Abstract
In yeast, fragmentation of amyloid polymers by the Hsp104 chaperone allows them to propagate as prions. The prion-forming domain of the yeast Sup35 protein is rich in glutamine, asparagine, tyrosine, and glycine residues, which may define its prion properties. Long polyglutamine stretches can also drive amyloid polymerization in yeast, but these polymers are unable to propagate because of poor fragmentation and exist through constant seeding with the Rnq1 prion polymers. We proposed that fragmentation of polyglutamine amyloids may be improved by incorporation of hydrophobic amino acid residues into polyglutamine stretches. To investigate this, we constructed sets of polyglutamine with or without tyrosine stretches fused to the non-prion domains of Sup35. Polymerization of these chimeras started rapidly, and its efficiency increased with stretch size. Polymerization of proteins with polyglutamine stretches shorter than 70 residues required Rnq1 prion seeds. Proteins with longer stretches polymerized independently of Rnq1 and thus could propagate. The presence of tyrosines within polyglutamine stretches dramatically enhanced polymer fragmentation and allowed polymer propagation in the absence of Rnq1 and, in some cases, of Hsp104.
Some proteins can undergo polymerization coupled with conformational rearrangement. This results in the formation of amyloid fibers distinguished by regular cross-β-sheet structure. Such fibers tend to aggregate, forming amyloid plaques or intracellular inclusions. Amyloids are considered to be the cause of >30 diseases of man and animals, many of which are neurodegenerative, including Alzheimer, Parkinson, and Huntington diseases (1). Amyloid diseases are noninfectious, with the exception of prion diseases related to PrP protein. Prion diseases include Creutzfeldt-Jacob disease, sheep scrapie, and other transmissible spongiform encephalopathies (2).
Amyloids were also discovered in fungi, in which they define useful or potentially useful phenotypes rather than diseases. In most cases, fungal amyloids are heritable and transmissible via cell fusions, which allows them to be considered as prions. In the yeast Saccharomyces cerevisiae, three prion-based genetic determinants have been described: [PSI+], [URE3], and [PIN+] (3–6). [PSI+] reflects polymerization of the translation termination factor Sup35 (eRF3). It shows a nonsense suppressor phenotype due to reduced termination of translation. [URE3] relates to polymerization of the Ure2 protein, which controls nitrogen metabolism. Sup35 and Ure2 both have an N-terminal prion-forming domain and a functional C-terminal domain. Sup35 also has a middle domain, which presumably acts as a spacer. [PIN+] relates to polymerization of the Rnq1 protein with unknown function (7). It has no phenotype of its own, but its presence allows [PSI+] induction de novo, presumably via cross-seeding of Sup35 polymerization with Rnq1 prion polymers (8–11).
Propagation of yeast prions requires the Hsp104 chaperone (12). A substantial body of evidence suggests that Hsp104 fragments prion polymers, thus increasing their number and accelerating polymerization (13–17). It is not clear whether other chaperones are required for this process. All yeast prions exhibit “strain” variation, i.e. heritable differences in phenotype and polymerization efficiency (18–20). “Strong” [PSI+] variants are distinguished by efficient nonsense suppression and high mitotic stability, whereas “weak” variants show weak suppression and low stability. The key physical property defining prion variation is the different propensity of polymers, dependent on their fold, to be recognized and fragmented by Hsp104 (14). More frequent fragmentation results in smaller but more numerous polymers. This defines higher mitotic stability of a prion and its more efficient polymerization. In the case of Sup35, the latter means lower levels of its monomers and more efficient nonsense suppression. The fragmentation may be so inefficient that it does not allow stable polymer propagation. We observed that the majority of the polymers formed by overproduced Sup35 in [PIN+] cells cannot propagate via fragmentation and exist due to continuous appearance, being seeded with the Rnq1 prion particles (10).
The polymerization domains of yeast prion proteins are distinguished by specific amino acid composition, being highly enriched in glutamine and/or asparagine. Other residues frequently found in prion domains, particularly in those of Sup35 from various yeast species, are tyrosine and glycine. In contrast, charged residues are very rare in them. It appears that such an amino acid composition of prion domains is more important for prion formation than their precise amino acid sequence because the sequences of amino acids in the Ure2 and Sup35 prion domains may be randomized without disrupting their ability to form prions (21, 22). Plain polyglutamine (polyQ)3 fragments of sufficient length can also cause amyloid formation. In particular, expansion of the polyQ fragments in human huntingtin and eight other proteins leads to their amyloid polymerization and respective diseases (23). In yeast, proteins with long polyQ domains also form aggregates (24, 25) composed of amyloid polymers (10). Polymer analysis is more reliable and informative than the study of aggregates because often the size of aggregates does not correlate with the size of polymers (14, 26). Study of the polyQ polymers led to the conclusion that they are not heritable because of their poor recognition and fragmentation by Hsp104 (10). Chaperone recognition is likely to require surface-exposed hydrophobic residues, whereas glutamine is not hydrophobic.
We proposed that the recognition of polyQ polymers by chaperones may be improved by incorporation of tyrosine, which represents the most frequent hydrophobic residue of the Sup35 prion-forming domain. To investigate this, we created two sets of plasmid constructs encoding fusions of polyQ or polyQ with dispersed tyrosine (polyQY) polypeptides to the Sup35 middle and C-terminal domains (referred to as Sup35MC). Analysis of these constructs showed that the presence of tyrosine residues dramatically improves polymer fragmentation. Unexpectedly, we found that polymers of proteins with a polyQ stretch longer than 70 residues are also fragmented and can propagate.
EXPERIMENTAL PROCEDURES
Strains and Genetic Methods—Yeast strain 74-D694/ΔS35 was obtained from 74-D694 (MATa ura3-52 leu2-3,112 trp1-289 his3-Δ200 ade1-14) [PIN+] (12) by disruption of the chromosomal SUP35 gene with TRP1 insertion and introduction of centromeric plasmid pRS315-Sup35C (27) or pRS313-Sup35C (described below) encoding the Sup35 C-terminal domain (referred to as Sup35C) to support viability. Yeast cells were grown at 30 °C in rich YPD (1% yeast extract (Oxoid), 2% peptone (Sigma), 2% glucose), YPD-red (1% yeast extract, 2% peptone, 4% glucose), or synthetic (0.67% yeast nitrogen base and 2% glucose supplemented with the required amino acids) medium.
Plasmid and Gene Replacement Construction—The fusion construct series were based on the pSBSE plasmid derived from pEMBLyex4-SUP35 (28). pEMBLyex4-SUP35 is a multicopy plasmid carrying the SUP35, URA3, and LEU2-d genes. LEU2-d was removed from it by deleting the 2263-bp ClaI fragment. The region of SUP35 encoding amino acids 1–124 was removed and replaced with the sequence GATCCCCGGGGATCCGAGCTCAAGGCGCC containing cloning sites SmaI, BamHI, SacI (Ecl136II), and EheI (NarI) but lacking the translation start site. Polyglutamine-encoding DNA constructs were synthesized using three pairs of complementary oligonucleotides (see supplemental Fig. 1). The “terminator” pair (AGTACTGATCAGCATGTCTGGCC and CTGGCCAGACATGCTGACTAGTA) encoded the start of translation and contained cloning sites Sau3AI and ScaI (underlined). “Elongator” pairs (AGCAACAACAGCAAC and CTGTTGCTGTTGTTG; AGCAACAATATCAAC and CTGTTGATATTGTTG) encoded QQQQQ or QQQYQ sequence. Every pair formed a duplex with protruding complementary 5′-AG and 5′-CT ends. Each elongator pair was mixed with the terminator pair at a 10:1 proportion. The mixture was treated with kinase and ligated, resulting in long polyoligomers with random distances between terminator fragments. Ligated DNA was cut with Sau3AI and ScaI, size-fractionated on agarose gel, and ligated to the BamHI and Ecl136II sites of the pSBSE plasmid. This created a fusion of the synthetic polyglutamine-encoding fragment to Sup35MC encoded by the plasmid. The ligation mixture was used to transform Escherichia coli. Clones of transformants were tested by PCR for the length of the polyglutamine-encoding inserts. Selected clones were verified by sequencing of the inserts. In this way, we obtained a series of multicopy yeast plasmids (pnQ/QY-Sup35MC) expressing fusion proteins with the sequence MSG-(QQQ[Q/Y]Q)m-QSQGA-(Sup35MC). Surprisingly, some plasmids encoded proteins with the sequence MSG-(QQQ[Q/Y]Q)m-PQGA-(Sup35MC), with P instead of QS. These constructs were also used for studies because the difference did not appear to be significant. The plasmid yex4MetQ66MC35 encoding 66Q and the plasmid and procedure used for RNQ1 disruption with HIS3 were described previously (10). To disrupt HSP104 with LEU2, HSP104 (from -558 to +801) was cloned into the SmaI and SalI sites of the pBC-KS+ plasmid (Stratagene) and disrupted with LEU2 between BglII and HindIII sites. For HSP104 disruption in the chromosome, this plasmid was cut with SmaI and SacI and used to transform yeast. RNQ1 and HSP104 disruptions were confirmed by immunoblot analysis of cell lysates for Rnq1 and Hsp104.
Preparation of Yeast Cell Lysates—Yeast cultures were grown in liquid medium to A600 = 1.5. The cells were harvested; washed in water; and lysed by glass beads in 25 mm Tris-HCl (pH 7.4), 100 mm NaCl, 1 mm dithiothreitol, and 1% Triton X-100. To prevent proteolytic degradation, 10 mm phenylmethylsulfonyl fluoride and Complete™ protease inhibitor mixture (Roche Applied Science) were added. Cell debris was removed by centrifugation at 1500 × g for 4 min.
Electrophoresis and Blotting—These were performed as described previously (29). Protein loads were equalized for each gel. Protein monomers of lysates were analyzed on 9% SDS-polyacrylamide gels without boiling of the samples. For separation of amyloid polymers, we used horizontal 1.8% agarose gels in Tris acetate/EDTA buffer with 0.1% SDS. Lysates were then incubated in 0.5× Tris acetate/EDTA, 2% SDS, 5% glycerol, and 0.05% bromphenol blue for 5 min at 37 °C. After electrophoresis, the proteins were transferred from gels to Immobilon-P polyvinylidene difluoride sheets (Millipore Corp.) by vacuum blotting overnight (agarose gels) or electrophoretically (acrylamide gels). Bound antibody was detected using the Amersham Biosciences ECL system. It should be noted that SDS in the non-boiled samples increases degradation of Sup35 monomers. This can result in the absence of Sup35 monomer bands on semidenaturing detergent-agarose gels (14, 29). All semidenaturing detergent-agarose gel electrophoresis (SDD-AGE) experiments were repeated at least two times, and typical images are presented.
RESULTS
Incorporation of Tyrosine Residues Improves Fragmentation of Polyglutamine Polymers—Two sets of plasmid constructs encoding polyglutamine domains fused to Sup35MC were created as described under “Experimental Procedures” (Fig. 1A and supplemental Fig. 1). The encoded fusion proteins are designated hereafter as nQ and nQY, where n is the length of the polyQ/QY tract (e.g. 70Q or 76QY). Despite the use of the SUP35 promoter, the expression levels of these proteins per gene copy were significantly lower than the native Sup35 level. The expression of 70Q from a centromeric plasmid in a strain with disrupted chromosomal SUP35 did not ensure efficient translation termination and resulted in nonsense suppression (data not shown). Multicopy plasmids encoding the polyQ/QY proteins provided expression levels ∼2- or 3-fold higher compared with wild-type SUP35 (data not shown), and these plasmids were used in further work.
FIGURE 1.
Polymerization of polyQ/QY proteins in [PIN+] cells. A, schematic representation of the polyQ/QY-Sup35MC proteins. The amino acid numbering of native Sup35 is given in parentheses. M, middle domain; C, C-terminal domain. In B–D, the indicated polyQ/QY proteins were expressed in [PIN+] cells instead of endogenous Sup35. B, cell phenotypes on YPD-red medium and synthetic medium lacking adenine (9th day of growth). Sup35MC served as the control. In C and D, cells were grown in YPD medium, and cell lysates were analyzed by electrophoresis. Gel blots were stained with antibody to Sup35M, which reveals polyQ/QY fusion proteins. C, monomer analysis by SDS-PAGE. The samples were not boiled to show the levels of only monomeric polyQ/QY proteins. D, polymer analysis by SDD-AGE. The molecular mass standards titin (4000 kDa) and nebulin (700 kDa) are indicated.
The strain 74-D694/ΔS35 used in this work carried disrupted chromosomal SUP35 and a centromeric plasmid encoding Sup35C to support viability. Because of the ade1-14 UGA mutation, cells of this strain require adenine and accumulate red pigment related to impaired adenine biosynthesis. Polymerization of the Sup35 fusion variants should reduce efficiency of translation termination and allow suppression of ade1-14. Such cells become independent of adenine and acquire white or pink color corresponding to high or intermediate efficiency of suppression and Sup35 polymerization, respectively.
The plasmids encoding the polyQ/QY proteins were introduced into 74-D694/ΔS35 [PIN+] to replace the resident plasmid encoding Sup35C. The transformants producing 131Q, 76QY, and 120QY proteins showed a suppressor phenotype (white color) on YPD medium, which correlated with decreased levels of these proteins in soluble fraction (Fig. 1, B and C). This confirmed that suppression was due to reduction of the soluble fraction of these proteins. The cells producing 46QY and 50QY showed suppression on adenine omission medium, but not on YPD medium. We presume that the growth in the absence of adenine was due to a small fraction of these cells, as it occurs with [PIN+] cells overproducing Sup35, which are also red on YPD medium but grow in the absence of adenine (10). All transformants showed slight phenotypic instability and segregated a small proportion of clones differing from the majority by the suppressor phenotype. This was manifested in altered colony color and, only in the case of cells producing non-polymerizing constructs 25Q and 30QY, in the ability to grow on adenine-deficient medium. The reason for instability is unknown but is unrelated to alteration in the levels of soluble polyQ/QY proteins (data not shown). For these reasons, we further relied mainly on biochemical, rather than phenotypic, evidence for protein polymerization.
SDD-AGE analysis of lysates of transformants showed the presence of polymers for 51Q, 46QY, and proteins with longer polyQ/QY tracts (Fig. 1D). Unlike the prion ([PSI+]) polymers of Sup35, these polymers appeared rapidly and without cell selection for suppression, being observed at 30 cell generations after transformation, a minimum time required to produce analyzable biomass from a single transformed cell. The size of polymers was much smaller in the polyQY fusion series. This indicates better fragmentation of the polymers of proteins with polyglutamine domains containing tyrosine, which was the basic assumption for this work.
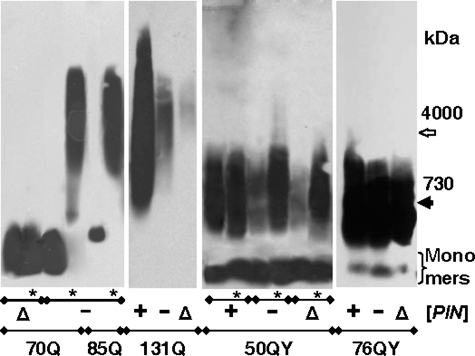
[PIN+] Effects on Polymer Appearance—It was shown previously that polymerization (10) and aggregation of polyQ proteins (24, 25) requires the [PIN+] prion. Here, polymerization of the polyQ/QY proteins showed a varying dependence on [PIN+]. [PIN+] accelerated but was not required for polymerization of the 70Q and 85Q fusion proteins: in [pin-] cells, the polymers were not observed at 30 generations after transformation but appeared after ∼100 generations (Fig. 2). It is noteworthy that the cells with polymers remained [pin-] because Rnq1 was soluble after subsequent shuffle of the 70Q- and 85Q-expressing plasmids for a plasmid encoding Sup35C (data not shown). In the Δrnq1 cells, the polymers of 70Q (Fig. 2) and 85Q (data not shown) proteins did not appear even at 100 generations. The dependence of polymer appearance on [PIN+] decreased with extension of the polyQ/QY tract and was less pronounced for polyQY proteins. A small amount of 131Q polymers was observed in the [pin-] and Δrnq1 cells at 30 generations after transformation. The 50QY protein polymerized in the [pin-] and Δrnq1 cells, but polymerization efficiency was much higher after 100 generations. Polymerization of 76QY proceeded equally efficiently in the [PIN+], [pin-], and Δrnq1 cells (Fig. 2).
FIGURE 2.
Dependence of polymer appearance on the [PIN] status. Plasmids encoding the 70Q, 85Q, 131Q, 50QY, and 76QY proteins were introduced into the [PIN+], [pin-], and Δrnq1 cells, and transformants were grown for 30 or 100 (*) generations. Cell lysates were analyzed by SDD-AGE with blot immunostaining for Sup35M. Molecular mass standards are indicated in kilodaltons.
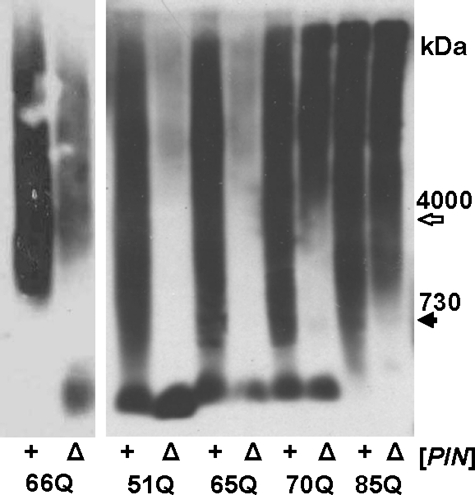
Propagation of Polymers of the 70Q and 85Q Proteins Does Not Depend on [PIN+]—The elimination of [PIN+] by deletion of RNQ1 did not cause the disappearance of the 70Q and 85Q polymers (Fig. 3), in contrast to our previous data that such deletion blocks propagation of 66Q polymers (10). It should be noted that the 66Q protein constructed in our earlier work differed by the presence of 11 additional amino acids at the N terminus (MPSHFGGSETS) and by higher intracellular levels due to the use for its expression of the strong MET17 promoter. To solve this discrepancy, we compared the effects of the RNQ1 deletion on polymers of the 51Q, 65Q, 70Q, 85Q, and old 66Q fusion proteins. The proportion of polymers observed after the RNQ1 deletion was highly dependent on the length of the polyQ stretch. The polymers of the 51Q protein were almost completely eliminated, whereas the polymers of 85Q were barely affected (Fig. 3). Polymerization of 66Q in the Δrnq1 cells was significantly reduced, but not abolished. The cell phenotypes confirmed the polymerization data: the shift in colony color to red, which indicates decreased polymerization, was observed for the 66Q, 51Q, and 65Q proteins, but not for 70Q and 85Q proteins (supplemental Fig. 2A). Thus, the polymers of polyQ proteins can propagate in yeast in the absence of [PIN+], but this depends on the length of their polyQ domain.
FIGURE 3.
Effects of the RNQ1 deletion on propagation of polyQ polymers. The indicated polyQ proteins were expressed in [PIN+] cells (+) and yielded polymers. Then, the RNQ1 gene was disrupted in these cells (Δ). Cell lysates were analyzed by SDD-AGE with blot immunostaining for Sup35M.
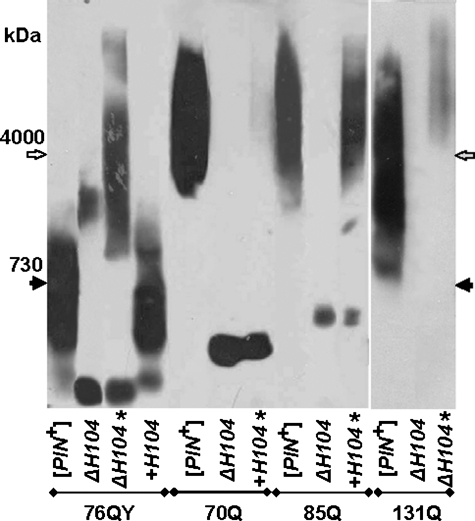
Effects of HSP104 Deletion on Propagation of Polymers—Because deletion of HSP104 eliminates [PIN+], the effects of this deletion were studied only for the polymers that propagate independently of [PIN+]. HSP104 deletion blocked propagation of polymers of the 70Q, 85Q, and 131Q proteins (Fig. 4). However, a small proportion of 131Q polymers appeared after prolonged growth. Reintroduction of HSP104 into the centromeric pRS313-HSP104 plasmid caused gradual recovery of polymerization of polyQ proteins. After 100 generations of growth, polymerization of 85Q was restored fully, whereas 70Q polymers were barely detectable. Thus, polymerization of proteins with shorter polyQ tracts is re-established more slowly. The HSP104 disruption also interfered with polymerization of polyQY proteins. The amount of 76QY polymers was greatly reduced at 30 cell generations after the disruption. Efficient polymerization of 76QY was restored at 100 generations, but the polymers were much larger than in the presence of Hsp104. This suggests that, in the absence of Hsp104, the fragmentation of 76QY polymers was significantly reduced but was sufficient for their propagation (see “Discussion”). Reintroduction of HSP104 returned the 76QY polymers to their original size. 120QY also polymerized in the Δhsp104 cells, whereas 50QY did not form polymers even after 100 cell generations (data not shown), which could be related to its shorter polyQY tract.
FIGURE 4.
Effects of the HSP104 deletion. [PIN+] cells were transformed with plasmids encoding the 76QY, 70Q, 85Q, and 131Q proteins. In these cells, HSP104 was disrupted (ΔH104) and then reintroduced into a centromeric plasmid (+H104). Cell lysates were prepared at 30 or 100 (*) generations after transformation and analyzed by SDD-AGE with blot immunostaining for Sup35M. Molecular mass standards are indicated.
Guanidine Hydrochloride Effects—Guanidine hydrochloride (GdnHCl) inhibits Hsp104 activity (15, 16) and can fully block fragmentation of the Sup35 prion polymers, which increases the Sup35 polymer size by 2-fold per generation (14). The size of the 70Q polymers also increased upon cell growth in the presence of 2 mm GdnHCl, although to a much lesser extent (Fig. 5A). The lack of a significant increase in polymer size may be due to technical reasons. The 70Q polymers are large, and their largest fraction may be preferentially lost, i.e. trapped in the cell pellet, broken with glass beads, and incompletely transferred from agarose gel. The 76QY polymers in the presence of GdnHCl increased in size significantly, although by <2-fold per generation (Fig. 5A). This may reflect an incomplete block of fragmentation if the additional fragmentation activity unrelated to Hsp104 is not affected by GdnHCl. To check this, the Δhsp104 cells producing 76QY polymers were grown in the presence of GdnHCl. After two generations of growth, no effect was detected on either the size of polymers or the proportion of monomers (Fig. 5A). This confirms that the additional fragmentation activity is insensitive to GdnHCl.
FIGURE 5.
Effects of GdnHCl. A, yeast cells, either wild-type or disrupted for HSP104 (ΔH104), producing 70Q and 76QY in a polymer form were grown in YPD medium containing 3 mm GdnHCl for the indicated number of generations, and cells with 76QY were then grown for 2 h in the absence of GdnHCl. B, 70Q and 85Q polymers were eliminated in the Δrnq1 strain by growing single cells into colonies in the presence of 3 mm GdnHCl. Cell lysates were analyzed by SDD-AGE with blot staining for Sup35M. The agarose gel concentration was 1.2% for the cells with 70Q and 76QYΔH104 (A) and 1.8% in other cases. Molecular mass standards are indicated.
Prion-like Properties of the Q70 and Q85 Proteins—The data presented show that many of the polyQ/QY polymers have the distinguishing features of prions, being, in contrast to non-heritable amyloids, susceptible to fragmentation. However, being a prion also implies the availability of a stable non-prion state, which may be “infected” with a prion. For 70Q and 85Q, the stable non-polymerizing state was unobservable in [PIN+], unstable in [pin-], but stable in Δrnq1 cells (Figs. 2 and 5B). The polymers of 70Q and 85Q in Δrnq1 cells were also stable and curable by growth in the presence of GdnHCl (Figs. 3 and 5B), which resulted in a deeper red color of colonies (supplemental Fig. 2B). To confirm the prion properties of 85Q, we transformed the Δrnq1 cells containing this protein in a monomer form with the lysate of Δrnq1 cells containing 85Q polymers following the method of Tanaka and Weissman (30). Cell lysates of 20 randomly selected transformants were tested by SDD-AGE, and 12 of them contained 85Q polymers. However, in the control transformation with the lysate containing non-polymerizing Q85, two of 20 transformants also contained polymers (data not shown). These results show infectivity of the Q85 polymers. However, they also reveal that the stability of the non-polymerizing state of Q85 is low compared with natural prion-forming proteins.
The polyQY proteins did not have stable alternative states in the tested genetic backgrounds. 76QY and 120QY polymerized even in the Δhsp104 cells. 50QY was stable as a monomer only in Δhsp104 cells, but it never polymerized in these cells (data not shown).
DISCUSSION
PolyQ and PolyQY Proteins Readily Form Polymers in Vivo—Polyglutamine- and some glutamine-rich domains confer upon proteins an ability to polymerize in vivo. Expansion of polyglutamine stretches in some human proteins causes their polymerization, aggregation, and disease, e.g. Huntington disease (23). Glutamine-rich domains define prion properties of yeast proteins. Here, we undertook a systematic study of polymerization of polyglutamine proteins to model both yeast prion formation and huntingtin polymerization. It was shown previously that polyglutamine can polymerize in yeast, but these polymers cannot propagate independently of [PIN+] because of poor polymer fragmentation (10). We proposed that efficient fragmentation requires the presence within the polymerization domain of hydrophobic residues, which could attract chaperone-fragmenting machinery. To test this, we created fusions of polyQ and polyQY tracts of different length to Sup35MC and studied their polymerization in yeast.
All hybrid proteins larger than 51Q and 46QY were able to polymerize. In contrast to Sup35 prion ([PSI+]) polymers, which appear rarely and can be obtained only upon selection for the suppressor phenotype, the polymerization of polyQ/QY proteins started rapidly (Table 1) and did not require cell selection. The ease of polymer appearance could be related to simplicity of the polyQ/QY primary structure. It is presumed that, in the Sup35 amyloid structure, the adjacent protein molecules are arranged as parallel in-register β-strands (22, 31, 32). However, the restriction for register is likely to be inapplicable for the uniform polyQ/QY sequences. This increases greatly the number of possible variants of relative location of adjacent molecules in a polymer and is likely to cause (i) faster polymer appearance and (ii) less precise copying of the initial amyloid fold and its plasticity, i.e. an ability to change gradually between different forms. In agreement with the latter, we failed to find stable variants differing by polymer size for any tested polyQ/QY polymers.
TABLE 1.
Ability of polyQ/QY proteins to polymerize in vivo RNQ1 deletion was made either before the introduction of the polyQ/QY-encoding plasmids or after polyQ/QY polymer appearance (Δ after p.a.). Generations indicate the number of cell generations from the beginning of production of the indicated proteins. ND, not determined; +/–, reduced amount of polymers.
|
Protein |
Genetic background |
|||||||
|---|---|---|---|---|---|---|---|---|
|
[PIN+] |
[pin–] |
Δrnq1 |
Δhsp104 |
|||||
| 30 generations | 30 generations | 100 generations | 30 generations | 100 generations | Δ after p.a. | 30 generations | 100 generations | |
| 70Q | + | – | + | – | – | + | – | – |
| 85Q | + | – | + | – | – | + | – | – |
| 131Q | + | +/– | ND | +/– | ND | ND | – | +/– |
| 50QY | + | +/– | + | – | + | + | – | – |
| 76QY | + | + | + | + | + | + | +/– | + |
[PIN+] accelerated polymer appearance for polyQ and 50QY, but was not required for it. These polymers appeared at 100 generations in [pin-] cells, whereas in [PIN+] cells, polymers were observed at 30 generations (Table 1) and possibly were present at much earlier points unavailable for analysis. The propensity to polymerize was higher for the polyQY proteins and increased with the size of the polyQ/QY tract, which was manifested in faster acquisition of the polymer state and lesser dependence on [PIN+] and deletions of RNQ1 and HSP104. For example, polymers of 76QY and 120QY were observed at 30 generations independently of the [PIN] status and were able to propagate in the Δhsp104 background.
Not only the prion form of Rnq1, but the very presence of this protein facilitated the appearance of polyQ and 50QY polymers because they appeared faster in the [pin-] cells than in the Δrnq1 cells. This observation suggests interaction, presumably co-polymerization of these proteins at initial steps of the polyQ and 50QY polymer appearance. As a likely scenario, the first appearing polyQ/Y polymers are poorly fragmented, and incorporation of Rnq1 improves their recognition by Hsp104. With time, the polymer fold evolves, through the imprecise copying mentioned above, into the forms better recognizable by Hsp104 and not requiring Rnq1.
Tyrosine Residues Enhance Polymer Fragmentation—Incorporation of tyrosine residues into the polyQ domains greatly decreased the size of the respective polymers, which suggests their improved fragmentation. It may appear that the relation between polymer size and fragmentation is ambiguous because the size of polymers should also depend on the speed of polymerization. However, thorough mathematical analysis of prion polymerization in yeast by Tanaka et al. (33) shows the lack of such dependence. This conclusion, although not directly formulated by the authors, easily follows from their model, which gives expressions for the number of polymers, y = αγ/R2 - R/β, and the total number of molecules contained in polymers, z = α/R - R2/βγ. Here, α, β, γ, and R are the rate constants for Sup35 synthesis, polymer growth, polymer division, and growth of yeast cells, respectively. The average size of polymers (number of Sup35 molecules/polymer) equals the quotient of these values, z/y. Using the above expressions for z and y, one obtains z/y = R/γ. Thus, the size of polymers is defined solely by fragmentation (γ) and does not depend on polymerization (β). Then, a decreased size of polyQY polymers means their increased fragmentation. This confirms our starting assumption that hydrophobic residues serve as determinants for chaperone recognition and Hsp104-related fragmentation.
Hydrophobic residues themselves do not guarantee Hsp104 recognition because they usually fold inside of a protein structure. In line with this idea, we observed that most of the Sup35 polymers seeded by the prion form of Rnq1 are poorly fragmented and thus should have their hydrophobic residues hidden (10). The content of such residues in the polyQY proteins (20% Tyr) is similar to that in the Sup35 N-terminal domain (16% Tyr and 8% others). The difference in fragmentation of the polyQ and polyQY polymers suggests that at least some tyrosines of the polyQY proteins are exposed. Tyrosines may be placed on the polymer surface via a self-selection (microevolution) process. Presumably, the polymer folds with exposed tyrosines appear with low frequency, inversely related to the proportion of exposed tyrosines. However, these folds have an increased propagation potential. When polymers with different folds are present in one cell, the one that allows faster multiplication of prion particles should increase its proportion and eventually displace others (20, 33). Faster multiplication requires reasonably high fragmentation efficiency. Thus, a feature that improves polymer fragmentation, such as exposed tyrosine residues, would be selected.
Polyglutamine Polymers Are Fragmented and Show Prion-like Properties—Here, we have shown that polyQ stretches longer than 70 residues allow proteins to form polymers able to propagate in the absence of Rnq1. Propagation of such polymers suggests that they are fragmented. This contradicts our assumption that polyQ polymers should not be recognized by chaperones because their polymerization domains lack hydrophobic residues. This contradiction may be solved by proposing the following mechanisms for fragmentation of the polyQ polymers. (i) Chaperones can recognize, although inefficiently, the amyloid structures lacking hydrophobic residues; (ii) polymerization of the polyglutamine region interferes with proper folding of the adjacent Sup35 middle domain (referred to as Sup35M), which becomes a target for chaperone(s); and (iii) other cellular glutamine-rich proteins that contain hydrophobic residues co-polymerize with polyglutamine domains and attract fragmentation. In support of the latter opportunity, we have observed that Rnq1 and Sup35 co-polymerize efficiently with 70Q and 85Q.4
Previously, we described two categories of yeast amyloids, heritable (prion) and non-heritable, the propagation of which depends on [PIN+] (10). The polymers of 70Q and 85Q proteins should belong to the heritable category, being [PIN+]-independent, but formally they are not prions because in wild-type cells they lack a stable non-polymerizing state, which could be infected. This formal restriction is essentially removed in the Δrnq1 cells, in which 70Q and 85Q proteins are relatively stable in the non-polymerizing form. The polymer state of these proteins was curable by cell growth in the presence of GdnHCl, and the polymers of 85Q were able to infect cells producing non-polymerizing 85Q. Thus, with some reservations, the 85Q polymers may be considered as prions. PolyQY polymers are efficiently fragmented and heritable, but again they may not be considered as prions because of the lack of a stable monomer state. Such polymers should be categorized as replicating amyloids, which are close to, but formally distinct from, prions.
Polymer Fragmentation in the Absence of Hsp104—Propagation of natural yeast prions, i.e. [PSI+], [URE3], and [PIN+], strictly requires Hsp104 because the propagation involves polymer fragmentation performed by this chaperone. However, polymers of 76QY and 120QY propagated in the absence of Hsp104. This may be due to either of two mechanisms: (i) polymer fragmentation independent of Hsp104 and (ii) highly efficient formation of these polymers de novo. However, the second mechanism may be excluded. With it, the efficiency of polymer formation should not depend on time, which contradicts the data. The deletion of HSP104 almost eliminated 76QY polymers, as observed at 30 cell generations after deletion, but polymerization was restored to nearly the original level at 100 generations. Apparently, the original 76QY polymer fold could not propagate without Hsp104 due to poor fragmentation but, after some time, rearranged to allow better fragmentation. Then, two opportunities are available: the fragmentation is performed by a protein, presumably, chaperone, or it occurs because of intracellular mechanic shearing forces. In the first case, it may be proposed that the new amyloid fold was distinguished by further increase in the proportion of exposed tyrosines. In the second, we may expect self-selection for the most fragile amyloid fold. The feasibility of a mechanic shearing mechanism is difficult to estimate because it is unknown whether the shearing forces are sufficient to break amyloids. Two following observations are more compatible with Hsp104-independent fragmentation performed by a chaperone. Such fragmentation does not occur or is very inefficient for polyQ polymers, which may be related to tyrosines being a chaperone recognition target. Fragmentation improves with the length of polyQY stretch, which should alleviate the access of chaperones to the amyloid stem, but should not affect its fragility.
The size of 76QY polymers in the absence of Hsp104 was increased by 4–6-fold, which suggests that fragmentation was several times less efficient than in the presence of Hsp104. In contrast to Hsp104, the additional fragmentation was insensitive to GdnHCl, which suggests that Hsp104 is the only target inhibited by GdnHCl related to prion polymer fragmentation.
Implications for Huntington and Related Diseases—In human diseases related to expansion of polyglutamine tracts in some proteins, longer tracts define earlier onset and faster progression of these diseases. The results of this work illuminate the reasons for this. We observed that shorter polyQ stretches (51Q and 65Q) allow polymerization only in a non-heritable, [PIN+]-dependent mode, whereas 70Q and longer stretches can polymerize in a prion mode. The latter means fundamentally faster kinetics of polymerization because fragmentation multiplies amyloid polymerization seeds. Also, the ability of polyQ polymers to appear independently of other amyloids and the ease of their appearance increased with polyQ size. For human diseases, such properties should define earlier disease onset for longer polyQ stretches. For both yeast Sup35 and animal PrP prions, the initial appearance of prion is a very infrequent event. The polyQ polymers appear incomparably faster than Sup35 prion polymers. This suggests that the appearance of polyQ polymers in human cells should be restricted not by the primary seed formation, but by the ability of a cell to counteract the amyloid polymerization. It should be noted that, in contrast to the PrP protein, which forms polymers extracellularly, the intracellular huntingtin polymers replicating in the “prion” mode are not infectious because the spread of polymerization is restricted by cell borders. Such polymers may be regarded as “intracellular prions.”
Supplementary Material
This work was supported by the Wellcome Trust, Russian Foundation for Basic Research Grant 08-04-00062, Russian Foundation for Basic Research/U. S. Civilian Research & Development Foundation Joint Grant 07-04-91105/2859, and International Science and Technology Center Grant 2750. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: polyQ, polyglutamine or polyglutamine-Sup35 middle and C-terminal domain fusion protein; polyQY, polyglutamine-tyrosine (4:1) fusion protein; SDD-AGE, semidenaturing detergent-agarose gel electrophoresis; GdnHCl, guanidine hydrochloride.
I. M. Alexandrov, A. B. Vishnevskaya, M. D. Ter-Avanesyan, and V. V. Kushnirov, unpublished data.
References
- 1.Chiti, F., and Dobson, C. M. (2006) Annu. Rev. Biochem. 75 333-366 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B., Scott, M. R., DeArmond, S. J., and Cohen, F. E. (1998) Cell 93 337-348 [DOI] [PubMed] [Google Scholar]
- 3.Wickner, R. B. (1994) Science 264 566-569 [DOI] [PubMed] [Google Scholar]
- 4.Derkatch, I. L., Bradley, M. E., Zhou, P., Chernoff, Y. O., and Liebman, S. W. (1997) Genetics 147 507-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N., and Ter-Avanesyan, M. D. (1996) EMBO J. 15 3127-3134 [PMC free article] [PubMed] [Google Scholar]
- 6.Patino, M. M., Liu, J. J., Glover, J. R., and Lindquist, S. (1996) Science 273 622-626 [DOI] [PubMed] [Google Scholar]
- 7.Sondheimer, N., and Lindquist, S. (2000) Mol. Cell 5 163-172 [DOI] [PubMed] [Google Scholar]
- 8.Derkatch, I. L., Bradley, M. E., Hong, J. Y., and Liebman, S. W. (2001) Cell 106 171-182 [DOI] [PubMed] [Google Scholar]
- 9.Derkatch, I. L., Uptain, S. M., Outeiro, T. F., Krishnan, R., Lindquist, S. L., and Liebman, S. W. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12934-12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salnikova, A. B., Kryndushkin, D. S., Smirnov, V. N., Kushnirov, V. V., and Ter-Avanesyan, M. D. (2005) J. Biol. Chem. 280 8808-8812 [DOI] [PubMed] [Google Scholar]
- 11.Vitrenko, Y. A., Gracheva, E. O., Richmond, J. E., and Liebman, S. W. (2007) J. Biol. Chem. 282 1779-1787 [DOI] [PubMed] [Google Scholar]
- 12.Chernoff, Y. O., Lindquist, S. L., Ono, B., Inge-Vechtomov, S. G., and Liebman, S. W. (1995) Science 268 880-884 [DOI] [PubMed] [Google Scholar]
- 13.Kushnirov, V. V., and Ter-Avanesyan, M. D. (1998) Cell 94 13-16 [DOI] [PubMed] [Google Scholar]
- 14.Kryndushkin, D. S., Alexandrov, I. M., Ter-Avanesyan, M. D., and Kushnirov, V. V. (2003) J. Biol. Chem. 278 49636-49643 [DOI] [PubMed] [Google Scholar]
- 15.Jung, G., and Masison, D. C. (2001) Curr. Microbiol. 43 7-10 [DOI] [PubMed] [Google Scholar]
- 16.Ferreira, P. C., Ness, F., Edwards, S. R., Cox, B. S., and Tuite, M. F. (2001) Mol. Microbiol. 40 1357-1369 [DOI] [PubMed] [Google Scholar]
- 17.Shorter, J., and Lindquist, S. (2004) Science 304 1793-1797 [DOI] [PubMed] [Google Scholar]
- 18.Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G., and Liebman, S. W. (1996) Genetics 144 1375-1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlumpberger, M., Prusiner, S. B., and Herskowitz, I. (2001) Mol. Cell. Biol. 21 7035-7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradley, M. E., Edskes, H. K., Hong, J. Y., Wickner, R. B., and Liebman, S. W. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 Suppl. 4, 16392-16399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross, E. D., Baxa, U., and Wickner, R. B. (2004) Mol. Cell. Biol. 24 7206-7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross, E. D., Edskes, H. K., Terry, M. J., and Wickner, R. B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12825-12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landles, C., and Bates, G. P. (2004) EMBO Rep. 5 958-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krobitsch, S., and Lindquist, S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1589-1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meriin, A. B., Zhang, X., He, X., Newnam, G. P., Chernoff, Y. O., and Sherman, M. Y. (2002) J. Cell Biol. 157 997-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, Y., Wu, Y. X., Jung, G., Tutar, Y., Eisenberg, E., Greene, L. E., and Masison, D. C. (2005) Eukaryot. Cell 4 289-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urakov, V. N., Valouev, I. A., Kochneva-Pervukhova, N. V., Packeiser, A. N., Vishnevsky, A. Y., Glebov, O. O., Smirnov, V. N., and Ter-Avanesyan, M. D. (2006) BMC Mol. Biol. 7 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ter-Avanesyan, M. D., Kushnirov, V. V., Dagkesamanskaya, A. R., Didichenko, S. A., Chernoff, Y. O., Inge-Vechtomov, S. G., and Smirnov, V. N. (1993) Mol. Microbiol. 7 683-692 [DOI] [PubMed] [Google Scholar]
- 29.Kushnirov, V. V., Alexandrov, I. M., Mitkevich, O. V., Shkundina, I. S., and Ter-Avanesyan, M. D. (2006) Methods 39 50-55 [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, M., and Weissman, J. S. (2006) Methods Enzymol. 412 185-200 [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, M., Chien, P., Yonekura, K., and Weissman, J. S. (2005) Cell 121 49-62 [DOI] [PubMed] [Google Scholar]
- 32.Shewmaker, F., Wickner, R. B., and Tycko, R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19754-19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka, M., Collins, S. R., Toyama, B. H., and Weissman, J. S. (2006) Nature 442 585-589 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.