Abstract
Sco1 is implicated in the copper metallation of the CuA site in Cox2 of cytochrome oxidase. The structure of Sco1 in the metallated and apo-conformers revealed structural dynamics primarily in an exposed region designated loop 8. The structural dynamics of loop 8 in Sco1 suggests it may be an interface for interactions with Cox17, the Cu(I) donor and/or Cox2. A series of conserved residues in the sequence motif 217KKYRVYF223 on the leading edge of this loop are shown presently to be important for yeast Sco1 function. Cells harboring Y219D, R220D, V221D, and Y222D mutant Sco1 proteins failed to restore respiratory growth or cytochrome oxidase activity in sco1Δ cells. The mutant proteins are stably expressed and are competent to bind Cu(I) and Cu(II) normally. Specific Cu(I) transfer from Cox17 to the mutant apo-Sco1 proteins proceeds normally. In contrast, using two in vivo assays that permit monitoring of the transient Sco1-Cox2 interaction, the mutant Sco1 molecules appear compromised in a function with Cox2. The mutants failed to suppress the respiratory defect of cox17-1 cells unlike wild-type SCO1. In addition, the mutants failed to suppress the hydrogen peroxide sensitivity of sco1Δ cells. These studies implicate different surfaces on Sco1 for interaction or function with Cox17 and Cox2.
Cytochrome c oxidase (CcO)2 is the terminal enzyme of the energy-transducing, electron transfer chain within the mitochondrial inner membrane (IM). The enzyme contains two copper centers important for its function (1). One center is the binuclear copper site (CuA) residing in the Cox2 subunit. The second center is the CuB site in the Cox1 subunit that forms a heterobimetallic site with heme a3. The formation of the copper sites in the two mitochondrially encoded subunits occurs within the mitochondrial intermembrane space (IMS) by a series of accessory proteins Cox11, Cox17, and Sco1 (2). Cox17 is a soluble Cu(I)-binding protein largely localized within the IMS. In yeast, CuCox17 appears to specifically transfer Cu(I) to the CuA site via the IM-tethered Sco1 and to the CuB site via the IM-anchored Cox11 (3).
The role of Cox17 in formation of the CuA site was initially implicated from the isolation of SCO1 as a high copy suppressor of cox17-1 respiratory deficient cells (4). Yeast lacking Sco1 are devoid of CcO activity and show greatly attenuated Cox2 protein levels (5, 6). Cu(I) bound to Cox17 can be transferred to Sco1 through an apparent transient protein-mediated complex. The C57Y mutation in the cox17-1 strain precludes Cu(I) transfer to Sco1, but not Cox11, consistent with each transfer occurring through a specific protein complex (3). Although attempts to isolate the Cox17·Sco1 complex have failed, a mass spectrometry study with human Sco1 and Cox17 revealed a low abundance ion consistent with a complex (7). Copper metallation of Sco1 and Cox11 is an intermediate step in the transfer of Cu(I) to Cox2 and Cox1, respectively. One preliminary study reported an interaction of Sco1 with Cox2 (8), but no direct interaction has been reported for Cox11 and Cox1.
Unlike yeast, human cells have two functional Sco molecules that are required for viability (9). Mutations in either human SCO1 or SCO2 lead to decreased CcO activity and early death. Studies with immortalized fibroblasts from SCO1 and SCO2 patients suggest that Sco1 and Sco2 have non-overlapping but cooperative functions in CcO assembly (9). Sco1 and Sco2 localize to the IM and are tethered by a single transmembrane helix. A globular domain of each exhibiting a thioredoxin fold protrudes into the IMS (10–13). A single Cu(I) binding site exists within the globular domain consisting of two cysteinyl residues within a CX3C motif and a distal conserved histidine (14, 15). Mutation of the Cys or His residues abrogates Cu(I) binding and leads to a non-functional CcO complex (14, 16). The structure predicts that the single Cu(I) ion coordinated to Sco1 is solvent-exposed and poised for a ligand exchange transfer reaction (17). An additional important aspect of human and yeast Sco1 function is the ability to bind a Cu(II) ion in a type II-like site with a higher coordination number than the trigonal Cu(I) site (16). It is unclear whether Sco1 transfers both Cu(I) and Cu(II) ions to build the mixed valent, binuclear CuA site in Cox2.
A single pedigree was reported with SCO1 mutations that result in fatal neonatal hepatopathy (18). The afflicted individuals were compound heterozygotes, with a nonsense mutation on one allele and a P174L missense mutation on the second allele. SCO1 patient fibroblasts exhibit CcO deficiency along with a severe cellular copper deficiency (19). This secondary copper deficiency occurring from enhanced cellular copper efflux arises from an apparent signaling role of the Sco proteins that is independent of respiration. The severe deficiency in CcO activity in SCO1 patient fibroblasts is partially rescued by overexpression of the P174L mutant protein, but the cellular copper deficiency is only rescued by overexpression of SCO2. The P174L substitution is adjacent to the second Cys in the Cu(I)-binding CX3C motif in Sco1, yet the mutant protein retains the ability to bind Cu(I) or Cu(II) when expressed in bacteria. The molecular defect in the P174L mutant Sco1 is an impaired ability to be copper metallated by Cox17 (7, 20). The defect is attributed to an attenuated interaction with Cox17 (20), in addition to a modest structural defect that attenuates the Cu(I) binding affinity (7). Defective Cox17-mediated copper metallation of Sco1, and subsequent failure of CuA site maturation, is the basis for the inefficient assembly of the CcO complex in SCO1 patient fibroblasts.
The role of Sco2 in human CcO assembly remains unclear. Whereas metallation of human Sco1 is dependent on Cox17 when expressed in the highly chelating environment of the yeast cytoplasm, metallation of Sco2 occurs independently of Cox17 (16). Sco2 is suggested to modulate the redox or metallation state of Sco1 (19). Patient mutations in Sco2 pedigrees map close to either the Cu(I)-binding CX3C motif (E140K) or the His ligand (S225F). From the structure of human Sco2, the prediction is that the mutations either attenuate Cu(I) binding or destabilize the tertiary fold (13). One study confirmed that E140K substitution perturbed copper binding (21).
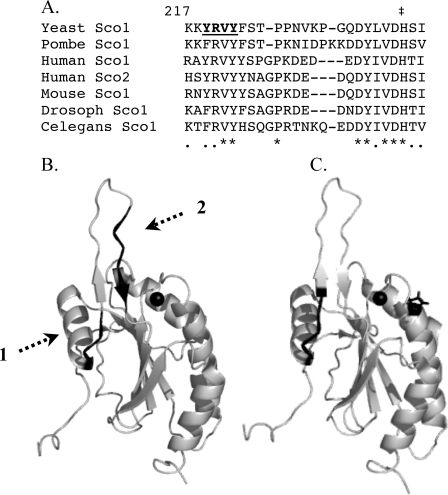
The structures of the metal-free human Sco1 and Cu1Sco1 are similar with only one loop (loop 8) showing significant rearrangements (17). The movement of this loop orients the Cu(I) binding His residue at the base of the loop in the proper orientation for metal binding. Although the copper binding site is somewhat disordered in the apo-conformation, especially the loop 8 segment, the site is largely preformed poised for Cu(I) binding (7, 11). The structural dynamics of loop 8 suggests it may be an important interface for interactions with Cox17 and/or Cox2. Within the loop 8 segment, a series of conserved residues map to the leading edge of loop 8 and residues preceding the Cu(I) binding His. The tip of loop 8 is devoid of conserved residues and shows length variation in Sco proteins from differing species (Fig. 1A).
FIGURE 1.
Mutagenized loop 8 segment of Sco proteins. Panel A, sequence alignment of Sco protein in the loop 8 region with numbering in reference to the yeast Sco1. The underlined residues in bold are the four residues that when mutated result in loss of function. Panel B, the two conserved segments of loop 8 are highlighted in black. The mutated residues lie on the leading edge of loop 8 (shown by the dashed arrow 1), whereas the conserved residues adjacent to the Cu(I) binding His residue (shown by ‡ in panel A) are in black with a dashed arrow 2. The Cu(I) ion is shown as a black sphere. Panel C, the YRVY motif important for Cox2 interaction is shown in black along with the Pro mutated in human Sco1 (P174L) that abrogates Cu(I) metallation by Cox17.
To elucidate the significance of loop 8 in the function of Sco1, we carried out mutational analysis of conserved residues in the loop and evaluated the consequences on metallation by CuCox17 and interaction with the Cox2 target. We show presently that a series of functionally important residues at the leading edge of loop 8 define an interaction site for Cox2 but do not modulate the Cox17 metallation reaction.
MATERIALS AND METHODS
Yeast Strains—Saccharomyces cerevisiae strains used in this study include the haploid BY4741 wild-type strain (Mat-a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), and the isogenic sco1Δ strain (Mat-a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, sco1:: KanMX4), both obtained from Research Genetics. The haploid W303-1A cox17Δ strain (Mat-α, ade2-1, his3-1, 15 leu2-3, 112 trp1-1, ura3-1, cox17::TRP1) was generously provided by Dr. Alexander Tzagoloff.
Growth Media—Yeast strains cultured in liquid medium were pre-grown in synthetic complete medium containing 2% glucose as a carbon source. Pre-cultures were then inoculated into synthetic complete medium containing 2% raffinose, and grown to early stationary phase. Yeast plate cultures were grown on synthetic complete medium or rich medium (yeast extract and peptone) containing 2% glucose or a 2% glycerol, 2% lactate mixture (Gly/Lac) as carbon sources. For growth in exogenous copper, plate culture medium was supplemented with 1 (synthetic complete) or 8 mm (rich medium) CuSO4.
Yeast Vectors—A pRS413 (YCp) vector expressing yeast wild-type SCO1 with a C-terminal HA tag under the control of the MET25 promoter and CYC1 terminator was utilized as a template for generating mutants. All mutants were generated by site-directed mutagenesis using the Stratagene QuikChange kit. Additional vectors utilized in this study include a pRS426 (YEp) vector expressing yeast COX19 with a C-terminal Myc tag under control of the MET25 promoter and CYC1 terminator, as well as a pRS426 (YEp) vector expressing COX17 under the control of its own promoter and terminator. COX19-Myc was cut as a BamHI/SalI fragment out of pRS316 (22) and subcloned into pRS426. The COX17 open reading frame including 500 bp of upstream and 500 bp of downstream flanking sequence was cut as a KpnI/SacI fragment from pRS316 (23) and subcloned into pRS426. pLacIII (YCp) vectors expressing COX17 (23) or a CYB2-C57Y COX17 under control of the MET25 promoter and CYC1 terminator were also used. The CYB2-C57Y COX17 was cut from pRS316 (23) as a KpnI/SacI fragment and subcloned into pLacIII. SCO1 Cys mutants were expressed in the pRS424 (YEp) vector under control of the SCO1 promoter and terminator (14). Wild-type hSCO1 cloned into the Gateway-modified retroviral expression vector pLXSH was used as a PCR template for cloning hSCO1 into a yeast vector. The 3′ oligonucleotide PCR primer removed the stop codon from the hSCO1 open reading frame and added a coding sequence for a C-terminal Myc tag. hSCO1-Myc PCR product was cloned into pRS426 under the control of the MET25 promoter and the CYC1 terminator. The hSCO1 P174L mutant allele was generated by site-directed mutagenesis using the Stratagene QuikChange kit. Sequencing was used to confirm all cloning and mutagenesis.
Protein Purification—The Escherichia coli expression vector pHis-Parallel2 containing His6-tagged-soluble SCO1 was used as a template to generate the 4 non-functional SCO1 mutants (14). Mutant alleles were generated by site-directed mutagenesis using the Stratagene QuikChange kit. All mutants were verified by sequencing. pHis-Parallel2 vectors containing wild-type or mutant His6-soluble SCO1 were transformed into competent BL21(pLysS) E. coli. Transformants were pre-grown at 37 °C to A600 = 0.6, induced by the addition of 0.45 mm isopropyl 1-thio-β-d-galactopyranoside, and incubated for an additional 4 h at 30 °C. For purification of the copper-loaded protein, CuSO4 was added to the culture medium 15 min prior to induction, to a final concentration of 1 mm. Cells were harvested, washed, and then resuspended in PBS containing 10 mm imidazole and 1 mm DTT. Cells were lysed by freeze-thawing followed by repeated sonication. Lysates were cleared by centrifugation at 50,000 × g for 30 min at 4 °C. Supernatants were loaded onto 10-ml nickel-NTA Superflow (Qiagen) columns. After loading, columns were washed three times: first with PBS containing 10 mm imidazole and 1 mm DTT, followed by washes with PBS containing 20 mm imidazole and 1 mm DTT, and 2 times with PBS containing 20 mm imidazole and 1 mm DTT. Purified His-tagged Sco1 proteins were eluted with PBS containing 250 mm imidazole and 1 mm DTT. Purified proteins were concentrated to the desired volume in a 5000-Da cut-off Vivaspin 20 (VivaScience) spin column. To monitor protein purification, purification fractions (load, flow-through, washes, and elution) were analyzed by SDS-PAGE on a 15% polyacrylamide gel and visualized by Coomassie staining. Copper concentrations were determined for the same purification fractions using a PerkinElmer Life Sciences AAnalyst 100 atomic absorption spectrophotometer. Pure, concentrated protein was subjected to 12 h dialysis in PBS containing 1 mm DTT at 4 °C using a Slide-A-Lyzer dialysis cassette with a 3500-Da cut-off membrane. Wild-type His-tagged Sco1 protein was purified side by side (in parallel) with each mutant protein. Proteins were quantified by amino acid analysis on a Hitachi L-8800 analyzer after hydrolysis in 5.7 n HCl containing 0.1% phenol in vacuo at 110 °C.
CuCox17 protein used for in vitro transfer assays was purified as described previously (3). Apo-His-Sco1 was generated by incubation of pure protein with KCN for 30 min, followed by desalting into PBS using Bio-Gel P6 resin (Bio-Rad). In vitro Cu(I) transfer assays were performed as previously described (3).
Spectroscopy—Copper concentrations in protein samples were measured using a PerkinElmer Life Sciences AAnalyst 100 atomic absorption spectrophotometer or a PerkinElmer Optima 3100XL inductively coupled plasma optical emission spectrometer. Absorption spectra were recorded on a Beckman DU640 UV-visible spectrophotometer. Luminescence was measured on a PerkinElmer Life Sciences LS55 spectrometer with an excitation wavelength of 300 nm, monitoring emission from 350 to 700 nm. Excitation and emission slit widths were set at 5 and 15 nm, respectively, using a 350-nm band pass filter.
Isolation of Mitochondria and Oxidase Activity—Mitochondria were isolated according to the method described previously (24) in the presence of 1 mm phenylmethylsulfonyl fluoride. Mitochondrial protein concentrations were quantified by Bradford assay (25). CcO enzymatic activity in isolated mitochondria (5–10 μg of protein) was quantified by monitoring the oxidation of 32 μm reduced equine heart cytochrome c at 550 nm in 40 mm KH2PO4, pH 6.7, 0.5% Tween 80.
Immunoblotting Analysis—40 μg of mitochondrial protein was loaded onto a 15% polyacrylamide gel, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Membranes were probed with the indicated primary antibody and visualized with ECL reagents (Pierce), following incubation with a horseradish peroxidase-conjugated secondary antibody. Mouse monoclonal anti-HA and anti-Myc antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antisera to the mitochondrial OM porin (Por1) was obtained from Molecular Probes, Inc. (Eugene, OR).
Hydrogen Peroxide Treatment of Yeast Cells—Cells were grown in synthetic complete medium containing 2% glucose at 30 °C, to mid-exponential phase. Hydrogen peroxide was added to a final concentration of 1 or 6 mm, followed by a 2-h incubation at 30 °C. Cells were then serially diluted and plated into rich medium plates containing 2% glucose (YPD) (26).
RESULTS
Mutational Analysis of Loop 8 in Yeast Sco1—A comparison of Sco1 proteins from diverse eukaryotes reveals two conserved segments in the prominently solvent-exposed loop 8 that exhibits structural dynamics between the metallated and apo-conformers. One segment consisting of 233QDYLVDH239 (numbering based on the yeast Sco1 sequence) is adjacent to the Cu(I) binding His239 residue (Fig. 1B). We showed previously that neither Gln233 nor Asp234 was important for Sco1 function, but Asp238 was functionally important and influenced Cu(II) binding to Sco1 (16). Presently, we focused on the conserved sequence motif 217KKYRVYF223 preceding loop 8 (Fig. 1B). To elucidate the significance of loop 8 in the function of Sco1, we carried out mutational analysis of conserved residues in this segment.
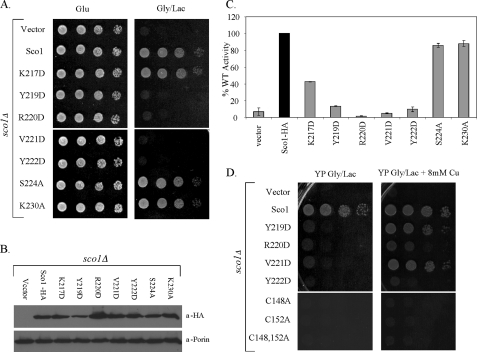
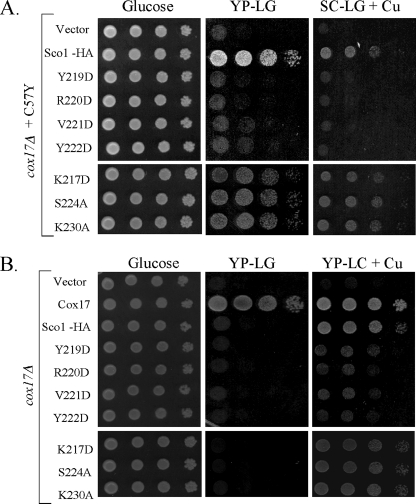
Single codon mutations were generated at seven positions from the loop 8 base toward the loop tip. Asp substitutions were made at most positions to alter the chemical functionality at that position. The rationale was that if the conserved motif served as an interaction site for Cox17 and/or Cox2, a change in chemical functionality would have a more dramatic effect than an Ala substitution. Three of the Sco1 substitution mutants were functional when expressed in sco1Δ cells (Fig. 2A). The K217D, S224A, and K230A alleles enabled sco1Δ cells to propagate on glycerol/lactate medium and assemble a functional CcO enzyme (Fig. 2, A and B), although CcO oxidase activity was attenuated with the K217D mutant cells. Growth of yeast cells on glycerol/lactate medium can occur in cells with >30% of wild-type CcO activity. In contrast, four Sco1 mutants failed to rescue glycerol/lactate growth in the sco1Δ cells (Fig. 2A). Cells harboring Y219D, R220D, V221D, and Y222D mutant Sco1 proteins did not propagate on glycerol/lactate medium and had attenuated CcO activities (Fig. 2C). The substitutions did not destabilize the protein. The four mutant proteins were stably expressed (Fig. 2B), suggesting that the function of Sco1 was compromised by the substitutions. The non-functional mutants have no dominant negative effects when expressed in wild-type yeast cells (data not shown).
FIGURE 2.
Functionality of Sco1 mutants. Panel A, serial dilution of BY4741 sco1Δ cells containing low copy plasmid SCO1 encoding wild-type or mutant alleles of Sco1-HA as indicated on glucose and glycerol/lactate medium (synthetic complete medium). Panel B, steady-state levels of Sco1-HA and Sco1-HA mutants were assessed by immunoblotting of mitochondrial fractions from corresponding strains. Mitochondrial fractions were probed with anti-HA antibody. Por1 was used as a mitochondrial marker and loading control. Panel C, CcO enzymatic activities on mitochondria isolated from mutant strains. Activities are expressed as percent of CcO activity in wild-type mitochondria. Panel D, growth test of sco1Δ cells harboring various sco1 alleles for copper suppression. Non-functional Sco1-HA mutants were tested for suppression of their phenotype by growth on glycerol/lactate (rich medium) supplemented with 8 mm CuSO4 (glucose loading control not shown). Copper-binding Sco1 mutants (C148A, C152A, and C148A,C152A) were used as negative controls. Serial dilutions of cells were plated.
Because Sco1 is implicated in copper metallation of Cox2, we tested whether the lack of glycerol/lactate growth in sco1Δ cells expressing Y219D, R220D, V221D, and Y222D mutants was suppressed by supplemented copper salts (Fig. 2D). The respiratory deficiency of cells lacking Cox17 is weakly suppressed by high exogenous copper salts in rich growth medium (4). Supplemental copper salts cannot confer growth of sco1Δ cells (4), but partial growth was restored in sco1Δ cells harboring Y219D, R220D, and V221D, but not Y222D mutant alleles when plated on rich medium with glycerol/lactate as carbon sources. With copper-supplemented synthetic medium with glycerol/lactate, only the V221D mutant Sco1 enabled partial growth (data not shown).
The only non-functional patient mutation in human Sco1 identified to date is the P174L substitution adjacent to the Cu(I)-binding CX3C motif (18). The molecular defect in P174L Sco1 appears confined to the interaction with Cox17, and elevated Cox17 levels can restore Cu(I) transfer in vitro (20). We tested whether overexpression of COX17 would enhance glycerol/lactate growth of sco1Δ cells harboring the nonfunctional mutants (supplemental Fig. S1). The presence of elevated Cox17 levels enhanced growth only of sco1Δ cells containing the V221D mutant consistent with the suppression by exogenous copper. Overexpression of the Cox17-related IMS protein Cox19 failed to confer growth in combination with any of the four mutant alleles (data not shown).
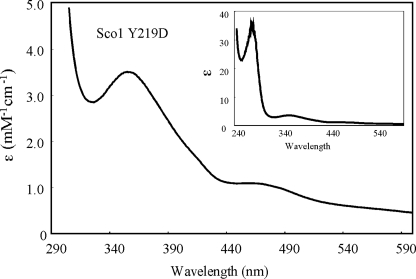
Respiratory-deficient Loop 8 Mutants Bind Cu(I)/Cu(II) Normally and Are Not Impaired in Cu(I) Metallation by Cox17—The four non-functional Sco1 mutants were recombinantly expressed in bacteria to evaluate their copper binding properties. N-terminal truncates of the Sco1 mutants were expressed and purified as soluble proteins, as previously described (14, 16). Sco1 purified from E. coli cultures supplemented with exogenous copper salts results in the isolation of Cu(I)-bound Sco1 (14, 16). The Cu(I) complex with wild-type Sco1 is stable to dialysis. Purification of the Y219D, R220D, V221D, and Y222D mutant proteins followed by dialysis revealed that each mutant retained the ability to bind Cu(I) (Table 1). In addition, each mutant Sco protein retained the ability to bind Cu(II) as shown by the S-Cu(II) charge transfer bands (Fig. 3 and Table 1). Thus, the mutations did not abrogate copper binding.
TABLE 1.
Copper binding stoichiometries of Sco1 mutants
Recombinant soluble His-Sco1 mutant proteins were purified from E. coli cultured in medium containing supplemental copper. Copper per protein stoichiometries were measured from protein samples as purified, following concentration, as well as 12 h dialysis. Cu(II) binding was assessed by comparing A360 of mutants to A360 of wild-type His-Sco1 at the same protein concentrations.
|
Cu/protein |
WT | Y219D | R220D | V221D | Y222D |
|---|---|---|---|---|---|
| Sco1 | Sco1 | Sco1 | Sco1 | Sco1 | |
| Elution | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 |
| Concentration | 1.0 ± 0.1 | 1.1 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 | 0.8 ± 0.1 |
| Post-dialysis | 0.9 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 | 0.7 ± 0.1 | 0.7 ± 0.1 |
| ε Cu(II)/WT ε Cu(II) | 1.0 | 1.0 | 0.9 | 0.8 | 1.1 |
FIGURE 3.
Absorption spectroscopy of CuSco1 (Y219D). The absorption spectrum of recombinant, purified His-tagged CuSco1 Y219D reveals the normal Cu(II) chromophore transitions in the visible region with maxima at 360 and 480 nm. All mutants purified exhibited transitions representative of the wild-type protein, confirming the ability of the mutants to bind Cu(II) normally. The inset shows the entire absorption spectrum of the Cu-Sco1 (Y219D).
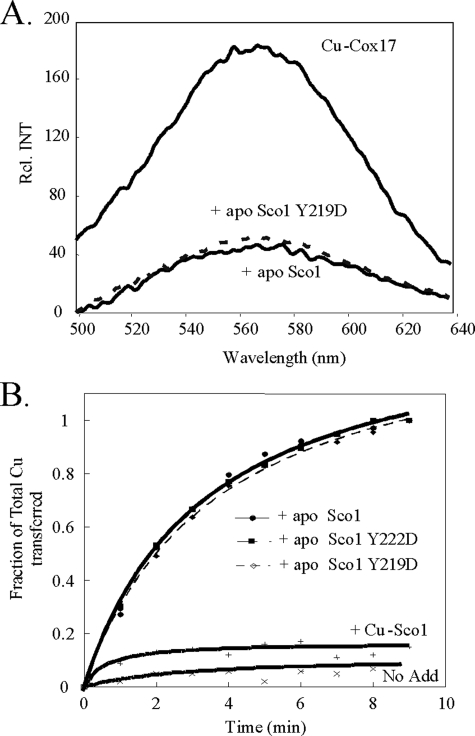
A series of assays were performed to assess whether the non-functional mutations abrogated a functional interaction with Cox17. We demonstrated previously that specific Cu(I) transfer from Cox17 to Sco1 occurs in both in vitro and in vivo reactions (3). We used the in vitro Cu(I) transfer assay to assess the efficiency of Cox17-mediated Cu(I) transfer to Sco1. Incubation of CuCox17 with apo-Sco1 results in a time-dependent transfer of Cu(I) to Sco1 as assessed by the loss of emission of the Cu(I)-thiolate coordination in Cox17 (Fig. 4A) (3). The emission of CuCox17 is stable when incubated with CuSco1 or heterologous molecules. We showed previously that CuCox17 with a C57Y substitution, which does not alter Cu(I) binding, fails to transfer Cu(I) to apo-Sco1. In contrast, incubation of wild-type CuCox17 with equimolar Y219D and Y222D mutant Sco1 molecules showed similar kinetics of Cu(I) transfer as the wild-type Sco1 suggesting that the mutations did not attenuate Cu(I) transfer from Cox17 (Fig. 4B). The V221D Sco1 mutant also exhibited wild-type kinetics of Cu(I) transfer from CuCox17 (data not shown). This in vitro assay suggests that the mutant and wild-type Sco1 molecules have an equivalent affinity for Cu(I).
FIGURE 4.
Cox17-mediated Cu(I) transfer to apo-Sco1 and mutant Sco1 molecules. Panel A, CuCox17 (45 mm copper) was incubated with 50 mm wild-type or mutant apo-Sco1 for 10 min at room temperature. The luminescence of CuCox17 before and after the addition of either wild-type apo-Sco1 or the Y219D mutant is shown. The loss of luminescence observed upon addition of apo-Sco1 Y219D was representative of all other mutants tested. Panel B, the kinetics of Cox17-mediated copper transfer to apo-Sco1 is shown. The loss of luminescence was monitored upon addition of either wild-type or mutant apo-Sco1. The total copper transferred was taken as the total loss of luminescence, whereas the fraction of total copper transferred was calculated by loss of luminescence at any given time point, over total loss of luminescence. The controls include CuCox17 alone (No Add.), as well as the addition of metallated, Cu-Sco1 (+Cu-Sco1). The Sco1 V221D mutant (not shown) demonstrated similar kinetic loss of luminescence.
Loop 8 Mutants Are Impaired in Sco1 Function with Cox2—Overexpression of Sco1 was shown to partially suppress the respiratory deficiency of cox17-1 mutant cells, but suppression of cox17Δ cells required the presence of supplemental CuSO4 in addition to overexpression of SCO1 (4). To bypass Cox17, high copy Sco1 must facilitate its own metallation and that of Cox11 for the subsequent metallation of Cox1 and Cox2, respectively. We tested whether Y219D, R220D, V221D, and Y222D Sco1 mutants retained the ability to suppress the respiratory defect of cox17-1 cells. If the mutations abrogated Cox17-mediated metallation, the prediction is that the mutants would be functional as the wild-type Sco1. Alternatively, if the mutations abrogate the function of Sco1 in CuA site formation in Cox2, then overexpression of the mutant proteins would fail to confer respiratory growth to cox17-1 cells. Episomal wild-type SCO1 transformants of cox17-1 cells enabled the cells to propagate on glycerol/lactate medium, whereas episomal mutant SCO1 alleles encoding each of the four substitutions failed to confer growth (Fig. 5). These results suggest that the mutations attenuate the interaction of Sco1 with Cox2.
FIGURE 5.
Suppression of cox17-1 and cox17Δ cells. Panel A, SCO1 mutant alleles were tested for their ability to suppress the cox17-1 growth defect on glycerol/lactate medium. W303-1A cox17Δ cells harboring plasmid-encoded Cyb2-C57Y Cox17 were transformed with wild-type or mutant SCO1 alleles and subsequently assessed for growth on glucose (synthetic complete) and glycerol/lactate (synthetic complete + 1 mm CuSO4, and rich, YP glycerol/lactate). Serial dilutions of cells were plated. Panel B, the same Sco1 mutants were tested as in panel A in cox17Δ cells cultured in synthetic complete glucose, and YP-glycerol/lactate + 8 mm CuSO4.
Multiple attempts were made to document a physical interaction of Sco1 and Cox2. The sco1Δ cells expressing the loop 8 mutants lack stable Cox2, therefore immunoprecipitation was attempted from mitochondria of wild-type cells co-expressing the mutant proteins. We observed variable results with wild-type Sco1 limiting our ability to reproducibly quantify the levels of Sco1 interacting with Cox2 with either the HA-tagged wild-type or mutant Sco1 molecules. Detergent-solubilized Sco1 fractionates as a high mass complex, but the size of the complex is independent of the presence of Cox2 (14). The same conclusion was reached in monitoring the migration of Sco1 on blue-native PAGE after in vitro import of Sco1 translated in a rabbit reticulocyte lysate with [35S]cysteine in mitochondria of wild-type cells compared with cox2Δ cells. In both cases, Sco1 migrated as a ∼300-kDa major complex (data not shown). Thus, the more direct assays to quantify a Sco1-Cox2 interaction were not informative.
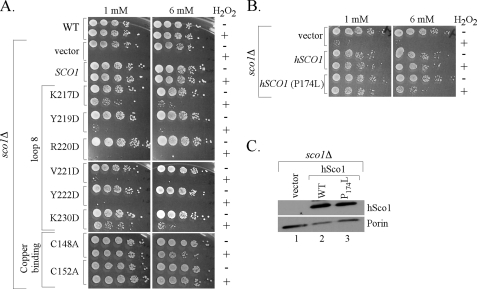
Cells lacking Sco1 are sensitive to hydrogen peroxide (28). We demonstrated that peroxide sensitivity of sco1Δ cells arises from the transient accumulation of a pro-oxidant heme a3:Cox1 stalled intermediate (26). The peroxide sensitivity of sco1Δ cells is suppressed by the addition of either wild-type or Cys mutant alleles of Sco1 that fail to bind Cu(I). We proposed that the lack of Sco1 destabilizes Cox2 sufficiently that it fails to interact with Cox1 leading to peroxide sensitivity. We tested the loop 8 Sco1 mutants for their ability to suppress hydrogen peroxide sensitivity of sco1Δ cells (Fig. 6A). Whereas the wild-type Sco1 protein effectively suppresses the hydrogen peroxide sensitivity of sco1Δ cells, the addition of the Y219D, R220D, V221D, and Y222D alleles failed to confer peroxide resistance. Curiously, two mutant alleles, K217D and K230D, that confer glycerol/lactate growth in sco1Δ cells are partially compromised in their ability to confer peroxide resistance. In contrast, as shown previously, respiratory-defective mutants Sco1 compromised in Cu(I) binding (C148A and C152A mutant Sco1) are fully competent to suppress hydrogen peroxide sensitivity (Fig. 6A). Cys mutants of Sco1 were shown to retain the ability to bind Cox2 (8). Suppression of the hydrogen peroxide sensitivity of sco1Δ cells by Sco1 variants is dependent on the Sco1 transmembrane domain. Replacement of the transmembrane segment by the transmembrane from Cox11 failed to generate a chimera active in suppression of peroxide sensitivity (data not shown).
FIGURE 6.
Sensitivity of sco1Δ transformants to hydrogen peroxide. SCO1 mutant alleles were tested for their sensitivity to hydrogen peroxide exposure. Panel A, BY4741 sco1Δ cells expressing wild-type or mutant Sco1-HA were grown to mid-exponential phase and incubated with (+) or without (–) 1 or 6 mm H2O2 for 2 h at 30 °C. Respective serial dilutions were plated onto rich medium containing 2% glucose (YPD) for growth assessment. Panel B, BY4741 sco1Δ cells expressing hSco1 or hSco1 P174L mutant were grown and treated as in A. Panel C, steady-state levels of hSco1 and hSco1 P174L were assessed by immunoblotting mitochondrial fractions from corresponding strains. Mitochondrial fractions were probed with Myc antisera to detect human Sco1-Myc. Por1 was used as a mitochondrial marker and loading control.
Human Sco1 that is nonfunctional in yeast CcO assembly is also able to suppress the peroxide sensitivity of yeast sco1Δ cells (26). The P174L mutation in human Sco1 that attenuates copper transfer from Cox17 (20) confers resistance to yeast sco1Δ cells to hydrogen peroxide (Fig. 6B). Both wild-type and P174L Sco1 proteins are equivalently expressed (Fig. 6C).
DISCUSSION
The conversion of apo-Sco1 to the Cu(I) conformer is accompanied by only structural rearrangements of a protruding loop that orients the Cu(I) binding His residue (17). The prominent exposure of loop 8 in the Sco1 structure was consistent with a candidate role as an interaction interface with either its Cu(I) donor Cox17 and/or its Cu(I) target Cox2. We show presently that aspartate substitutions at four conserved positions at the start of loop 8 compromise the function of yeast Sco1. Cells harboring Y219D, R220D, V221D, and Y222D mutant Sco1 proteins failed to restore glycerol/lactate growth or CcO activity in sco1Δ cells. The mutant proteins are stably expressed and are competent to bind Cu(I) and Cu(II) normally.
These conserved resides at the leading edge of loop 8 appear to define a function with Cox2 but are less important for Cox17. The respiratory defect of the Y219D, R220D, V221D, and Y222D mutant cells was not rescued by overexpression of COX17, unlike the P174L mutant Sco1 that is compromised in Cu(I) transfer by Cox17. However, elevated Cox17 levels had a modest stimulatory effect with the V221D mutant Sco1. In vitro Cu(I) transfer studies with CuCox17 and purified mutant proteins suggest that Cox17-mediated metallation of Sco1 proceeds normally with the four mutant Sco1 proteins. Two lines of evidence suggest that the mutant proteins are impaired in the function with its target Cox2. The two assays indirectly report on the transient Sco1-Cox2 interaction. The first assay involves the known suppression of the respiratory deficiency of cox17-1 cells by high copy Sco1 (4). The mechanism of suppression is likely the ability of high levels of Sco1 protein to bypass the impaired copper metallation of Sco1 by the C57Y mutant Cox17 and subsequent Sco1-mediated metallation of Cox2. With the first assay we show that the Y219D, R220D, V221D, and Y222D substitutions abrogate the ability of Sco1 to suppress the respiratory defect of cox17-1 cells. The high copy Y219D, R220D, V221D, and Y222D mutant Sco1 proteins fail to suppress even in cox17-1 cultures supplemented with 1 mm exogenous copper.
The second assay involves the hydrogen peroxide sensitivity of sco1Δ cells that can be suppressed by certain non-functional Sco1 mutants (26). Copper-binding yeast Sco1 mutants (C148A and C152A mutants) and wild-type human Sco1 confer peroxide resistance to sco1Δ cells without supporting CcO biogenesis. We anticipate that in the absence of Sco1, Cox2 is compromised in its interaction with Cox1 allowing accumulation of the pro-oxidant heme a3:Cox1 intermediate. Heme a/a3 insertion likely occurs in Cox1 prior to the addition of Cox2 in the biogenesis of CcO. Cox2 packs onto the partially accessible face of Cox1 that may be the site of hemes a/a3 insertion prior to Cox2 docking. CcO assembly mutants that block Cox2 addition to the preassembly Cox1 containing hemes a/a3 are peroxide sensitive. With the second assay, we observe that the loop 8 mutants fail to reverse hydrogen peroxide sensitivity of sco1Δ cells. The inability of the Y219D, R220D, V221D, and Y222D mutant Sco1 proteins to reverse the peroxide sensitivity of sco1Δ cells cannot arise from the non-functional status of these mutants as the non-functional C148A and C152A Sco1 mutants compromised in Cu(I) binding do restore peroxide resistance. The K217D and K230D mutants that restore limited CcO activity in sco1Δ cells also confer limited peroxide resistance.
The P174L Sco1 patient mutation is more impaired in its function with Cox17 than Cox2, whereas the four loop 8 mutants are more compromised with Cox2. Consistent with this conclusion is the observation that P174L Sco1 confers peroxide resistance to sco1Δ cells unlike the Y219D, R220D, V221D, and Y222D mutant Sco1 proteins. Pro174 in human Sco1 corresponds to yeast Pro153. Pro153 and loop 8 segment encompassing Tyr219–Tyr222 are on distinct faces of Sco1 (Fig. 1C) leading to the possibility that Sco1 uses distinct interfaces for interaction with Cox17 and Cox2. However, some overlap may exist as the V221D mutant Sco1 that is compromised in its interaction with Cox2, also shows a modest enhancement in function with high copy Cox17. The entire loop 8 does not appear to be an interaction interface as the loop tip shows no sequence conservation and significant length variation occurs (Fig. 1A).
A neutral surface on Sco1 was predicted previously to be an interface for Cox2 without supporting evidence. The surface implicated involved human Sco1 Thr261 and residues 169–172 of the CPDV sequence (11). Pro174 is also prominent on that surface and based on the present studies, we predict that the neutral surface mentioned is more important in the transient interaction with Cox17 than Cox2. In a study comparing Cu(I) transfer to human Sco1 from CuCox17 versus Cu(I) acetonitrile, differential perturbations in residues 258–262 were observed in 1H-15N-HSQC spectra. Those residues are spacially close to the surface defined by residues 169–174 (7). Thus, the surface of human Sco1 consisting of residues 169–174 and 258–261 may be important for Cox17 interaction. Because the residues implicated in this study for Cox2 function (minimally residues 241–245 of human Sco1) lie on an adjacent face of Sco1, an intriguing possibility exists that a tertiary complex of Cox17, Sco1, and Cox2 may form for the Cu(I) transfer steps. The weak suppression seen by overexpression of Cox17 with the V221D Sco1 mutant in sco1Δ cells may arise from a stabilizing effect of Cox17 on the V221D Sco1-Cox2 transient interaction.
A significant challenge remains to obtain a soluble Cox2 domain for detailed analysis of the copper transfer reaction. In the absence of available Cox2 for in vitro studies, one cannot be certain that the transfer of Cu(I) from Sco1 to Cox2 proceeds directly without the participation of another assembly factor such as Cox20 that is implicated as a Cox2 chaperone (27).
This work was supported, in whole or in part, by National Institutes of Health Grant ES 03817 (NIEHS) (to D. R. W.). This work was also supported by the Center of Excellence in Molecular Hematology core facility Grant DK P30 072437 for FPLC chromatography and the United Mitochondrial Disease Foundation (to P. A. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: CcO, cytochrome c oxidase; IM, inner membrane; IMS, intermembrane space; PBS, phosphate-buffered saline; DTT, dithiothreitol; HA, hemagglutinin.
References
- 1.Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Hakashima, R., Yaono, R., and Yoshikawa, S. (1995) Science 269 1069–1074 [DOI] [PubMed] [Google Scholar]
- 2.Cobine, P. A., Pierrel, F., and Winge, D. R. (2006) Biochim. Biophys. Acta 1763 759–772 [DOI] [PubMed] [Google Scholar]
- 3.Horng, Y. C., Cobine, P. A., Maxfield, A. B., Carr, H. S., and Winge, D. R. (2004) J. Biol. Chem. 279 35334–35340 [DOI] [PubMed] [Google Scholar]
- 4.Glerum, D. M., Shtanko, A., and Tzagoloff, A. (1996) J. Biol. Chem. 271 20531–20535 [DOI] [PubMed] [Google Scholar]
- 5.Schulze, M., and Rodel, G. (1988) Mol. Gen. Genet. 211 492–498 [DOI] [PubMed] [Google Scholar]
- 6.Krummeck, G., and Rödel, G. (1990) Curr. Genet. 18 13–15 [DOI] [PubMed] [Google Scholar]
- 7.Banci, L., Bertini, I., Ciofi-Baffoni, S., Leontari, I., Martinelli, M., Palumaa, P., Sillard, R., and Wang, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lode, A., Kuschel, M., Paret, C., and Rodel, G. (2000) FEBS Lett. 448 1–6 [DOI] [PubMed] [Google Scholar]
- 9.Leary, S. C., Kaufman, B. A., Pellechia, G., Gguercin, G.-H., Mattman, A., Jaksch, M., and Shoubridge, E. A. (2004) Hum. Mol. Genet. 13 1839–1848 [DOI] [PubMed] [Google Scholar]
- 10.Balatri, E., Banci, L., Bertini, I., Cantini, F., and Cioffi-Baffoni, S. (2003) Structure 11 1431–1443 [DOI] [PubMed] [Google Scholar]
- 11.Williams, J. C., Sue, C., Banting, G. S., Yang, H., Glerum, D. M., Hendrickson, W. A., and Schon, E. A. (2005) J. Biol. Chem. 280 15202–15211 [DOI] [PubMed] [Google Scholar]
- 12.Abajian, C., and Rosenzweig, A. C. (2006) J. Biol. Inorg. Chem. 11 459–466 [DOI] [PubMed] [Google Scholar]
- 13.Banci, L., Bertini, I., Ciofi-Baffoni, S., Gerothanassis, I. P., Leontari, I., Martinelli, M., and Wang, S. (2007) Structure 15 1132–1140 [DOI] [PubMed] [Google Scholar]
- 14.Nittis, T., George, G. N., and Winge, D. R. (2001) J. Biol. Chem. 276 42520–42526 [DOI] [PubMed] [Google Scholar]
- 15.Beers, J., Glerum, D. M., and Tzagoloff, A. (2002) J. Biol. Chem. 277 22185–22190 [DOI] [PubMed] [Google Scholar]
- 16.Horng, Y.-C., Leary, S. C., Cobine, P. A., Young, F. B. J., George, G. N., Shoubridge, E. A., and Winge, D. R. (2005) J. Biol. Chem. 280 34113–34122 [DOI] [PubMed] [Google Scholar]
- 17.Banci, L., Bertini, I., Calderone, V., Ciofi-Baffoni, S., Mangani, S., Martinelli, M., Palumaa, P., and Wang, S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8595–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valnot, I., Osmond, S., Gigarel, N., Mehaye, B., Amiel, J., Cormier-Daire, V., Munnich, A., Bonnefont, J. P., Rustin, P., and Rotig, A. (2000) Am. J. Hum. Genet. 67 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leary, S. C., Cobine, P. A., Kaufman, B. A., Guercin, G. H., Mattman, A., Palaty, J., Lockitch, G., Winge, D. R., Rustin, P., Horvath, R., and Shoubridge, E. A. (2007) Cell Metab. 5 9–20 [DOI] [PubMed] [Google Scholar]
- 20.Cobine, P. A., Pierrel, F., Leary, S. C., Sasarman, F., Horng, Y. C., Shoubridge, E. A., and Winge, D. R. (2006) J. Biol. Chem. 281 12270–12276 [DOI] [PubMed] [Google Scholar]
- 21.Foltopoulou, P. F., Zachariadis, G. A., Politou, A. S., Tsiftsoglou, A. S., and Papadopoulou, L. C. (2004) Mol. Genet. Metab. 81 225–236 [DOI] [PubMed] [Google Scholar]
- 22.Rigby, K., Zhang, L., Cobine, P. A., George, G. N., and Winge, D. R. (2007) J. Biol. Chem. 282 10233–10242 [DOI] [PubMed] [Google Scholar]
- 23.Maxfield, A. B., Heaton, D. N., and Winge, D. R. (2004) J. Biol. Chem. 279 5072–5080 [DOI] [PubMed] [Google Scholar]
- 24.Diekert, K., de Kroon, A. I., Kispal, G., and Lill, R. (2001) Methods Cell Biol. 65 37–51 [DOI] [PubMed] [Google Scholar]
- 25.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 26.Khalimonchuk, O., Bird, A., and Winge, D. R. (2007) J. Biol. Chem. 282 17442–17449 [DOI] [PubMed] [Google Scholar]
- 27.Hell, K., Tzagoloff, A., Neupert, W., and Stuart, R. A. (2000) J. Biol. Chem. 275 4571–4578 [DOI] [PubMed] [Google Scholar]
- 28.Banting, G. S., and Glerum, D. M. (2006) Eukaryot. Cell 5 568–578 [DOI] [PMC free article] [PubMed] [Google Scholar]