporine, z-VAD-fmk was added to prevent further progression of apoptosis. (B) Effects on the formation of apoptotic nuclei as detected with Hoechst staining. No z-VAD-fmk was added to allow further progression of apoptosis. The percentages are means of three independent experiments, each with 300–400 cells per data point. The error bars show SD for these three experiments. An unpaired Student's t test was used for statistical analysis of data collected after 90 min with staurosporine.
A Multimeric Complex Containing Mff Protein
Two computer algorithms (COILS and PAIRCOIL) identify a segment within Mff with high coiled coil-forming propensity. To test whether Mff can form homodimers, we transfected HEK293 cells with myc-tagged and GFP-tagged Mff expression constructs. Twelve hours after transfection, these cells were lysed, and proteins were immunoprecipitated with anti-myc antibodies. Lysates from cells that overexpress both fusion proteins show coprecipitation of GFP-tagged Mff with myc-tagged Mff, whereas samples from cells expressing GFP-tagged Mff but not myc-tagged Mff show no precipitation of GFP-tagged Mff (Figure 8A). We conclude that Mff is able to multimerize in vivo.
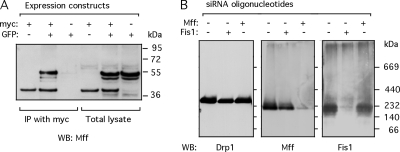
Figure 8.
Multimers of Mff and endogenous Mff complexes. (A) Coimmunoprecipitations to determine whether Mff can form homomultimers. HEK293 cells were transfected with myc- and GFP-tagged Mff expression constructs, the transfected cells were lysed, and the lysates were incubated with anti-myc antibody coupled to protein A beads and immunoprecipitated. The blot was probed with Mff antibody, showing coimmunpreciptation of GFP-tagged Mff with anti-myc antibody when the GFP and myc-tagged constructs are cotransfected, but not when the GFP-tagged construct is transfected alone. The lanes with total lysates verify that the constructs are expressed. The constructs used for these experiments encode isoform 8, which lacks exons 5, 6, and 7 (Figure 1). (B) Size of native Mff complex determined with Blue Native Gel electrophoresis. Detergent lysates from cells transfected with scrambled, Fis1 siRNA, and Mff siRNA oligonucleotides were subjected to BNGE, blotted, and probed with Drp1, Mff, and Fis1 antibodies.
To determine the size of the endogenous complex formed by Mff, HeLa cells were lysed with buffer containing 0.1% N-dodecyl-β-d-maltoside. The lysates were size fractionated with BNGE, Western blotted and probed with anti-Mff, anti-Drp1 and anti-Fis1 antibodies. High molecular weight size markers show that Mff is present in a 200-kDa complex, very different from the 320-kDa Drp1 complex, which most likely consists of Drp1 tetramers (Figure 8B). The size of the Mff complex is, however, only slightly different from the size of the Fis1 complex, which was also ∼200 kDa. To verify that Mff and Fis1 are in different protein complexes, we subjected lysates of Mff and Fis1 siRNA transfected cells to BNGE. We found that Mff siRNA does not alter the size or abundance of the Fis1 complex, nor does Fis1 siRNA alter the size or abundance of the Mff complex (Figure 8B). The size of the Mff complex does suggest that it contains other proteins, but not Fis1 or Drp1. We conclude that Mff is part of a novel membrane complex that contributes to fission independent of the Fis1 complex.
DISCUSSION
Our results show that Mff is a tail-anchored protein that affects the fission of mitochondria and peroxisomes. The inhibition of mitochondrial fission by Mff siRNA is most pronounced on uninduced fission, which occurs continually under normal growth conditions, but there is also an inhibitory effect on CCCP-induced fission and on cytochrome c release from mitochondria during apoptosis. The inhibitory effects seem to be specific for mitochondrial and peroxisomal fission, because other physiological functions, such as maintaining mitochondrial membrane potential, or membrane transport functions, such as secretion or endocytosis, are not notably impaired (data not shown). The effects of Mff and Fis1 siRNA on CCCP-induced fission and reversal of mitochondrial fragmentation induced by dominant-negative Mitofusins are weaker than those of Drp1 siRNA, but we could not tell whether this difference is due to partial redundancy of Fis1 and Mff or incomplete knockdown of these proteins. These results, nevertheless, confirm that Mff specifically contributes to fission. We conclude that Mff is a novel mitochondrial and peroxisomal fission protein.
Although Fis1 and Mff act in the same pathway, there is no obvious Mff homologue in yeast, nor have extensive genetic screens for suppressors of mitochondrial fusion mutants yielded a functional equivalent of Mff in yeast (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001). This suggests that Mff is a later evolutionary acquisition or that yeast has dispensed of its function. The effects of Mff siRNA on peroxisomes are consistent with the discovery of a small but important fraction of Mff on peroxisomes, similar to the dual localization of Fis1 (Koch et al., 2005; Kobayashi et al., 2007). Both proteins are localized through their carboxy-terminal transmembrane segments, as is commonly observed with other tail-anchored proteins (Borgese et al., 2007). We conclude that Mff and Fis1 are similarly localized, have similar topologies and act in the same pathway. Because Fis and Mff are equally important for mitochondrial and peroxisomal fission, but they are not in the same complex, it seems likely that they fulfill different functions in these processes.
The sequence of Mff gives some clues as to its function. Mff has two short repeats in the amino terminal half. These repeats are conserved in metazoans, suggesting that they might function as binding motifs, for example, recruiting other molecules to the mitochondrial outer membrane, similar to the functions of the TPR motifs of Fis1 (Jofuku et al., 2005; Wells et al., 2007; Zhang and Chan, 2007). The coiled coil domain and the transmembrane segment of Mff are also conserved, and the level of conservation is more than needed to maintain the alpha helical periodicity normally found in coiled-coil proteins, suggesting that these domains might also bind to other proteins. Moreover, Mff is in a protein complex that is much larger than expected for a homodimer. It therefore seems likely that Mff interacts with other proteins. Dimerization, as observed by us, may simply be a first step toward forming a larger complex on the surface of mitochondria.
Mitochondrial fission can be divided into several stages, including the initial constriction of mitochondrial tubules, the mobilization of cytosolic Drp1, its recruitment to mitochondria, the assembly of a scission complex, the actual scission event and disassembly of the scission complex. Some of these stages have been observed by light microscopy (Labrousse et al., 1999; Frank et al., 2001), others have been inferred from the localization or proposed functions of scission proteins. It is not yet known what causes the initial constriction of mitochondria, but this most likely entails other as yet unidentified proteins, because the diameter of undivided mitochondria is in the range of 500-1000 nm, whereas the largest diameter of Drp1 rings, as measured with the yeast Drp1 homologue Dnm1, is ∼100 nm (Ingerman et al., 2005). The final constriction and scission process are most likely driven by Drp1 GTP binding and hydrolysis, but other parts of the cycle might be controlled by ubiquitination (Karbowski et al., 2007). There are several plausible functions for Mff within this framework. Mff could, for example, recruit proteins that mediate the initial constriction process or it could act together with Fis1 to form the scission complex.
In conclusion, Mff siRNA affects mitochondrial and peroxisomal fission similar to the effects of Drp1 and Fis1 siRNA in mammalian cells. Mff is localized to the mitochondrial outer membrane, but it is not in a complex with Fis1, suggesting that Mff and Fis1 act at different stages of the fission process. We therefore propose that Mff is a novel molecular adaptor that helps to recruit or organize components of the mitochondrial outer membrane fission machinery alongside Fis1.
Supplementary Material
ACKNOWLEDGMENTS
We thank other members of the lab for helpful discussions and critical reading of the manuscript. We also thank Dr. Bernard Mathey-Prevot, Dr. Norbert Perrimon, and other members of the Drosophila RNA Screening Center at Harvard Medical School for the wonderful opportunities provided by the center, for help with the screen, and for help evaluating screening results. This work was supported by American Cancer Society grant RSG-01-147-01-CSM and National Institutes of Health grant GM-051866.
Abbreviations used:
- BNGE
Blue Native Gel electrophoresis
- CCCP
carbonylcyanide 3-chloro phenyl hydrazone
- TPR
tetratricopeptide.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1287) on March 19, 2008.
REFERENCES
- Alexander C., et al. OPA1, encoding a dynamin–related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Bard F., et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- Bhar D., Karren M. A., Babst M., Shaw J. M. Dimeric Dnm1–G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 2006;281:17312–17320. doi: 10.1074/jbc.M513530200. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J. M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N., Brambillasca S., Colombo S. How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Cerveny K. L., McCaffery J. M., Jensen R. E. Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell. 2001;12:309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Chang C. R., Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs J. T., Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C., et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Desagher S., Martinou J. C. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- Echeverri C. J., Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat. Rev. Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- Estaquier J., Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14:1086–1094. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Yokota S., Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Fekkes P., Shepard K. A., Yaffe M. P. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Fritz S., Weinbach N., Westermann B. Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell. 2003;14:2303–2313. doi: 10.1091/mbc.E02-12-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. E., Graumann J., Chan D. C. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., Kanazawa T., van der Bliek A. M. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. G., Fuller M. T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J. E. Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Jofuku A., Eura Y., Mihara K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem. Biophys. Res. Commun. 2003;301:891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- James D. I., Parone P. A., Mattenberger Y., Martinou J. C. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Jofuku A., Ishihara N., Mihara K. Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem. Biophys. Res. Commun. 2005;333:650–659. doi: 10.1016/j.bbrc.2005.05.154. [DOI] [PubMed] [Google Scholar]
- Kanazawa T., Zappaterra M. D., Hasegawa A., Wright A. P., Newman-Smith E. D., Buttle K. F., McDonald K. L., Mannella C. A., an der Bliek A. M. The C. elegans opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008;4((2)) doi: 10.1371/journal.pgen.1000022. e1000022doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R. J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Tanaka A., Fujiki Y. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 2007;313:1675–1686. doi: 10.1016/j.yexcr.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M. A., Schrader M. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 2003;278:8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- Koch A., Yoon Y., Bonekamp N. A., McNiven M. A., Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N., Ohkuni K., Kitagawa K., McCaffery J. M., Shaw J. M., Okamoto K. The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol. Biol. Cell. 2006;17:3756–3767. doi: 10.1091/mbc.E06-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S. A., Kaiser J. T., Chen H., McCaffery J. M., Chan D. C. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Labrousse A. M., Zapaterra M., Rube D. A., van der Bliek A. M. C. elegans dynamin-related protein drp-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Jeong S. Y., Karbowski M., Smith C. L., Youle R. J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gould S. J. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 2003;278:17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J. M., Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M., Shaw J. M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Pinedo C., Guio-Carrion A., Goldstein J. C., Fitzgerald P., Newmeyer D. D., Green D. R. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc. Natl. Acad. Sci. USA. 2006;103:11573–11578. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J. E., Nunnari J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Olesen C., Larsen N. J., Byskov A. G., Harboe T. L., Tommerup N. Human FATE is a novel X-linked gene expressed in fetal and adult testis. Mol. Cell. Endocrinol. 2001;184:25–32. doi: 10.1016/s0303-7207(01)00666-9. [DOI] [PubMed] [Google Scholar]
- Pfeffer S. R. Unsolved mysteries in membrane traffic. Annu. Rev. Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- Praefcke G. J., McMahon H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Santel A., Frank S., Gaume B., Herrler M., Youle R. J., Fuller M. T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Santel A., Fuller M. T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schrader M., Yoon Y. Mitochondria and peroxisomes: are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays. 2007;29:1105–1114. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S. M., Yaffe M. P., Jensen R. E. Mgm1p, a Dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K. A., Yaffe M. P. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B. D., Schmid S. L. A molecular motor or a regulator? Dynamin's in a class of its own. Biochemistry. 2003;42:1369–1376. doi: 10.1021/bi027062h. [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Koutsopoulos O. S., Okamoto K., Ryan M. T. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. Mdv1p is a WD repeat protein that interacts with the Dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Okreglak V., Naylor K., Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A. M. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M., Meyerowitz E. M. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- van der Bliek A. M., Redelmeier T. E., Damke H., Tisdale E. J., Meyerowitz E. M., Schmid S. L. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J. Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. C., Picton L. K., Williams S. C., Tan F. J., Hill R. B. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J. Biol. Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. D., Wagner J. A., Gorsich S. W., McCaffery J. M., Shaw J. M., Nunnari J. The Dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa S., Lee J. S., Ishimoto A. Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J. Biol. Chem. 1998;273:32353–32359. doi: 10.1074/jbc.273.48.32353. [DOI] [PubMed] [Google Scholar]
- Yoon Y., Krueger E. W., Oswald B. J., McNiven M. A. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chan D. C. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. USA. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H. M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.