Abstract
The highly conserved signature N-terminal peptide of histone protein H3 plays crucial roles in gene expression controls. We investigated the conformational changes of the peptide caused by lysine dimethylation and acetylation of the histone H3 N-terminal tail by molecular dynamics and replica-exchange molecular dynamics simulations. Our results suggest that the most populated structures of the modified H3 N-terminal peptides are very similar to those of the wild-type peptide. Thus, the modifications introduce marginal changes to the most favorable conformations of the peptides. However, the modifications have significant effects on the stabilities of the most populated states that depend on the modifications. Whereas dimethylation of lysine 4 or lysine 9 alone tends to stabilize the most populated states, double dimethylation and acetylation of both lysine 4 and lysine 9 reduce both the helical conformation and the stability of the most populated states significantly. The calculated melting temperatures showed that the doubly acetylated peptide has the lowest melting temperature (Tm = 324 K), which is notably lower than the melting temperatures of the other four peptides (Tm ≈ 346–350 K). In light of the existing experimental evidence, we propose that the changes in the dynamics of the modified variants contribute to their different functional roles.
INTRODUCTION
In eukaryotic cells, DNA molecules exist in the form of chromatins and are packed into nuclei in a hierarchical manner through which the nucleosomes serve as the basic building blocks of chromatins. In each nucleosome core particle (NCP), DNA wraps around a disk-like histone hetero-octamer composed of eight subunits including one (H3-H4)2 tetramer and two H2A-H2B dimers. Sequence comparison indicates that the histone proteins are highly conserved among the eukaryotic cells. They contain a large portion of basic amino acids, mainly lysines and arginines (1). A growing body of evidence has shown that the functions of the NCP (e.g., DNA packing and gene transcription) are closely correlated to the amino acid modifications of the histones (2).
Posttranslational modifications, such as phosphorylation, methylation, and acetylation, are important mechanisms in cellular signaling (3). These modifications occur commonly on certain natural amino acids. For example, lysines can be methylated or acetylated; serines and threonines can be phosphorylated. These modifications are crucial in changing the physiochemical properties and, sometimes, the structure of the associated proteins. These changes often result in the initiation of important processes such as signal transduction. In other cases, such modifications can lead to significant changes in chemical properties of the proteins and are often utilized for proteins to communicate and interact with other molecules.
Interesting posttranslational modifications are frequently found on lysines, which can be methylated or acetylated through the replacement of the H from the ɛ-NH3 group with methyl or acetyl groups. Studies have shown that modifications on histones are highly correlated to the control of gene expression. For example, methylation of the Lys-4 of histone protein H3 is observed to be significantly more abundant in actively expressed chromatin regions than in other regions (4), whereas methylation of other lysines (Lys-9 or Lys-27) is correlated with repression of gene expression (5,6). At the molecular level, these chemical modifications are thought to change the chemical properties of the amino acids and the associated local regions of the proteins. For example, the wild-type (WT) lysine carries one positive net charge under physiological conditions, but its ɛ-NH2 group can be capped with a methyl group in the case of monomethylation. Although methylation does not remove the net charge, the bulky methyl group reduces hydrophilicity. On the other hand, acetylation removes the net charge because the replacement of the H by the acetyl group changes the charge distribution of the residue (7). Smart and McCammon (8) conducted Monte Carlo/stochastic dynamics simulations and found that the phosphorylation of serine stabilizes the N-termini of α-helices by favoring electrostatic interactions between the phosphate and the helix backbone.
In this study, we investigated the effects of methylation and acetylation on the structure and dynamics of an 18-residue variant of the histone H3 N-terminal peptide (WT sequence: ARTKQ TARKS TGGKA PGG). This peptide is almost identical to the H3 N-terminal peptide except for the last two glycines. The peptide is a signature peptide of histone protein H3; it is very specific to nucleosome H3 protein N-terminal and is highly conserved through evolution. We chose to study this peptide because of its functional significance and the availability of the nuclear magnetic resonance (NMR) structure of the peptide in complex with the mouse HP1 protein chromodomain (PID: 1GUW) (9). The peptide is important for the binding of heterochromatin protein 1 (HP1) to nucleosomes and is used to study the binding mechanism experimentally (9). HP1 plays roles in modulating the gene silencing and in the formation of heterochromatin through interactions with H3 protein N-termini and other nucleus proteins, and the HP1 protein family plays roles in gene regulation, DNA replication, and nuclear architecture, as reviewed by Eissenberg and Elgin (10). Experiments have demonstrated that H3 N-terminal peptides with a dimethylated Lys-9 can bind to HP1 in competition with the nucleosome (presumably through binding to histone tails) (11). The same effects have not been observed for either the WT or the dimethylated Lys-4 peptides. Methylation of Lys-9 has been found to be essential for the binding of HP1 (12). Furthermore, Lys-4 and Lys-9 are in the dimethylated state in the NMR structure (PDB code: 1GUW) formed as a complex of HP1 and the H3 N-terminal peptide (9).
Recent studies on the binding mechanism of the ING2 plant homeodomain to histone H3 with trimethylated Lys-4 have shown that the binding affinity is greatly affected by an intermolecular hydrogen bond and complementary surface interactions (13). In the crystal structure, the H3 peptide with trimethylated Lys-4 has an extended β-strand conformation, and the residues sit in two grooves connected by a narrow channel. The grooves perfectly match the peptide with the trimethylated Lys-4 but not the WT peptide or the peptides carrying other types of modifications. The focus of this article is on the induced changes in conformation and dynamics caused by posttranslational modifications and their roles in interacting with other proteins.
METHOD
System setup
Our study used the AMBER 8 package (14), and the peptides were represented by the AMBER ff03 force field parameter set developed by Duan et al. (15). We used the generalized Born model developed by Onufriev et al. (16) to mimic the solvent effects in the simulations. The sequence of simulated peptide is: ARTXQ TARXS TGGKA PGG, where X can be either the WT or the modified (dimethylated or acetylated) lysine. In theory, there potentially can be nine types of modified peptides by taking different states of X. Here we focus on those that are present in the histones and have been shown to play important roles in the functions of NCPs. Simulations on five states of the peptide were performed, including the WT with unmodified natural lysines, dimethylated Lys-4 (DIM4), dimethylated Lys-9 (DIM9), acetylated Lys-4, Lys-9, and Lys-14 (ACK), and the doubly dimethylated (dDIM) peptide where both Lys-4 and Lys-9 were dimethylated.
The force field parameters for methylated and acetylated lysines were obtained by following the procedure used in the AMBER force field ff03 (15) development. The charges were obtained by fitting to the electrostatic potentials of dipeptides (ACE-ACK-NME and ACE-DIM-NME) using the RESP method (17). The electrostatic potentials were calculated using the B3LYP/cc-pvtz quantum mechanical method in ether with the PCM model (18). The structures were optimized with HF/6-31G** before calculation of the electrostatic potentials, and the main chain dihedral angles were constrained to the α-helix (−60.0, −40.0) and β-sheet (−120.0, 140.0) conformations. All quantum mechanical calculations were performed using the Gaussian 03 program (19). The partial charges are summarized in Table 1. The backbone torsion parameters are the same as those of the natural lysine residue in the ff03 force field (15).
TABLE 1.
Partial charges of the acetylated lyine (ACK) and the dimethylated lysine (MLY)
| MLY
|
ACK
|
||||
|---|---|---|---|---|---|
| Atom name | Atom type | Charge | Atom name | Atom type | Charge |
| N | N | –0.4775 | N | N | –0.4773 |
| H | H | 0.2888 | H | H | 0.2829 |
| CA | CT | 0.0125 | CA | CT | –0.0069 |
| HA | H1 | 0.0852 | HA | H1 | 0.0939 |
| CB | CT | –0.0051 | CB | CT | –0.0050 |
| HB2 | HC | 0.0239 | HB2 | HC | 0.0175 |
| HB3 | HC | 0.0239 | HB3 | HC | 0.0175 |
| CG | CT | 0.0164 | CG | CT | –0.0017 |
| HG2 | HC | 0.0088 | HG2 | HC | 0.0066 |
| HG3 | HC | 0.0088 | HG3 | HC | 0.0066 |
| CD | CT | 0.0047 | CD | CT | 0.0291 |
| HD2 | HC | 0.0279 | HD2 | HC | 0.0142 |
| HD3 | HC | 0.0279 | HD3 | HC | 0.0142 |
| CE | CT | –0.0626 | CE | CT | 0.0171 |
| HE2 | HP | 0.1056 | HE2 | H1 | 0.0614 |
| HE3 | HP | 0.1056 | HE3 | H1 | 0.0614 |
| NZ | N3 | 0.0250 | NZ | N | –0.4866 |
| HZ1 | H | 0.3027 | HZ1 | H | 0.3173 |
| CH1 | CT | –0.2097 | C | C | 0.6496 |
| CH2 | CT | –0.2097 | O | O | –0.5936 |
| C | C | 0.6767 | CF | C | 0.6210 |
| O | O | –0.6003 | O24 | O | –0.5686 |
| H23 | HP | 0.1368 | C25 | CT | –0.4533 |
| H24 | HP | 0.1368 | H26 | HC | 0.1275 |
| H25 | HP | 0.1368 | H27 | HC | 0.1275 |
| H26 | HP | 0.1368 | H28 | HC | 0.1275 |
| H27 | HP | 0.1368 | |||
| H28 | HP | 0.1368 | |||
MD simulation
For each peptide, we performed simulations at two different temperatures: normal temperature simulations at 300 K to study the properties at physiological temperature and higher-temperature simulations at T = 360 K to study the structural preference and to investigate the response to the temperature change. Five independent trajectories for each peptide at each temperature were simulated. All the simulations were 100 ns long for a total simulation time of 5 μs. The initial structure was based on the NMR structure of the mouse HP1 protein chromodomain complex (PID: 1GUW) (9). For each peptide, the starting conformations were the same for all five trajectories. Different initial velocities were assigned according to the Maxwell-Boltzmann distribution to explore different regions of the conformation space.
The energy was minimized with 500 steps of the steepest descent gradient method. The temperature was controlled with Berendsen's weak coupling algorithm (20) with a coupling constant of 2.0 ps. Although the ensemble generated by the temperature-control method is not known, the distribution of potential energies resembles microcanonical sampling for the same system. The protein interior dielectric constant was set to 1.0, and the exterior solvent dielectric constant was set to 78.5. The salt concentration was set to 0.2 M. Conformations were saved every 10.0 ps. SHAKE (21) was applied to constrain the bonds connecting hydrogen atoms. To avoid unstable dynamics, a smaller step size (1.0 fs) was used whenever necessary at T = 360 K. The cutoff for nonbonded interactions and GB pairwise summation was set to be 12.0 Å.
Replica exchange MD (REMD) simulation
The multisander module in an AMBER 8 package was used for REMD simulations. To achieve a 20% acceptance rate, we simulated 10 replicas at optimized target temperatures: 260 K, 287 K, 306 K, 326 K, 347 K, 370 K, 394 K, 420 K, 447 K, 476 K, and 507 K. The systems were first equilibrated for 1.0 ns to reach targeted temperatures, and then exchange was attempted every 2.0 ps (2000 MD steps). The simulation time step was set to 1.0 fs. The REMD simulations were conducted for 120 ns, and the data from the last 100 ns were used for analysis. All the other parameters were set to be the same as for the MD simulations.
Clustering structures
In all the clustering processes, a root mean-square deviation (RMSD) cutoff of 2.5 Å was applied, and five trajectories for each peptide were combined to form the corresponding conformation ensembles. The samples from REMD simulation were taken at T = 306 K. The RMSD was calculated based on Cα atom coordinates. Structures with pairwise RMSD below cutoff were clustered in the same group, and the cluster center was selected as the representative structure.
RESULTS
We first examined the effects of modifications by comparing the dihedral angle distributions and the secondary structures. Then the sampled conformations were clustered to find the most populated structures. We will focus on the differences among the modified and WT peptides.
Secondary structure distributions
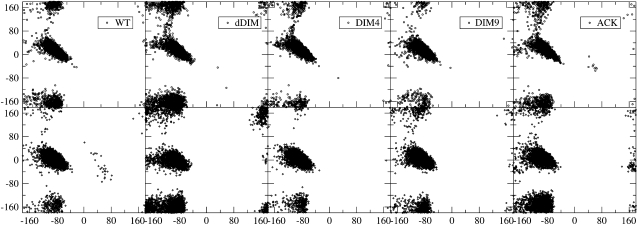
The main chain φ and ψ dihedral angles are the principal degrees of freedom for the backbone conformation that can be used to characterize the protein and peptide structures. The Ramachandran plots of Lys-4 and Lys-9 are shown in Fig. 1, and the (percentage) populations in the α-helix region are summarized in Table 2. It is interesting to note that, despite the minor differences, the MD and REMD simulation results are in qualitative agreement. Because the replica exchange MD simulations gave better convergence than the MD simulations of comparable length, the results presented here are primarily based on the data from REMD simulations at T = 306 K. In all cases, although both the WT and modified lysines consistently prefer α-helical regions, there were variations depending on the state of modifications. For example, the single dimethylations on either Lys-4 alone (DIM4) or Lys-9 (DIM9) alone had only marginal effects on the α-helix conformation in comparison to the WT. In comparison, when both Lys-4 and Lys-9 are dimethylated (dDIM), both Lys-4 and Lys-9 have a notably smaller α-helix preference (26.8% and 45.3% lower for Lys-4 and Lys-9, respectively), making them much more likely than other peptides to form extended β-sheet conformations as found in the NMR complex structures. This also suggests a profound cooperative effect. Similarly, in the acetylated (ACK) peptide, the α-helix populations are also decreased. In particular, Lys-9 in the acetylated peptide had a notably 27% smaller α-helix population than the WT. The data from MD simulations at T = 300 K also gave the same qualitative trend.
FIGURE 1.
Ramachandran plots of Lys-4 (upper row) and Lys-9 (lower row) in the five peptides. These data are calculated from simulation trajectories at T = 300 K.
TABLE 2.
α-Helix population of Lys-4 and Lys-9
|
α-Helix population (%)
|
Relative to WT
|
|||
|---|---|---|---|---|
| Peptide | Lys-4 | Lys-9 | Lys-4 | Lys-9 |
| From MD simulations at 300 K | ||||
| WT | 83 ± 18 | 93 ± 6 | 0 | 0 |
| DIM4 | 91 ± 6 | 93 ± 18 | 8 | 0 |
| DIM9 | 95 ± 5 | 86 ± 18 | 12 | –7 |
| dDIM | 63 ± 25 | 55 ± 38 | –20 | –38 |
| ACK | 88 ± 15 | 68 ± 39 | 5 | –25 |
| From REMD simulations at 306 K | ||||
| WT | 97 ± 2 | 95 ± 3 | 0 | 0 |
| DIM4 | 96 ± 3 | 97 ± 3 | –1 | 2 |
| DIM9 | 95 ± 2 | 85 ± 6 | –2 | –10 |
| dDIM | 71 ± 11 | 50 ± 18 | –26 | –45 |
| ACK | 90 ± 9 | 68 ± 9 | –7 | –27 |
Clustering result
The most populated structures of different peptides are similar
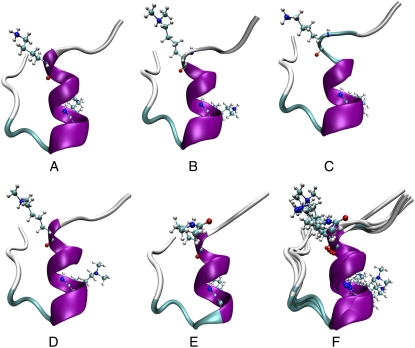
The representative structures of the most populated clusters for all five peptides are summarized in Fig. 2. Interestingly, these representative structures are very similar: all share a helical segment from Lys-4 to about Thr-11, and the rest of the peptides are in extended conformation. We compared the most populated structures and calculated the pairwise RMSD (Table 3). The most populated structures of the five peptides were highly similar from both MD and REMD simulations. The backbone RMSD is between 0.49 Å and 1.39 Å from the MD simulations at 300 K. From the REMD simulations at 306 K, the backbone RMSD ranges from 0.58 Å to 1.08 Å. The MD (300 K) and the REMD (T = 306 K) also yielded similar structures, and the largest pairwise backbone RMSD between the representative structures from these two types of simulation is within 1.68 Å.
FIGURE 2.
Most populated structures. (A) WT. (B) Dimethylated residue 4 (DIM4). (C) Dimethylated residue 9 (DIM9). (D) Dimethylated residues 4 and 9 (dDIM). (E) Acetylated residues 4 and 9 (ACK). (F) Superimposed.
TABLE 3.
Pairwise RMSD between the most populated structures
| WT | DIM4 | DIM9 | dDIM | ACK | |
|---|---|---|---|---|---|
| From MD at 300 K | |||||
| WT | 0 | ||||
| DIM4 | 1.18 | 0 | |||
| DIM9 | 1.23 | 0.49 | 0 | ||
| dDIM | 1.14 | 0.64 | 0.69 | 0 | |
| ACK | 0.84 | 1.29 | 1.39 | 1.24 | 0 |
| From REMD at 306 K | |||||
| WT | 0 | ||||
| DIM4 | 0.58 | 0 | |||
| DIM9 | 0.85 | 0.77 | 0 | ||
| dDIM | 0.81 | 0.82 | 0.73 | 0 | |
| ACK | 1.04 | 1.06 | 1.08 | 0.88 | 0 |
The units are angstroms. The largest RMSD between the REMD and MD structures is 1.68 Å.
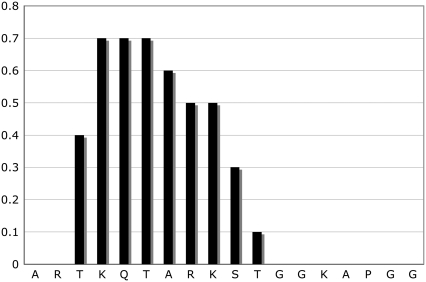
To compare with experimental results, we searched the Protein Data Bank (PDB) (22) for the experimentally solved structures of the WT peptide. The BLAST (23) search against PDB dataset returned 23 hits with an expectation value less than 0.001. Thirteen of them were NCP structures with undetermined structure of the N-terminal, which is the aligned region of our target sequence. The other 12 entries were the H3 N-terminal peptides bound by other proteins. Thus, the experimental apo- structure of the peptide remains unknown. Because there is no experimentally determined structure for this peptide in its free state, we resorted to secondary structure prediction tools that could provide indirect evidence about the structures. There are many sophisticated secondary structure prediction programs and servers. The secondary structures are shown in Fig. 3, as predicted by the PredictProtein server hosted by Columbia University Bioinformatics Center (24). We also examined other servers and secondary structure prediction programs, and the results are consistent. In these predictions, the fragment from Thr-3 to Ser-10 is remarkably more helical than the rest, in qualitative agreement with our simulation results on the WT peptide. Although the secondary structure prediction is applicable only to the WT peptide, our simulations on the modified peptides indicate that they behave in a manner similar to the WT. For instance, the lysines (both WT and modified lysines) in the most populated clusters (Fig. 2) all point away from the helix and in very similar orientations. In this type of conformation, the side chains of lysines stay on the surface, and they do not have strong interactions with other residues.
FIGURE 3.
α-Helix conformation score from secondary structure prediction. The prediction is conducted with the server hosted by the Columbia University Bioinformatics Center (http://www.predictprotein.org).
All five peptides exhibited similar behavior at different temperatures
We performed a set of simulations at higher temperature (T = 360 K) with the same protocol to study the responses of the peptide to the temperature changes. Because of the enhanced dynamics at T = 360 K that enabled better sampling, the results at this temperature are more reliable than those at T = 300 K, as demonstrated by considerably more transitions among the most populated states and other states.
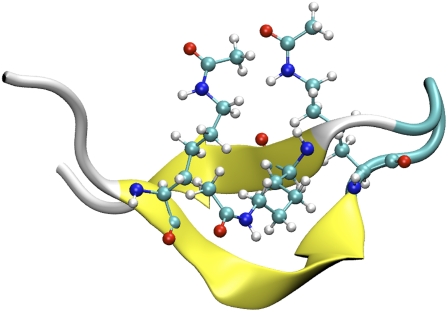
At T = 360 K, the most populated structures identified from the clustering analysis are also similar, with the ACK peptide being the only exception. For the ACK peptide, the most populated structure at 360 K resembles a β-hairpin conformation, as shown in Fig. 4 (to be discussed later), which is different from the helical structure identified from the 300 K simulations. However, the second most populated structure is helical, which is close to the most populated structures of other peptides. Thus, the peptides fold into very similar structures at two different temperatures, albeit with decreased population at higher temperature. This is reasonable because entropy plays a more significant role at higher temperature, such that the unfolded states are more favored. The population of the most populated structures at T = 360 K decreased to ∼20% of the corresponding population at T = 300 K.
FIGURE 4.
Representative structure for the most populated cluster of the peptide ACK at T = 360 K. The three acetylated lysines formed a cluster, which is different from the structures shown in Fig. 2.
Different peptides exhibited different stabilities
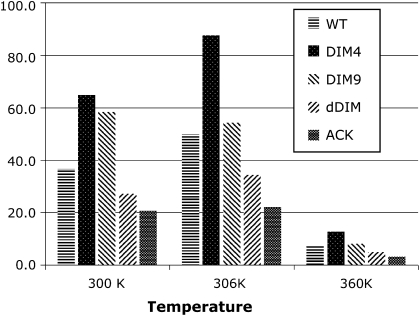
Although the most populated structures of the five peptides are similar, we found that they have very different stabilities. We summarized the populations and percentages of the most populated clusters in Table 4 and Fig. 5. The populations vary significantly for five peptides, indicating the considerable difference in stabilities. It is important to note that the 300 K MD and the 306 K REMD simulations yielded comparable results. This is rather encouraging, suggesting that the sampling by the MD simulations was adequate. In comparison, the 360 K MD data are notably different from the other two because of the elevated temperature. Nevertheless, the three sets of simulations gave qualitatively consistent results. The DIM4 peptide has the most stable conformation, whereas the ACK and dDIM peptides have the least, largely because of their reduced tendency to form helical structures. Because the dDIM peptide is found to bind the HP1 in the extended form, our simulation suggests that the reduced helical tendency may facilitate the binding. In comparison, DIM4 is more likely to maintain its helical structure.
TABLE 4.
Populations and percentages of the most populated structure clusters
| Population*
|
Percentage
|
|||||
|---|---|---|---|---|---|---|
| Peptide | 300 K† | 360 K† | 306K‡ | 300 K† | 360 K† | 306K‡ |
| WT | 1842 | 366 | 2509 | 36.8 | 7.3 | 50.2 |
| DIM4 | 3242 | 630 | 4365 | 64.8 | 12.6 | 87.3 |
| DIM9 | 2915 | 410 | 2713 | 58.3 | 8.2 | 54.3 |
| dDIM | 1364 | 243 | 1716 | 27.3 | 4.9 | 34.3 |
| ACK | 1030 | 165 | 1102 | 20.6 | 3.3 | 22.0 |
Total number of structures is 5000 for each peptide at each temperature.
Data at 300 K and 360 K are from conventional MD simulations.
Data at 306 K are from replica-exchange MD simulations.
FIGURE 5.
Populations of the largest clusters. The 306 K data are from REMD simulation (center group), and the other two groups are from MD simulations. The total population is 5000.
Based on the REMD simulation data, we calculated heat-capacity profiles and the melting temperatures for each peptide. Among the five variants, the ACK peptide has the lowest melting temperature of Tm = 324 K. The other four peptides have comparable Tm values, ranging from Tm = 346 K for dDIM and Tm = 347 K for WT to Tm = 350 K for DIM4 and DIM9 peptides. These results all suggest that the modifications indeed changed the structural stabilities.
Instead of changing the structures directly, the posttranslational modifications play roles in altering the stability of the most populated structures. In particular, the double dimethylations on both Lys-4 and Lys-9 changed properties much more significantly. This might have contributed to the stronger binding of the dDIM peptide to HP1 proteins. Because the peptide in the HP1-bound form is in the extended conformation, whereas the free peptide tends to be in the helical conformation, induced conformational changes are necessary to form the complex with HP1. Thus, peptides that are structurally more stable would be less favorable to the binding. Similarly, when the HP1 protein is present in the chromatin environment and searches for its binding sites, it selectively binds to the H3 N-terminal tails that more easily undergo the required conformational changes. Because of the difficulty of changing a helix into a strand in the bound state as a result of steric hindrance and the fact that HP1 has several β-strands and a long tail, which could interact with and stabilize the H3 N-terminal tail only if the tail is a β-strand, we propose that binding of HP1 to H3 N-terminal tails is facilitated by the enhanced flexibility of the modified tails.
DISCUSSION AND CONCLUSIONS
The H3 N-terminal sequence is a signature of histone protein H3. We searched the sequence against the National Center for Biotechnology Information's nonredundant database (http://www.ncbi.nlm.nih.gov/BLAST) and received 100 hits, all of them H3 N-terminal sequences except three unnamed protein products (23). This means that this sequence is very specific to the nucleosome H3 protein N-terminal and is very much conserved through evolution. Clearly, this underscores the fact that the peptide must be very important for all the eukaryotic cells to keep H3 N-terminal sequences unaltered. Thus, it is crucial to understand its structure and dynamics, which could help to provide information on gene regulation.
There are many discussions about the significant roles of the histone H3 N-terminal tails. Yet, only a few studies on the structures are available. Some view the tail as intrinsically disordered (25), which may help to interpret the tail's complex roles. If the H3 N-terminal is intrinsically disordered, it could change conformation easily according to the binding enzymes or factors, providing a natural framework to explain the multiple roles of the terminal tails. However, the H3 N-terminal also has selections on the binding proteins, which is the hallmark of the “histone code” hypothesis, suggesting that the terminal has preference for structures. Our simulation results suggest that the modifications mainly influence the stability, not the structures.
In this study, we applied MD simulations to investigate the changes in structural and dynamic properties of the H3 N-terminal peptide caused by the posttranslational modifications. We focused on the differences among peptides under various modifications. Because the simulations and the analyses are consistent throughout all peptides, and both MD and REMD studies yield consistent results, our conclusions on the posttranslational modification effects are plausible.
Acetylation on lysine is highly correlated with gene expression activation. One hypothesis is that the acetylated lysine could reduce the compactness of the chromatin, such that the transcription factors can access the DNA in acetylated chromatin regions. In our simulation, the ACK peptide with acetylated lysines was the least stable among the five peptides and had the highest preference toward a stable β-hairpin conformation. A closer examination of the structures reveals that the three acetylated lysines were packed closely to form a cluster. In comparison, lysines (or methylated lysines) point to different directions in other peptides. This is likely related to the fact that acetylation removes the net charge and increases the hydrophobicity. The reduced net charge of the acetylated lysines also weakens the binding interaction toward the DNA backbone as in the chromatin environment. Thus, the three acetylated lysines can form a hydrophobic cluster either by themselves to stabilize the β-hairpin-like conformation or with other hydrophobic residues, which could prevent the binding to the DNA backbone. They may also bind to the hydrophobic portion of the proteins that are associated with gene expression activation.
The simulation results showed that the modification of lysine(s) could alter the structure of the H3 N-terminal peptide marginally, and different modification states exhibit very similar conformational preferences. However, our model cannot fully describe the real situations of the nucleosome-protein interactions because there are many other factors in the nucleus that could affect those interactions. For example, we did not simulate the complete NCP system in our current study because of limitations of the computing resources. Rather, as an essential step to understanding the nucleosome dynamics, our study focuses on the intrinsic properties of H3 N-terminal peptides with implications for many important cellular processes, such as histone-DNA interactions, binding to other nucleosome proteins (i.e., HP1), heterochromatin formation, gene silencing and expression, and chromatin packing.
Acknowledgments
This work was supported by research grants from the National Institutes of Health (Nos. GM64458 and GM67168 to Y.D.). Usage of the Visual Molecular Dynamics graphics package is gratefully acknowledged.
Editor: Ron Elber.
References
- 1.Davey, C. A., D. F. Sargent, K. Luger, A. W. Maeder, and T. J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319:1097–1113. [DOI] [PubMed] [Google Scholar]
- 2.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature. 403:41–45. [DOI] [PubMed] [Google Scholar]
- 3.Clark, R. S., H. Bayir, and L. W. Jenkins. 2005. Posttranslational protein modifications. Crit. Care Med. 33:S407–S409. [DOI] [PubMed] [Google Scholar]
- 4.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA. 96:14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science. 300:131–135. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama, J.-i., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. S. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 292:110–113. [DOI] [PubMed] [Google Scholar]
- 7.Ramsden, M. J., F. S. R. Blake, and N. J. Fey. 1997. The effect of acetylation on the mechanical properties, hydrophobicity, and dimensional stability of Pinus sylvestris. Wood Sci. Technol. 31:97–104. [Google Scholar]
- 8.Smart, J. L., and J. A. McCammon. 1999. Phosphorylation stabilizes the N-termini of α-helices. Biopolymers. 49:225–233. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 416:103–107. [DOI] [PubMed] [Google Scholar]
- 10.Eissenberg, J. C., and S. C. R. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204–210. [DOI] [PubMed] [Google Scholar]
- 11.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 410:120–124. [DOI] [PubMed] [Google Scholar]
- 12.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 410:116–120. [DOI] [PubMed] [Google Scholar]
- 13.Peña, P. V., F. Davrazou, X. Shi, K. L. Walter, V. V. Verkhusha, O. Gozani, R. Zhao, and T. G. Kutateladze. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 442:100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Case, D. A., T. A. Darden, I. T. E. Cheatham, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, K. M. Merz, B. Wang, D. A. Pearlman, M. Crowley, S. Brozell, V. Tsui, H. Cohlke, J. Mongan, V. Homak, G. Cui, P. Beroza, C. Schafmeister, J. W. Caldwell, W. S. Ross, and P. A. Kollman. 2004. AMBER 8. University of California, San Francisco.
- 15.Duan, Y., C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang, and P. A. Kollman. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24:1999–2012. [DOI] [PubMed] [Google Scholar]
- 16.Onufriev, A., D. Bashford, and D. A. Case. 2004. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 55:383–394. [DOI] [PubMed] [Google Scholar]
- 17.Cornell, W. D., P. Cieplak, C. I. Bayly, and P. A. Kollman. 1993. Application of RESP charges to calculate conformational energies, hydrogen-bond energies, and free-energies of solvation. J. Am. Chem. Soc. 115:9620–9631. [Google Scholar]
- 18.Tomasi, J., B. Mennucci, and E. Cancès. 1999. The IEF version of the PCM solvation method: an overview of a new method addressed to study molecular solutes at the QM ab initio level. Theochem-J. Mol. Struct. 464:211–226. [Google Scholar]
- 19.Frisch, M. J., G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. J. A. Montgomery, T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez, and J. A. Pople. 2003. Gaussian 03. Gaussian, Inc., Pittsburgh, PA.
- 20.Berendsen, H. J. C., J. P. M. Postma, W. F. v. Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 21.Ryckaert, J.-P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 22.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen, J. C., X. Lu, E. D. Ross, and R. W. Woody. 2006. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281:1853–1856. [DOI] [PubMed] [Google Scholar]
- 26.Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman. 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J. Phys. Chem. 97:10269–10280. [Google Scholar]