Abstract
Shape-dependent local differentials in cell proliferation are considered to be a major driving mechanism of structuring processes in vivo, such as embryogenesis, wound healing, and angiogenesis. However, the specific biophysical signaling by which changes in cell shape contribute to cell cycle regulation remains poorly understood. Here, we describe our study of the roles of nuclear volume and cytoskeletal mechanics in mediating shape control of proliferation in single endothelial cells. Micropatterned adhesive islands were used to independently control cell spreading and elongation. We show that, irrespective of elongation, nuclear volume and apparent chromatin decondensation of cells in G1 systematically increased with cell spreading and highly correlated with DNA synthesis (percent of cells in the S phase). In contrast, cell elongation dramatically affected the organization of the actin cytoskeleton, markedly reduced both cytoskeletal stiffness (measured dorsally with atomic force microscopy) and contractility (measured ventrally with traction microscopy), and increased mechanical anisotropy, without affecting either DNA synthesis or nuclear volume. Our results reveal that the nuclear volume in G1 is predictive of the proliferative status of single endothelial cells within a population, whereas cell stiffness and contractility are not. These findings show that the effects of cell mechanics in shape control of proliferation are far more complex than a linear or straightforward relationship. Our data are consistent with a mechanism by which spreading of cells in G1 partially enhances proliferation by inducing nuclear swelling and decreasing chromatin condensation, thereby rendering DNA more accessible to the replication machinery.
INTRODUCTION
In vivo structuring processes such as embryonic development (1) and wound healing (2) are considered to be largely driven by local differentials in cell proliferation determined by cell shape. In endothelial cells (ECs), cell shape regulation of proliferation is an essential mechanism underlying endothelial barrier repair (3) and the formation of new blood vessels (angiogenesis) that occurs during normal growth and neoplastic processes (4). However, although it has long been recognized that entrance into the S phase is highly controlled by cell shape (5), the mechanisms by which this regulation is exerted remain unclear.
Studies using micropatterned islands to control the shape of single ECs in culture showed that increasing spreading while keeping both extracellular matrix contact area and growth factor signaling constant was sufficient to promote proliferation, whereas preventing cells from spreading had the opposite effect (6,7). These findings revealed that, in addition to biochemical mitogenic cues from the extracellular matrix and growth factors, biophysical signals associated with cell spreading were required for cell cycle progression. Later work identified that this biophysical signaling was related to the internal mechanical tension (prestress) developed within the actin cytoskeleton (CSK). Indeed, both cytoskeletal stiffness (8) and contractility (9–12) (both indicators of cytoskeletal mechanical tension (13)) increased during spreading in parallel with proliferation in different cell types including ECs, and inhibition of actin polymerization or of actin-myosin contractility in ECs blocked cell cycle progression beyond the G1 phase (14). Moreover, recent work showed that mechanical stresses transmitted through the CSK determined proliferation in sheets of ECs (15). However, there is evidence suggesting that the role of cytoskeletal tension in transducing cell shape signals to the cell cycle is more complex than initially hypothesized. In fact, it has been shown that reducing myosin-dependent contractility by inhibiting its upstream regulator, Rho kinase (ROCK), in ECs did not prevent cell cycle progression (16), and inverse (rather than direct) relationships between stiffness and proliferation have been reported in other cell types (8,17). Collectively, these previous findings suggest that the relationship between DNA synthesis and cytoskeletal tension might be highly nonlinear or not straightforward, and that other signals might mediate the regulation of cell proliferation exerted by cell spreading.
In addition to a rise in prestress, cell spreading induces changes in nuclear shape in ECs (6,18) and other cell types (19,20). These modifications in nuclear shape (which could also be mediated by prestress (21)) have been associated with changes in the expression of different genes, including those that control the cell cycle (19,20,22). Furthermore, decreases in nuclear size, as well as marked increases in the regions with high chromatin condensation (heterochromatin), have been observed to correlate with differentiation (23,24), which is commonly associated with quiescence or reduced growth. Conversely, less heterochromatin and bigger nuclei have been observed in proliferating nonmalignant cells (23,25–27). Based on these earlier observations, we hypothesize that nuclear volume regulation is a key mechanism by which cell shape controls cell cycle progression in ECs.
The aim of this work was to assess whether changes in nuclear volume and cell mechanical properties can account for the control of proliferation exerted by cell shape in ECs. To this end, single ECs were cultured on micropatterned adhesive islands to independently control different geometrical aspects of cell shape: spreading (cell projected area), pointedness (presence of corners), and elongation (ratio between cell length and width). For each shape, we measured cell proliferation in relation to cell mechanics (assessed by probing two complementary properties: compressive cell stiffness and contractility), actin CSK organization, nuclear volume and the apparent chromatin condensation. We found that both nuclear volume and the apparent chromatin condensation (but not cell stiffness, contractility, or actin spatial organization) had a shape dependence tightly coupled to proliferation rates. Our data are consistent with a mechanism by which cell spreading enhances DNA synthesis by inducing nuclear swelling, thereby decreasing chromatin condensation, rendering DNA more accessible to the replication machinery and ultimately promoting DNA synthesis.
MATERIALS AND METHODS
Substrate fabrication
Micropatterned substrates containing fibronectin-coated islands were fabricated using a modification of a previously described protocol (28). Stamps of poly(dimethylsiloxane) (PDMS) (Sylgard 184, Dow Corning, Midland, MI) containing raised structures with the desired geometry (circle/square), spreading (300, 900, and 2500 μm2), and elongation (1,6) were cast from silicon nitride substrates previously etched with focused ion beam technology (FEI, Hillsboro, OR). PDMS stamps were silanized overnight with (tridecafluoro-1,1,2,2 tetrahydrooctyl)-1-trichlorosilane after a 1-min exposure to an oxygen plasma, soaked in a phosphate-buffered saline (PBS) solution containing 25 μg/ml fibronectin (Sigma, St. Louis, MO) for 1 h, and placed in conformal contact with bacteriological polystyrene petri dishes (BD, Franklin Lakes, NJ) for 1 min. The remaining unstamped regions of the petri dishes were rendered nonadhesive by immersion in a PBS solution containing 1% Pluronic F108 (BASF, Mount Olive, NJ) for 1 h. For traction measurements, cells were micropatterned on flexible polyacrylamide gels using membrane patterning technology (10). Briefly, a master containing raised features in photoresist was fabricated with photolithography from high-resolution masks (CAD/Art services, Bandon, OR) printed on transparency sheets. PDMS membranes were obtained by spin-coating PDMS prepolymer on the photoresist master, curing at 70°C for 90 min, and peeling from the master. After storage in ethanol, the membranes were dried and placed in conformal contact with the prepared polyacrylamide substrates. A drop of type I collagen (200 μg/ml) was added to the membrane for 2 h to coat the gel through the membrane holes. After rinsing the collagen three times and removing the membrane, the gel was soaked in bovine serum albumin (10 mg/ml in PBS, Sigma) for 30 min and rinsed again three times to render the remaining parts of the gel nonadhesive to cells. Gels were stored in PBS until cell plating.
Cell culture and reagents
Lung human microvascular endothelial cells (HMVEC-L, Clonetics, East Rutherford, NJ) were cultured in EGM-2MV medium supplemented with 0.04% hydrocortisone, 0.4% human fibroblast growth factor, 0.1% vascular endothelial growth factor, 0.1% R3-IGF-1, 0.1% ascorbic acid, 0.1% human endothelial growth factor, 0.1% gentamicin/amphotericin B, and 5% fetal bovine serum (Clonetics). Cells were used between passages 3 and 11. For all experiments, cells were trypsinized, plated on micropatterned petri dishes, and probed 24 h later. In actin depolymerization experiments, cells were incubated for 30 min with 1 μg/ml cytochalasin D (Sigma) before measurements.
Cell mechanical measurements with atomic force microscopy
The complex shear modulus (G* = G′ + iG″, where G′ is the elastic storage modulus, G″ is the viscous loss modulus, and i is the imaginary unit) of cells was measured with a custom-built atomic force microscope (AFM) attached to an inverted optical microscope, as described previously (29,30) (Fig. 1). G′ provides a measurement of stiffness, and is indicative of cytoskeletal internal tension or prestress (13). G″ is a measurement of energy dissipation due to internal friction. The ratio between viscous and elastic moduli (G″/G′), also known as relative energy loss or loss tangent, indicates whether the mechanical and structural behavior of the sample is more fluidlike (>1, disordered) or solidlike (<1, ordered) (31). The spring constant, k, of gold-coated cantilevers (nominal k = 0.01 N/m, semiincluded angle θ = 35°; Veeco, Woodbury, NY) was calibrated using the thermal fluctuations method (32). To keep pH at 7.4, air with 5% CO2 was perfused on the petri dish during measurements. For each cell, measurements were carried out at three locations in the cell periphery at room temperature (Fig. 2 D). In brief, for each cell location, we first recorded 10 force-indentation (F-δ) curves (where F = kd and d is the cantilever deflection) (2.5 μm amplitude, 1 Hz, maximum indentation δ ≈ 1 μm) to determine the contact point between the cell and the cantilever tip. Second, the tip was set at an operating indentation δ0 ≈ 0.5 μm and a small sinusoidal oscillation (50 nm amplitude, 1.6 Hz) was applied (Fig. 1). From these measurements, G* was calculated, with Fourier analysis using custom software in MATLAB (The Mathworks, Natick, MA), as (30)
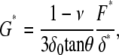 |
(1) |
where the * in F and δ denote their respective Fourier transforms and ν is the Poisson ratio (assumed to be 0.5). G* for each cell was taken as the average of the measured values over the three different locations. We ruled out any contribution of the underlying substrate on our mechanical calculations by computing the Young's modulus E from force-indentation curves as a function of indentation (33). G* data were corrected for the hydrodynamic force exerted by the medium on the cantilever (34). Cell topography images, which are very sensitive to the organization of the actin CSK (35), were obtained with a commercial AFM (Bioscope, Veeco) operated in contact mode at a scanning rate of 0.3 Hz, applying a constant force of ∼2 nN on the cells.
FIGURE 1.
Schematic description of AFM cell mechanical measurements. (Left) First, a force-indentation (F-δ) curve was obtained as the cantilever tip approached and contacted the cell (solid line) and retracted (dashed line). The F-δ approaching curve was used to determine the contact point between tip and cell (arrow). Once the contact point was determined, the cantilever was set at an operating indentation (δ0) of ∼500 nm and a low-amplitude (50 nm) and low-frequency (1.6 Hz) sinusoidal signal was applied. (Right) Corresponding sinusoidal force (upper) and indentation (lower) signals were used to calculate G* by Fourier analysis.
FIGURE 2.
Elastic storage modulus (G′) probed at the periphery of single ECs characterizes actin cytoskeletal stiffness. (A) Comparison of G′ measured in the cell periphery (black bars) and at the cell center (open bars) for nonelongated (elongation = 1) and elongated (elongation = 6) 2500-μm2-spread cells. ***, p < 0.001. (B and C) Relative changes induced by cytochalasin D (1 μg/ml) treatment in stiffness (B) and apparent F-actin content (C) in the cell periphery. Cytochalasin D significantly reduced F-actin content (for all shapes) and G′ (for 2500-μm2 cells) (p < 0.001). Solid and open symbols represent nonelongated and elongated cells, respectively. Data are presented as mean ± SE. (D) F-actin immunofluorescence images showing the absence of actin bundles (stress fibers) after cytochalasin D treatment in different cell shapes. White circles indicate the approximate measurement zones for the center (solid-line circles) and the cell periphery (dashed-line circles). Scale bar, 10 μm.
Traction microscopy measurements
Polyacrylamide gels were prepared as previously described (36) by mixing green fluorescent latex beads (0.2 μm diameter, 1:125 vol/vol solution of final mixture) with 7.5% acrylamide and 0.1% bis-acrylamide solutions in deionized water. The Young's modulus of the gels was of 5146 ± 409 (mean ± SE, as measured by AFM). For measurements, a gel with micropatterned cells on top was placed in an inverted optical microscope (TE2000, Nikon, Tokyo, Japan). A bright-field image of a patterned cell and a fluorescence image of the microbeads embedded close to the gel surface right below the cell were acquired with a 40× objective and a CCD camera (Orca, Hamamatsu, Morimoto, Japan). This procedure was repeated after detaching the cell by exposure to trypsin. Using the fluorescence microbead images recorded before and after cell detachment, the map of traction forces exerted by the cells was calculated with custom MATLAB software as previously described (36). Global cell contractility was assessed from the map of traction forces by calculating the elastic strain energy applied by the cell to the gel (36). To quantify mechanical anisotropy, the contraction moment matrix M (37) was rotated and oriented along its principal axes. This operation decomposes M in two components, M1 and M2. M1 corresponds to the contractile moment exerted along the main direction of force exertion, and M2 corresponds to the contractile moment exerted along the direction perpendicular to that of M1. Mechanical anisotropy was computed as M1/M2, where M1/M2 ≈ 1 indicates that the cell is equally contractile along the x and y directions and M1/M2 ≫ 1 indicates that the cell is only contracting along its main axis.
Cell and nuclear volume analysis
Cell volumes were computed from AFM images using the free WSxM software (Nanotec Electrónica, Tres Cantos, Spain) (38). For the smallest cells, we observed a large convolution between the cell and the cantilever tip in the final part of the AFM image due to the inclination of the cantilever tip with respect to the substrate. To correct for this artifact, the volume of the smallest cells was calculated as twice that of the first half of the image (which did not show convolution). All cell volume data were corrected for the underestimation due to the indentation caused by the applied force by adding Vi = δiAi to calculated values, where δi is the average indentation and Ai is the spreading area. To assess nuclear volume, nuclear DNA was stained with Hoechst 33342 (Molecular Probes, Carlsbad, CA) for 5 min, fixed with 4% formaldehyde in PBS and visualized with confocal microscopy (SP2 microscope, Leica, Wetzlar, Germany). Nuclear volume was computed from isointensity contours obtained from confocal sections covering the entire nuclear height and separated by 448 nm (39). Nuclear elongation was calculated as the ratio between the major and minor axes of the ellipse that best fit the nuclear contour with the largest area.
Apparent chromatin condensation and DNA content analysis
Nuclear DNA content was assessed by calculating the total fluorescence intensity emitted by each nucleus. The total fluorescence intensity was divided by the nuclear volume to obtain the average dye spatial density, which correlates with the average chromatin packing ratio (40) and with an increase in heterochromatin markers (hypoacetylation of histone H3 and H4) (22), and as such is indicative of chromatin condensation. To identify nuclei in the G1 phase, we plotted the histogram of the DNA content of the entire cell population (Supplementary Material, Fig. S1 in Data S1). In analogy to common flow cytometry data, the histogram showed two peaks corresponding to cells in G1 and G2, where the average DNA content of the G2 peak was approximately twice that of the G1 peak. The histogram was fitted to two Gaussian distributions. To select cells in G1, nuclei with DNA content higher than the center of the first Gaussian plus 1.5 standard deviations were discarded (Fig. S1 in Data S1). The DNA content of cells selected using this criterion were found to be independent of both cell spreading and anisotropy, thus indicating that our selection criteria did not include nuclei from cells in the S or G2 phase (which have more DNA content). All calculations were performed with custom made software in MATLAB.
Immunofluorescence microscopy of F-actin
Cells were fixed with 4% formaldehyde in PBS, permeabilized with 0.1% Triton X-100 in PBS, and incubated with phalloidin-tetramethylrhodamine (Sigma). To assess the spatial organization of the actin CSK, fluorescence images were taken with a 60× oil immersion objective. For each cell, the orientation of stress fibers was calculated by selecting four areas at the cell periphery and computing the orientation distribution function as previously reported (41), averaging the results for each cell, and averaging the results for all the cells of a given geometry. For a given angle range, the orientation distribution function can vary from 1 (all fibers aligned along that angle range) to 0 (no fibers aligned along that angle range).To assess the apparent F-actin content, we acquired fluorescence images with a 20× air objective, and added the total fluorescence intensity after subtracting the background. Immunofluorescence images were acquired with a CCD camera (Hamamatsu) attached to an inverted optical microscope (Eclipse TE 2000 microscope, Nikon).
DNA synthesis assay
The percentage of cells in the S phase was calculated for each shape and petri dish by measuring the incorporation of 5-bromo-2′-deoxyuridine (BrdU) into cellular DNA using an in situ cell proliferation kit (Roche Applied Science, Basel, Switzerland).
Statistical analysis
Data are shown as mean ± SE for a minimum of n = 6 cells for each particular shape and measurement. The effects of spreading and elongation on G′, strain energy, mechanical anisotropy, G″/G′, F-actin content, nuclear volume, nuclear elongation, apparent chromatin condensation, DNA content, and DNA synthesis were analyzed with two-way analysis of variance (ANOVA) tests. The effects of shape and cytochalasin D on G′ and F-actin content were analyzed with two-way ANOVA. All other reported comparisons were performed with two-tailed Student's t-tests.
RESULTS
The elastic storage modulus (G′) probed in the cell periphery characterizes the stiffness of the actin CSK
To assess the role of the actin CSK in our mechanical measurements, we first measured G′ for the largest cells (2500 μm2 spreading) both at the cell center and at the cell periphery (Fig. 2 A). G′ in circular cells was 20-fold stiffer in the cell periphery (rich in bundles of actin filaments or stress fibers) than at the cell center (with no visible stress fibers) (Fig. 2 D). In contrast, elliptic cells (which exhibited stress fibers both at the center and in the cell periphery) did not exhibit significant regional differences in G′. Second, we disrupted the actin CSK by inhibiting actin polymerization with cytochalasin D (Fig. 2, C and D). Actin depolymerization resulted in a marked decrease in G′ of the cell periphery for highly spread circular and elliptic cells (Fig. 2 B), whereas it had no statistically significant effect on cells with a very restricted spread area of 300 μm2 (which did not show stress fibers). Therefore, G′ probed in the cell periphery was determined mainly by the actin CSK, and is used henceforth as a representative value of cytoskeletal stiffness. To confirm that our mechanical measurements had contributions from the subcortical CSK, we calculated the Young's modulus, E, as a function of the indentation (Fig. S2 in Data S1). We found E independent of δ for δ > 200 nm, in good agreement with previous estimations of the thickness of the actin cortex in ECs (∼200 nm) (35). This held even for the thinnest cells (2500-μm2-spread circular and square cells), thereby confirming that our mechanical measurements were not affected by substrate stiffness. The global mechanical behavior of the cell was dominated by elastic stresses, as indicated by the low values of G″/G′ (0.15–0.3) (Fig. S3 in Data S1), by the independence of E from δ up to 1 μm, and by the fact that E was nearly threefold higher than G′ (3 corresponds to the elastic limit assuming a Poisson ratio of 0.5).
Spreading in nonelongated cells induced parallel increases in stiffness, nuclear volume, and proliferation
Increasing cell spreading from 300 to 2500 μm2 in circular cells induced a marked cell stiffening (50-fold), a 36% increase in nuclear volume of cells in G1, a 10-fold rise in proliferation rates, and a higher apparent F-actin content (Fig. 3 B). Spreading was also associated with the formation of an increasingly complex network of actin filaments (Fig. 3 A). Indeed, whereas a less organized actin CSK with no stress fibers was visible in 300-μm2-spread cells, 2500-μm2-spread cells showed an entangled filamentous network with actin bundles organized around the nucleus and oriented in different directions (Fig. 3 A). Largely spread cells also exhibited a lower G″/G′ (Fig. S3 in Data S1), probably reflecting the more solidlike (organized) CSK. It is interesting that pointedness (analyzed by comparing circular to square cells) did not have a significant effect on any of the measured parameters, except for a certain recruitment of actin bundles at the cell corners. This observation indicates that cytoskeletal stiffness at the cell periphery is not affected by this recruitment. We thus restricted our analysis of cell geometry to the study of cell spreading and elongation, and in further results, we pooled data for cells with square and circular shapes (elongation = 1) as well as for those with rectangular and elliptic shapes (elongation = 6).
FIGURE 3.
Cell spreading increases stiffness, apparent F-actin content, nuclear volume, and DNA synthesis (percent of cells in the S phase) in nonelongated ECs. (A) F-actin immunofluorescence (left column) and AFM deflection images (right column) of representative circular and square cells with different spreading. The AFM image of the 300-μm2 cell shows the first half of the image and its mirror reflection to account for the tip convolution effect (see methods for details). Scale bar, 10 μm. (B) Effect of spreading on G′, F-actin content, nuclear volume of cells in G1, and DNA synthesis for circular (circles) and square cells (squares). Data are presented as mean ± SE. The effect of spreading was statistically significant on all parameters (p < 0.05 for F-actin content, p < 0.01 otherwise).
For equal spreading, cell elongation induced a dramatic decrease in cell stiffness and contractility and an alignment of actin filaments, whereas nuclear volume and proliferation remained unaltered
Unlike in round and square cells (elongation = 1), actin bundles in elongated cells were parallel, showed virtually no entanglement or bending, and were preferentially oriented along the long axis of the cell (Fig. 4, A and B). This alignment of CSK filaments appeared to be specific to actin, since vimentin did not exhibit such preferential orientation (Fig. S4 in Data S1). Actin filament alignment in elongated cells was associated with a moderate increase in F-actin content (Fig. 4 C). In contrast, both G′ (Fig. 4 C) and G″/G′ (Fig. S3 in Data S1) of elongated cells were markedly lower than those of equally spread nonelongated cells (G′ was nine- and threefold lower in 2500-μm2- and 900-μm2-spread cells, respectively). These observations reveal that cell mechanical properties cannot be explained by the F-actin content alone, and that they greatly depend on the spatial organization of the actin CSK. To further confirm that elongation dramatically affects cell mechanics, we micropatterned cells on flexible polyacrylamide gels and measured their contractility by traction microscopy (Fig. 5). A representative example of ECs cultured on micropatterned gels is shown in Fig. 5 B. Cell contractility was assessed both globally, by quantifying the strain energy (Fig. 5 A), and locally, by computing the map of traction forces for each shape (Fig. 5 B, lower panels). In agreement with our G′ data, strain energy markedly increased with spreading and decreased with elongation (Fig. 5 A). The similar spatial distribution of traction forces (Fig. 5 B) and stress fibers (Fig. 4 B) suggests that stress fibers are directly involved in the traction forces developed by ECs on their underlying substrata. Cell mechanical anisotropy was unaffected by spreading but greatly increased with elongation (Fig. 5 A). The effect of cell elongation on mechanical anisotropy can clearly be visualized in the traction maps. Although nonelongated 2500-μm2-spread cells showed traction stresses distributed throughout their periphery, equally spread elongated cells exerted forces localized at the two cell tips and aligned along the long axis of the cell (Fig. 5 B). More important, whereas elongation had striking effects both in actin organization and in a comprehensive set of cell mechanical properties such as CSK stiffness, contractility, and anisotropy, it had no effect on either nuclear volume or DNA synthesis (Fig. 4 C).
FIGURE 4.
Cell elongation induces cell softening and stress fiber alignment, but does not affect nuclear volume or DNA synthesis. (A) F-actin immunofluorescence image (upper) and AFM deflection image (lower) of 2500-μm2 representative spread elongated elliptic cells. Scale bar, 10 μm. (B) Stress fiber orientation evaluated at the cell periphery for 2500-μm2-spread circular and elliptic cells. The drawings show the stress fiber orientation corresponding to 0° in the graphs. For a given angle range, the orientation distribution function can vary from 1 (all fibers aligned along that angle range) to 0 (no fibers aligned along that angle range). (C) Comparison of G′, apparent F-actin content, nuclear volume of cells in G1, and DNA synthesis between nonelongated cells (solid bars) and elongated cells (open bars) for 900-μm2 and 2500-μm2 spreading. Data are presented as mean ± SE. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
FIGURE 5.
Cell contractility increases with cell spreading and decreases with elongation. (A) Cell strain energy and mechanical anisotropy as a function of shape. Solid and open symbols represent nonelongated and elongated cells, respectively. The effect of spreading was significant on strain energy (p < 0.001) but not on mechanical anisotropy. The effect of cell elongation was significant in both cases (p < 0.05 for strain energy and p < 0.001 for mechanical anisotropy). Data are presented as mean ± SE. (B) Phase contrast images and corresponding traction force maps of micropatterned cells with different shapes. Color code indicates the intensity of traction force, whereas black arrows show the direction and also the relative intensity of force.
Proliferation strongly correlated with nuclear volume and apparent chromatin condensation, but not with cell mechanics
Our observation that all the mechanical properties measured here were dramatically affected by cell elongation, whereas DNA synthesis was not, indicates that cell mechanics alone is not sufficient to predict the proliferative status of an EC within a population. In contrast, the nuclear volume of cells in the G1 phase for all shapes strongly correlated with DNA synthesis rates (r = 0.84 (Fig. 6 A)) and largely conformed to a single master curve. This finding is in agreement with our hypothesis that nuclear volume is a key biophysical property mediating shape control of proliferation in ECs. To ascertain whether spreading-induced changes in nuclear volume were associated with a decrease in chromatin condensation, we measured the spatial density of the fluorescence intensity emitted by nuclear DNA stained with the Hoechst 33342 dye (apparent chromatin condensation). We found that the apparent chromatin condensation strongly and inversely correlated with changes in both nuclear volume (r = −0.95) and DNA synthesis (r = −0.82) (Fig. 6 B). As observed from the figures, an increase in nuclear volume in G1 was associated with a rise in DNA synthesis, although the relationship between these variables might not be linear. Although not conclusive, these strong correlations are consistent with a mechanistic relationship between cell spreading, nuclear swelling, chromatin decondensation, and G1-S transition. To assess the specificity of the coupling between proliferation and nuclear volume, we examined the correlation of DNA synthesis with two additional nuclear shape parameters: the nuclear/cytoplasmic volume ratio (previously associated with cell growth (42–44), ) and nuclear elongation. Unlike nuclear volume, neither of these two parameters showed significant correlations with DNA synthesis (Fig. 6, C and D), thereby suggesting that nuclear volume is the relevant nuclear shape parameter involved in conveying cell shape changes to the cell cycle.
FIGURE 6.
Nuclear volume and apparent chromatin condensation of cells in the G1 phase strongly correlate with cell proliferation. (A and B) DNA synthesis rates (percentage of the entire population of cells in the S phase) as a function of (A) the nuclear volume of cells in G1 and (B) the apparent chromatin condensation (average spatial density of stained DNA) of cells in G1 for all shapes. All correlation coefficients were statistically significant (p < 0.05). (C and D) DNA synthesis as a function of other nuclear geometrical factors: the average nuclear to cytoplasmic volume ratio (C) and nuclear elongation of cells in G1 (D). No significant correlations were observed. Solid and open symbols represent nonelongated and elongated cells, respectively. Triangles, 300 μm2 spreading; inverted triangles, 900 μm2 spreading; diamonds: 2500 μm2 spreading. Data are mean ± SE.
DISCUSSION
Even though the model based on cell tension was introduced more than a decade ago (45), the biophysical mechanisms underlying the mitogenic effects of cell spreading are still ill-defined. In this study, we used a comprehensive approach to examine the effect of cell shape on two major cell properties (cell mechanics and nuclear volume) in relation to cell proliferation. To control cell shape, we combined standard and novel micropatterning techniques to independently modify spreading, pointedness, and elongation. Our comprehensive approach revealed that only nuclear volume and chromatin condensation follow the same trend as DNA synthesis in response to alterations in cell shape. Indeed, cell elongation dramatically affected actin organization and cell mechanics, whereas DNA synthesis and nuclear volume remained unaltered. Our results show that none of the commonly measured cell mechanical parameters (stiffness, contractility, and mechanical anisotropy) alone is predictive of cell proliferation. We also report for the first time a strong and significant correlation between cell spreading, nuclear swelling, chromatin decondensation, and proliferation in single ECs.
Both stiffness and contractility reflect the cell mechanical state
In this work, two complementary techniques (AFM and traction microscopy) were used to assess the mechanical status of the cell. With AFM, cell stiffness is probed by applying compressive forces to the surface of the cell in the vertical (z) direction. In contrast, traction microscopy measures the forces that the cell applies to its underlying substratum in the horizontal (xy) plane. Despite the different nature of these measurements, both G′ and contractility data exhibited similar trends in response to changes in cell shape, i.e., an increase with cell spreading and a decrease with cell elongation. Consistent with our data, contractility increases with cell spreading (9–12) and decreases with elongation (12) had also been reported in other cell types. The similar behavior of G′ and contractility observed here supports the notion that both magnitudes are good indicators of the internal mechanical tension of the cell, and strongly suggests that the effects of cell elongation on cell mechanics were mainly due to changes in intracellular tension and largely independent of the experimental technique. Our data also show that the contractile forces exerted by elongated ECs are markedly anisotropic (Fig. 5 A). Given that blood vessels are known to present both elongated and nonelongated cells (46), it is likely that the marked effects of elongation on cell mechanics are important components of in vivo mechanotransduction processes such as those involved in blood flow (shear stress), the maintenance of the mechanical integrity of blood vessels, or angiogenesis.
Cytoskeletal stiffness, contractility, and mechanical anisotropy are largely governed by the spatial organization of the actin CSK rather than by the amount of F-actin or stress fibers alone
The mechanics of most cell types is known to be largely determined by the actin CSK. However, the relationship between cell shape and mechanics and the spatial organization of the actin CSK remains poorly defined. Our data reveal new insights into this relationship. We observed that actin polymerization and the formation of stress fibers were associated with stiffening and increased contractility in nonelongated spreading, whereas actin depolymerization and disassembly of stress fibers were associated with cell softening. These findings are consistent with data reported for other cell types (8,47) and support the generalized assumption that stress fibers are indicative of high stiffness and prestress. However, going against this common assumption, we found that neither stress fibers (highly present in elongated cells) nor F-actin content alone was indicative of increased cell stiffness and contractility, since cell elongation moderately increased F-actin content but markedly decreased stiffness and contractility. In addition, we observed a strong agreement between the spatial distribution of traction forces and stress fibers in both elongated and nonelongated cells. Altogether, these data show that the mechanics of the cell cannot be explained by the F-actin content or the formation of stress fibers alone, and strongly depends on the spatial organization and orientation of the actin CSK. A possible explanation for the reduced stiffness of elongated cells is the reduction in actin microfilament cross-linking caused by the parallel (rather than entangled) stress fibers of these cells (Fig. 4 A), in accordance with the behavior observed in reconstituted actin gels (48) and with mechanical models of semiflexible polymers (48). In addition, it has been proposed that a downregulation of mechanical tension is required to facilitate the outward polymerization of actin during cell spreading (49). In this scenario, the formation of very long actin filaments in elongated cells (along their long axis) will require a lower level of contractile activity (and thus stiffness) than in nonelongated cells. Consistent with this interpretation, the inhibition of Rho-induced contractility in epitheliocytes promoted actin polymerization at the cell edges and cell elongation (50).
Cell stiffness and contractility are not predictive of cell proliferation
In contrast to earlier studies on shape control of proliferation, we used different micropatterning techniques that independently controlled cell shape parameters other than spreading. A global statistical analysis of data obtained from all shape parameters revealed a strong positive correlation between spreading and DNA synthesis, thereby indicating that spreading per se is predictive of the proliferative state within a population. In contrast, although cell elongation had a very important effect on all the measured mechanical parameters (cytoskeletal stiffness, contractility, and mechanical anisotropy), it did not affect proliferation (Figs. 4 and 5). Specifically, elongation in 2500-μm2-spread cells induced ninefold and greater than threefold decreases in G′ and strain energy, respectively, yet DNA synthesis remained unaltered. Therefore, none of the mechanical parameters alone can be used as predictive of proliferative status (Figs. 4 and 5). In agreement with our findings, downregulation of cell contractility by inhibition of ROCK had no effect in cell cycle progression in ECs (16). Likewise, activating (17) or inhibiting (8) myosin light chain kinase (in fibroblasts and hepatocytes, respectively) decreased cell spreading and proliferation, but increased cell stiffness. Both cell stiffness and contractility have been widely used as indicators of internal cytoskeletal tension, which has been proposed to convey shape changes into the cell cycle machinery. Here, we show that the connection between these two mechanical indicators and proliferation is far more complex than a linear or direct relationship. Instead, our findings are consistent with a nonlinear and/or indirect (i.e., through a third signal) relationship between cell mechanics and proliferation. Indeed, nonlinear relations such as biphasic curves have been reported recently between mechanical signals and cellular processes such as myocyte differentiation (51) and stem cell commitment (52). A biphasic relation between DNA synthesis and cytoskeletal stiffness is also consistent with previous studies showing that proliferation was inhibited by treatments leading to very low (8) or very high (8,17) cell stiffness. However, our data indicate a relation more complex than a biphasic behavior, thereby suggesting that cytoskeletal mechanics could have indirect effects on proliferation mediated by other factors. These effects could be a mechanical regulation of the balance between the RhoA downstream effectors ROCK and mDia1 (16), a role of an alternative mechanical parameter not measured here, or, as discussed below, an effect of cytoskeletal tension on nuclear volume.
In addition to its role in cell mechanics, it is possible that the actin CSK is involved in the control of proliferation through other mechanisms. Indeed, we found that spreading induced an increase in proliferation, actin polymerization (F-actin content), and stress fiber formation (bundling), whereas elongation dramatically affected entanglement and bending of actin filaments but had little or no effect on proliferation, formation of stress fibers, and F-actin content. These observations indicate that the mechanisms regulating filament cross-linking and bending are not involved in the mitogenlike effects of spreading, whereas those mechanisms underlying either bundling or actin polymerization may be implicated. In support of the latter hypothesis, the actin polymerization regulator protein mDia1 was necessary to, but not sufficient for, the G1-S transition in ECs (16). In addition, actin polymerization may stimulate proliferation by increasing cyclin D1 levels, since there is evidence that cyclin D1 is regulated posttranscriptionally by cell shape, independent of mRNA levels (14), and that essential components of the synthetic machinery are associated with the actin CSK (53). A detailed knowledge of the role of the actin CSK in shape control of proliferation, however, awaits further investigation.
Nuclear volume in G1 is predictive of the proliferative status of single ECs
Like cell spreading, nuclear volume and the apparent chromatin decondensation of cells in the G1 phase were observed to strongly correlate both with DNA synthesis (Fig. 6) and with each other. These correlations between nuclear volume, chromatin condensation, and proliferation might in principle reflect unrelated downstream effects of cell spreading. However, and in agreement with our findings, inhibiting nuclear swelling blocked the transition into the S phase in HeLa cells (26), and overexpressing myosin light chain kinase in fibroblasts downregulated both proliferation and nuclear volume (17). In addition, an inverse relationship between nuclear volume and chromatin condensation similar to that observed in our study was reported in fibroblasts (20) and in epithelial cells (22,54). Therefore, these previous results, and our finding that the values of nuclear volume and chromatin condensation of cells that have not yet begun synthesizing DNA (in G1 phase) fit to a master curve with the DNA synthesis levels of the entire cell population, provides support for a mechanistic relationship between nuclear swelling, chromatin decondensation, the G1-S transition, and DNA synthesis. The role of nuclear volume could be important, even though its changes with spreading were small (from 1007 to 1323 μm3, 30% relative variation). Indeed, similar small relative changes in input biological signals are known to be sufficient to elicit dramatic biological responses, such as neutrophil migration in response to a chemoattractant gradient (55) or myocyte differentiation in response to narrow changes in substratum stiffness (51). Altogether these findings are consistent with our hypothesis that the nuclear volume of ECs in the G1 phase might be a key parameter mediating the effects of cell spreading in the transition to the S phase.
Biophysical model relating nuclear swelling and DNA synthesis
Due to the high concentration of macromolecules present in the nucleus, chromatin is embedded in an extremely crowded environment (56–58). Theoretical and experimental evidence shows that macromolecular crowding leads to volume exclusion (entropic) effects that increase macromolecular association (59–61). Conversely, it is expected that the reduction in macromolecular crowding caused by an increase in nuclear volume will reduce the entropic repulsions between different chromosomic regions, resulting in global chromatin decondensation. In support of this concept, macromolecular crowding and chromatin condensation were found to be associated in HeLa cells (61). Living cells could thus control chromatin organization by taking advantage of such a physical mechanism, in addition to biochemical processes such as histone acetylation or DNA methylation (22,23). Indeed, because chromatin is subjected to both biochemical and biophysical signals, it is likely that these two different cues are convoluted or complementary rather than acting in isolation. In support of this hypothesis, it has been recently reported that a downregulation of global histone acetylation (commonly associated with less condensed chromatin or heterochromatin) in mammary epithelial cells, initially observed as a downstream effect of binding to the extracellular matrix (ECM), could also be induced simply by changing cell shape in the absence of ECM signaling (22). Independent of the specific mechanism, chromatin decondensation could facilitate the entry into the S phase by rendering DNA more accessible to the replication machinery, since there is evidence that chromatin compaction constitutes a barrier for DNA replication (62). Indeed, a similar mechanism has been proposed for transcription regulation (12,54,61), as transcription and chromatin condensation are closely associated (20) and the degree of transcription and nuclear volume were shown to correlate in erythrocytes, lymphocytes, and erythroblasts (63–65). It is important to note that our findings and these previous data were obtained using cells cultured in two-dimensional substrata. When suitable micropatterning techniques become available in three-dimensional environments, it will be interesting to determine whether the strong correlation between nuclear volume, chromatin decondensation, and DNA synthesis still holds. In principle, however, we expect that the general mechanism described here could be valid also for three-dimensional cultures. Therefore, these and previous findings, although not conclusive, are consistent with a biophysical model by which a decrease in chromatin condensation (induced by nuclear swelling and/or other mechanisms) promotes DNA synthesis by rendering DNA more accessible to the replication machinery.
Regulation of nuclear volume by cell shape
Given the observed coupling between proliferation and nuclear volume, attention should be focused on understanding how nuclear volume is regulated by cell shape. Although the possible mechanisms remain largely unidentified, we can envision at least three of them. First, cell shape could affect nuclear volume through a link with cell volume, as has been reported for other cell types (42,66) However, we did not observe a straightforward relationship between nuclear and cell volume (data not shown), and the relationship between cell and nuclear size and the cell cycle remains controversial (42). Second, nuclear volume could be controlled by changes in biochemical signaling downstream of the ECM and growth factors given by spreading. Finally, and given the known mechanical coupling between the nucleus and the CSK (21,67), cell shape could regulate nuclear shape and volume through the mechanical tension and organization of the actin network and other CSK filaments. Indeed, reducing mechanical tension in the CSK decreased nuclear size in ECs (68). A high mechanical tension could thus stretch and enlarge the nucleus through direct mechanical distortion and/or through mechanotransduction processes (22,69,70). The transmission of mechanical stresses from the cytoskeleton to the nucleus, however, may not only depend on cytoskeletal tension, but also on how the nucleus is connected to cytoskeletal filaments and how these filaments are oriented. Indeed, the effect of these additional parameters might account for the lack of a direct relationship observed between cytoskeletal stiffness and nuclear volume. Nevertheless, whether this intricate force transmission between the cytoskeleton and the nucleus may help to explain the complex observed relationship between cytoskeletal tension, nuclear volume, and proliferation remains an open question. Additionally, it should be noted that none of these three mechanisms of nuclear volume regulation are mutually exclusive.
The development and maintenance of tissue architecture requires an exquisite and tight control of proliferation, which involves the interplay of microenvironmental cues including growth factors, extracellular matrix, and cell shape. Recently developed biophysical techniques can facilitate dissection of the mechanisms underlying the complex control of proliferation in culture. In this study, we controlled several aspects of cell shape through different micropatterning techniques to examine the role of major cell and nuclear biophysical properties (cytoskeletal stiffness, contractility, mechanical anisotropy, and nuclear volume) in mediating the mitogenlike effects of cell spreading in single ECs. We found that, unlike any of the commonly measured cell mechanical parameters, nuclear volume strongly correlated with DNA synthesis for all shapes. Our results show that the relationship between cell mechanics and proliferation is strongly nonlinear and/or indirect. Although not conclusive, our findings also support the hypothesis that nuclear swelling and chromatin decondensation in G1 could be part of a mechanism by which cell spreading promotes the G1-S transition in ECs. This mechanism may be important in in vivo physiological processes, including endothelial barrier repair and angiogenesis. In addition, similar mechanisms could underlie other shape-regulated processes, such as stem cell commitment (52,71).
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The authors acknowledge M. Rodríguez for technical assistance, Drs. C. Nelson, C. Mills, F. Bessueille, and M. Pla for help in defining the micropatterning technique, and Dr. X. Trepat for useful comments.
This work was supported in part by the Ministerio de Educación y Ciencia (NAN2004-09348-C04-04, SAF2005-00110), the Ministerio de Sanidad y Consumo (FIS-PI040929), and by a NANOTEC postdoctoral fellowship from the Generalitat de Catalunya (to J.A.).
Pere Roca-Cusachs and Jordi Alcaraz contributed equally to this work.
Editor: Gaudenz Danuser.
References
- 1.Ingber, D. E. 2006. Mechanical control of tissue morphogenesis during embryological development. Int. J. Dev. Biol. 50:255–266. [DOI] [PubMed] [Google Scholar]
- 2.Grose, R., and P. Martin. 1999. Parallels between wound repair and morphogenesis in the embryo. Semin. Cell Dev. Biol. 10:395–404. [DOI] [PubMed] [Google Scholar]
- 3.Lee, J. S. Y., and A. I. Gotlieb. 2003. Understanding the role of the cytoskeleton in the complex regulation of the endothelial repair. Histol. Histopathol. 18:879–887. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan, D., and J. Folkman. 1996. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 86:353–364. [DOI] [PubMed] [Google Scholar]
- 5.Folkman, J., and A. Moscona. 1978. Role of cell shape in growth control. Nature. 273:345–349. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. S., M. Mrksich, S. Huang, G. M. Whitesides, and D. E. Ingber. 1997. Geometric control of cell life and death. Science. 276:1425–1428. [DOI] [PubMed] [Google Scholar]
- 7.Dike, L. E., C. S. Chen, M. Mrksich, J. Tien, G. M. Whitesides, and D. E. Ingber. 1999. Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. In Vitro Cell. Dev. Biol. Anim. 35:441–448. [DOI] [PubMed] [Google Scholar]
- 8.Bhadriraju, K., and L. K. Hansen. 2002. Extracellular matrix- and cytoskeleton-dependent changes in cell shape and stiffness. Exp. Cell Res. 278:92–100. [DOI] [PubMed] [Google Scholar]
- 9.Wang, N., and D. E. Ingber. 1994. Control of cytoskeletal mechanics by extracellular matrix, cell shape, and mechanical tension. Biophys. J. 66:2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, N., E. Ostuni, G. M. Whitesides, and D. E. Ingber. 2002. Micropatterning tractional forces in living cells. Cell Motil. Cytoskeleton. 52:97–106. [DOI] [PubMed] [Google Scholar]
- 11.Tan, J. L., J. Tien, D. M. Pirone, D. S. Gray, K. Bhadriraju, and C. S. Chen. 2003. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 100:1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolic-Norrelykke, I. M., and N. Wang. 2005. Traction in smooth muscle cells varies with cell spreading. J. Biomech. 38:1405–1412. [DOI] [PubMed] [Google Scholar]
- 13.Wang, N., I. M. Tolic-Norrelykke, J. X. Chen, S. M. Mijailovich, J. P. Butler, J. J. Fredberg, and D. Stamenovic. 2002. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. Am. J. Physiol. Cell Physiol. 282:C606–C616. [DOI] [PubMed] [Google Scholar]
- 14.Huang, S., C. S. Chen, and D. E. Ingber. 1998. Control of cyclin D1, p27(Kip1), and cell cycle progression in human capillary endothelial cells by cell shape and cytoskeletal tension. Mol. Biol. Cell. 9:3179–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson, C. M., R. P. Jean, J. L. Tan, W. F. Liu, N. J. Sniadecki, A. A. Spector, and C. S. Chen. 2005. Emergent patterns of growth controlled by multicellular form and mechanics. Proc. Natl. Acad. Sci. USA. 102:11594–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammoto, A., S. Huang, K. Moore, P. Oh, and D. E. Ingber. 2004. Role of RhoA, mDia, and ROCK in cell shape-dependent control of the Skp2-p27(kip1) pathway and the G(1)/S transition. J. Biol. Chem. 279:26323–26330. [DOI] [PubMed] [Google Scholar]
- 17.Cai, S., L. Pestic-Dragovich, M. E. O'Donnell, N. Wang, D. Ingber, E. Elson, and P. De Lanerolle. 1998. Regulation of cytoskeletal mechanics and cell growth by myosin light chain phosphorylation. Am. J. Physiol. Cell Physiol. 44:C1349–C1356. [DOI] [PubMed] [Google Scholar]
- 18.Ingber, D. E., D. Prusty, Z. Q. Sun, H. Betensky, and N. Wang. 1995. Cell shape, cytoskeletal mechanics, and cell cycle control in angiogenesis. J. Biomech. 28:1471–1484. [DOI] [PubMed] [Google Scholar]
- 19.Thomas, C. H., J. H. Collier, C. S. Sfeir, and K. E. Healy. 2002. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA. 99:1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vergani, L., M. Grattarola, and C. Nicolini. 2004. Modifications of chromatin structure and gene expression following induced alterations of cellular shape. Int. J. Biochem. Cell Biol. 36:1447–1461. [DOI] [PubMed] [Google Scholar]
- 21.Maniotis, A. J., C. S. Chen, and D. E. Ingber. 1997. Demonstration of mechanical connections between integrins cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 94:849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Beyec, J., R. Xu, S. Y. Lee, C. M. Nelson, A. Rizki, J. Alcaraz, and M. J. Bissell. 2007. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp. Cell Res. In press. [DOI] [PMC free article] [PubMed]
- 23.Francastel, C., D. Schubeler, D. I. K. Martin, and M. Groudine. 2000. Nuclear compartmentalization and gene activity. Nat. Rev. Mol. Cell Biol. 1:137–143. [DOI] [PubMed] [Google Scholar]
- 24.Underwood, J. M., K. M. Imbalzano, V. M. Weaver, A. H. Fischer, A. N. Imbalzano, and J. A. Nickerson. 2006. The ultrastructure of MCF-10A acini. J. Cell. Physiol. 208:141–148. [DOI] [PubMed] [Google Scholar]
- 25.Grigoryev, S. A., T. Nikitina, J. R. Pehrson, P. B. Singh, and C. L. Woodcock. 2004. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J. Cell Sci. 117:6153–6162. [DOI] [PubMed] [Google Scholar]
- 26.Yang, L., T. L. Guan, and L. Gerace. 1997. Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol. 139:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yen, A., and A. B. Pardee. 1979. Role of nuclear size in cell-growth initiation. Science. 204:1315–1317. [DOI] [PubMed] [Google Scholar]
- 28.Tan, J. L., W. Liu, C. M. Nelson, S. Raghavan, and C. S. Chen. 2004. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 10:865–872. [DOI] [PubMed] [Google Scholar]
- 29.Roca-Cusachs, P., I. Almendros, R. Sunyer, N. Gavara, R. Farre, and D. Navajas. 2006. Rheology of passive and adhesion-activated neutrophils probed by atomic force microscopy. Biophys. J. 91:3508–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alcaraz, J., L. Buscemi, M. Grabulosa, X. Trepat, B. Fabry, R. Farre, and D. Navajas. 2003. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys. J. 84:2071–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams, D. S. 1992. Mechanisms of cell shape change: the cytomechanics of cellular response to chemical environment and mechanical loading. J. Cell Biol. 117:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butt, H. J., and M. Jaschke. 1995. Calculation of thermal noise in atomic force microscopy. Nanotechnology. 6:1–7. [Google Scholar]
- 33.Rico, F., P. Roca-Cusachs, N. Gavara, R. Farre, M. Rotger, and D. Navajas. 2005. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 72:021914. [DOI] [PubMed] [Google Scholar]
- 34.Alcaraz, J., L. Buscemi, M. Puig-de-Morales, J. Colchero, A. Baro, and D. Navajas. 2002. Correction of microrheological measurements of soft samples with atomic force microscopy for the hydrodynamic drag on the cantilever. Langmuir. 18:716–721. [Google Scholar]
- 35.Pesen, D., and J. H. Hoh. 2005. Micromechanical architecture of the endothelial cell cortex. Biophys. J. 88:670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavara, N., R. Sunyer, P. Roca-Cusachs, R. Farre, M. Rotger, and D. Navajas. 2006. Thrombin-induced contraction in alveolar epithelial cells probed by traction microscopy. J. Appl. Physiol. 101:512–520. [DOI] [PubMed] [Google Scholar]
- 37.Butler, J. P., I. M. Tolic-Norrelykke, B. Fabry, and J. J. Fredberg. 2002. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 282:C595–C605. [DOI] [PubMed] [Google Scholar]
- 38.Horcas, I., R. Fernandez, J. M. Gomez-Rodriguez, J. Colchero, J. Gomez-Herrero, and A. M. Baro. 2007. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 78:013705. [DOI] [PubMed] [Google Scholar]
- 39.Kubinova, L., J. Janacek, F. Guilak, and Z. Opatrny. 1999. Comparison of several digital and stereological methods for estimating surface area and volume of cells studied by confocal microscopy. Cytometry. 36:85–95. [DOI] [PubMed] [Google Scholar]
- 40.Mascetti, G., S. Carrara, and L. Vergani. 2001. Relationship between chromatin compactness and dye uptake for in situ chromatin stained with DAPI. Cytometry. 44:113–119. [DOI] [PubMed] [Google Scholar]
- 41.Kidoaki, S., T. Matsuda, and K. Yoshikawa. 2006. Relationship between apical membrane elasticity and stress fiber organization in fibroblasts analyzed by fluorescence and atomic force microscopy. Biomech. Model. Mechanobiol. 5:263–272. [DOI] [PubMed] [Google Scholar]
- 42.Umen, J. G. 2005. The elusive sizer. Curr. Opin. Cell Biol. 17:435–441. [DOI] [PubMed] [Google Scholar]
- 43.Gregory, T. R. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 76:65–101. [DOI] [PubMed] [Google Scholar]
- 44.Frazier, E. A. J. 1973. DNA synthesis following gross alterations of nucleocytoplasmic ratio in ciliate Stentor coeruleus. Dev. Biol. 34:77–92. [DOI] [PubMed] [Google Scholar]
- 45.Ingber, D. E., and J. Folkman. 1989. Mechanochemical switching between growth and differentiation during fibroblast growth factor-stimulated angiogenesis in vitro: role of extracellular matrix. J. Cell Biol. 109:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies, P. F. 1995. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 75:519–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doornaert, B., V. Leblond, E. Planus, S. Galiacy, V. M. Laurent, G. Gras, D. Isabey, and C. Lafuma. 2003. Time course of actin cytoskeleton stiffness and matrix adhesion molecules in human bronchial epithelial cell cultures. Exp. Cell Res. 287:199–208. [DOI] [PubMed] [Google Scholar]
- 48.Gardel, M. L., F. Nakamura, J. H. Hartwig, J. C. Crocker, T. P. Stossel, and D. A. Weitz. 2006. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. USA. 103:1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakatsuki, T., R. B. Wysolmerski, and E. L. Elson. 2003. Mechanics of cell spreading: role of myosin II. J. Cell Sci. 116:1617–1625. [DOI] [PubMed] [Google Scholar]
- 50.Omelchenko, T., J. M. Vasiliev, I. M. Gelfand, H. H. Feder, and E. M. Bonder. 2002. Mechanisms of polarization of the shape of fibroblasts and epitheliocytes: separation of the roles of microtubules and Rho-dependent actin-myosin contractility. Proc. Natl. Acad. Sci. USA. 99:10452–10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engler, A. J., M. A. Griffin, S. Sen, C. G. Bonnetnann, H. L. Sweeney, and D. E. Discher. 2004. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 166:877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engler, A. J., S. Sen, H. L. Sweeney, and D. E. Discher. 2006. Matrix elasticity directs stem cell lineage specification. Cell. 126:677–689. [DOI] [PubMed] [Google Scholar]
- 53.Hesketh, J. 1994. Translation and the cytoskeleton: a mechanism for targeted protein synthesis. Mol. Biol. Rep. 19:233–243. [DOI] [PubMed] [Google Scholar]
- 54.Dahl, K. N., A. J. Engler, J. D. Pajerowski, and D. E. Discher. 2005. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 89:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Onsum, M., and C. V. Rao. 2007. A mathematical model for neutrophil gradient sensing and polarization. PLoS Comput. Biol. 3:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook, P. R. 2002. Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 32:347–352. [DOI] [PubMed] [Google Scholar]
- 57.Medalia, O., I. Weber, A. S. Frangakis, D. Nicastro, G. Gerisch, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 298:1209–1213. [DOI] [PubMed] [Google Scholar]
- 58.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science. 291:843–847. [DOI] [PubMed] [Google Scholar]
- 59.Zimmerman, S. B., and A. P. Minton. 1993. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22:27–65. [DOI] [PubMed] [Google Scholar]
- 60.Ellis, R. J. 2001. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 26:597–604. [DOI] [PubMed] [Google Scholar]
- 61.Iborra, F. 2007. Can visco-elastic phase separation, macromolecular crowding and colloidal physics explain nuclear organisation? Theor. Biol. Med. Model. 4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demeret, C., Y. Vassetzky, and M. Mechali. 2001. Chromatin remodelling and DNA replication: from nucleosomes to loop domains. Oncogene. 20:3086–3093. [DOI] [PubMed] [Google Scholar]
- 63.Harris, H. 1974. Nucleus and the Cytoplasm. Clarendon Press, Oxford, UK.
- 64.Setterfield, G., R. Hall, T. Bladon, J. Little, and J. G. Kaplan. 1983. Changes in structure and composition of lymphocyte nuclei during mitogenic stimulation. J. Ultrastruct. Res. 82:264–282. [DOI] [PubMed] [Google Scholar]
- 65.Biggiogera, M., A. Trentani, T. E. Martin, and C. Pellicciari. 1999. Terminal differentiation of erythroblasts leads to RNP segregation and formation of heterogeneous ectopic RNP-derived structures. Histochem. Cell Biol. 112:473–477. [DOI] [PubMed] [Google Scholar]
- 66.Swanson, J. A., M. Lee, and P. E. Knapp. 1991. Cellular dimensions affecting the nucleocytoplasmic volume ratio. J. Cell Biol. 115:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu, S. H., J. X. Chen, J. P. Butler, and N. Wang. 2005. Prestress mediates force propagation into the nucleus. Biochem. Biophys. Res. Commun. 329:423–428. [DOI] [PubMed] [Google Scholar]
- 68.Sims, J. R., S. Karp, and D. F. Ingber. 1992. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J. Cell Sci. 103:1215–1222. [DOI] [PubMed] [Google Scholar]
- 69.Illi, B., S. Nanni, A. Scopece, A. Farsetti, P. Biglioli, M. C. Capogrossi, and C. Gaetano. 2003. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ. Res. 93:155–161. [DOI] [PubMed] [Google Scholar]
- 70.Alcaraz, J., C. M. Nelson, and M. J. Bissell. 2004. Biomechanical approaches for studying integration of tissue structure and function in mammary epithelia. J. Mammary Gland Biol. Neoplasia. 9:361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McBeath, R., D. M. Pirone, C. M. Nelson, K. Bhadriraju, and C. S. Chen. 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 6:483–495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








