Abstract
γ-Aminobutyric acid (GABA) binding to GABAA receptors (GABAARs) triggers conformational movements in the α1 and β2 pre-M1 regions that are associated with channel gating. At high concentrations, the barbiturate pentobarbital opens GABAAR channels with similar conductances as GABA, suggesting that their open state structures are alike. Little, however, is known about the structural rearrangements induced by barbiturates. Here, we examined whether pentobarbital activation triggers movements in the GABAAR pre-M1 regions. α1β2 GABAARs containing cysteine substitutions in the pre-M1 α1 (K219C, K221C) and β2 (K213C, K215C) subunits were expressed in Xenopus oocytes and analyzed using two-electrode voltage clamp. The cysteine substitutions had little to no effect on GABA and pentobarbital EC50 values. Tethering chemically diverse thiol-reactive methanethiosulfonate reagents onto α1K219C and α1K221C affected GABA- and pentobarbital-activated currents differently, suggesting that the pre-M1 structural elements important for GABA and pentobarbital current activation are distinct. Moreover, pentobarbital altered the rates of cysteine modification by methanethiosulfonate reagents differently than GABA. For α1K221Cβ2 receptors, pentobarbital decreased the rate of cysteine modification whereas GABA had no effect. For α1β2K215C receptors, pentobarbital had no effect whereas GABA increased the modification rate. The competitive GABA antagonist SR-95531 and a low, non-activating concentration of pentobarbital did not alter their modification rates, suggesting that the GABA- and pentobarbital-mediated changes in rates reflect gating movements. Overall, the data indicate that the pre-M1 region is involved in both GABA- and pentobarbital-mediated gating transitions. Pentobarbital, however, triggers different movements in this region than GABA, suggesting their activation mechanisms differ.
Ligand-gated ion channels (LGICs)2 are integral membrane proteins that mediate fast synaptic transmission between cells in the brain and at the neuromuscular junction. The type A γ-aminobutyric acid receptor (GABAAR) is the main inhibitory LGIC in the brain and is the target for a wide range of therapeutic agents such as benzodiazepines, barbiturates, and anesthetics. Barbiturates, such as pentobarbital (PB), have three distinct effects on GABAAR activity. At low concentrations, PB modulates GABA-mediated Cl- current (IGABA). At higher concentrations, PB directly activates the GABAAR in the absence of GABA, and at still higher concentrations, PB blocks channel activity (1). Little is known, however, about the structural rearrangements underlying these functional effects.
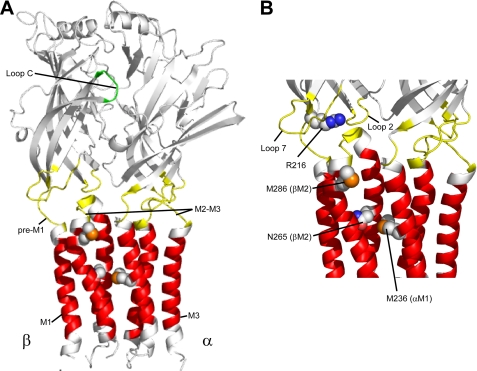
Single channel studies from mouse spinal neurons (2–4) and from rat hippocampal neurons (5) have shown that currents evoked by PB are similar in conductance as those evoked by GABA, suggesting that the open state structures stabilized by PB binding are similar to those stabilized by GABA. However, GABA and PB bind to distinct sites on the GABAAR (Fig. 1). The GABA binding site is located at the interfaces of the α1 and β2 subunits in the extracellular domain, whereas the PB/general anesthetics binding site(s) are believed to be located ∼50 Å below the GABA binding site in a water-accessible pocket located between the four transmembrane helices (M1–4) of the receptor (Fig. 1). Mutational analyses as well as photolabeling studies have identified positions in the GABAAR transmembrane helices that are important for mediating the effects of PB/anesthetics, with a proposed binding pocket involving residues in M1 (α1M236), M2 (β2N265), and M3 (β2M286) (6–8).
FIGURE 1.
Structural model of the GABAAR α1 and β2 subunits. A, the extracellular binding domain is colored in white. Domains believed to contribute to the GABA transduction mechanism (Loop 2, Loop 7, M2-M3 linker, and pre-M1) are highlighted in yellow. The Loop C region of the GABA binding site is highlighted in green. The transmembrane domains (M1, M2, and M3) are colored in red. Residues in the pre-M1 region (Arg-216) as well as residues forming the potential PB/general anesthetic binding site (Asn-265 and Met-286) in the β2 subunit and (Met-236) in the α1 subunit are shown in a space-filled format. The M4 transmembrane helix has been omitted for illustration purposes. B, detailed view of the interface between the ligand binding domain and the transmembrane domain.
The structural machinery associated with coupling agonist binding to channel gating in the Cys-loop family of LGICs likely involves distributed movements of, and interactions between, several discrete domains. Recent evidence suggests that binding of neurotransmitter in the extracellular domain triggers a series of molecular motions (conformational wave) that initiates in the ligand binding pocket, followed by movements in Loop 2, Loop 7 (Cys-loop), the pre-M1 region, the M2-M3 linker, and finally the transmembrane domains to gate the channel (9–11). Because PB- and GABA-activated channel open state structures are alike (3, 12) but PB and GABA bind to different sites, we were interested in determining whether gating motions induced by PB are similar to those induced by GABA.
We previously demonstrated that the α1 and β2 pre-M1 regions of the GABAAR, which connect the extracellular domain of each subunit with the transmembrane domain, undergo structural rearrangements during GABA activation (13). In this study, we measured PB-mediated changes in the accessibility of cysteines engineered into the pre-M1 region to monitor structural movements induced by activating concentrations of PB and compared these changes to those induced by GABA. Our data indicate that the pre-M1 region is part of a common gating pathway used by both ligands. PB, however, triggers different movements in this region than GABA, suggesting that structural transitions evoked and/or stabilized by PB binding/channel activation differ from those triggered by GABA.
EXPERIMENTAL PROCEDURES
Mutagenesis—Rat cDNAs encoding α1 and β2 GABAAR subunits were used for all molecular cloning and functional studies. Cysteine mutants were made as previously described (13).
Expression in Xenopus laevis Oocytes—Oocytes were prepared as previously described (14). Capped cRNAs encoding the α1, β2, α1K219C, α1K221C, β2K213C and β2K215C subunits in the vector pGH19 (15, 16) were transcribed in vitro using the mMessage mMachine T7 kit (Ambion, Austin, TX). Single oocytes were injected within 24 h with 27 nl of cRNA (10 ng/μl/subunit) in a ratio 1:1. Oocytes were incubated at 18 °C in ND96 (in mm: 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, and 5 HEPES, pH 7.2) supplemented with 100 μg/ml gentamycin and 100 μg/ml bovine serum albumin for 2–7 days before use.
Two-electrode Voltage Clamp—Oocytes were continuously perfused at a rate of ∼5 ml/min with ND96 while being held under two-electrode voltage clamp at -80 mV. The bath volume was ∼200 μl. Stock solutions of GABA (Sigma-Aldrich) and PB (Research Biochemicals, Natick, MA) were prepared fresh daily in ND96. Borosilicate electrodes (Warner Instruments, Hamden, CT) were filled with 3 m KCl and had resistances between 0.7 and 2 mΩ. Electrophysiological data were acquired with a GeneClamp 500 (Axon Instruments, Foster City, CA) interfaced to a computer with an ITC16 analog-to-digital device (Instrutech, Great Neck, NY) and recorded using Whole Cell Program 3.2.9 (kindly provided by J. Demspter, University of Strathclyde, Glasgow, Scotland).
Concentration Response Analysis—PB concentration responses were measured, and the resulting data were fit to the following equation: I = Imax/(1 + (EC50/[A])n), where I is the peak response to a given concentration of PB, Imax is the maximum amplitude of current, EC50 is the concentration of PB that evokes half-maximal response, [A] is the agonist concentration, and n is the Hill coefficient. At high PB concentrations, currents were partially blocked during PB application. Thus, peak PB currents were measured immediately after PB wash out, when a prominent tail current appears (21). GraphPad Prism 4 software (San Diego, CA) was utilized for data analysis and fitting.
Modification of Introduced Cysteine Residues by MTS Reagents—Three derivatives of methanethiosulfonate (CH3SO2X; MTS) were used to covalently modify the introduced cysteines: MTS-N-biotinylaminoethyl (X = SCH2CH2NH-biotin; MTSEA-biotin), MTS-ethyltrimethylammonium (X = SCH2CH2N(CH3)3+; MTSET+), and MTS-ethylsulfonate (X = SCH2CH2 SO3-; MTSES-) (Biotium, Hayward, CA). MTSET+ is positively charged whereas MTSES- is negatively charged at neutral pH. Stock solutions (100 mm) were made in DMSO for all MTS reagents, aliquoted into microcentrifuge tubes, and rapidly frozen on ice before storage at -20 °C. For each application of MTS reagent, a new aliquot was thawed, diluted in ND96 to the working concentration, and used immediately to avoid hydrolysis of the MTS compound. The final DMSO concentrations were ≤2%, which had no effect on PB-mediated current responses.
MTS modifications of the engineered cysteines were assayed by measuring changes in PB-evoked current (IPB). The effects of MTSEA-biotin, MTSET+, and MTSES- were studied using the following protocol: PB (EC40–60) current responses (10 s) were measured from oocytes expressing wild-type (α1β2) or mutant receptors and stabilized. Stability was defined as <10% variance of peak current responses to PB on two consecutive applications. After stabilization, the MTS reagent (2 mm) was bath-applied for 2 min, followed by a 5-min wash, and then IPB was measured at the same concentration as before the MTS treatment. The effect of the MTS application was calculated as: [((Iafter/Iinitial) -1) × 100], where Iafter is the peak PB current elicited after the MTS application and Iinitial is the peak current before MTS.
Rate of MTS Modification—The rates at which the various MTS reagents modified the engineered cysteines were determined by measuring the effect of sequential applications of low concentrations of MTS reagents on IGABA as described previously (17). The protocol is described as follows: EC40–60 GABA was applied for 10 s every 3–5 min until IGABA stabilized (<3% variance). After a 40-s ND96 wash out, MTS reagents were applied for 5–20 s, and the cell was washed for an additional 2.5–4.5 min. The procedure was repeated until IGABA no longer changed, indicating that the reaction had proceeded to apparent completion. Concentration of MTS reagent and time of application varied as follows: α1K219C: MTSEA-biotin, 10 μm, 20 s; α1K221C: MTSEA-biotin, 10 μm, 20 s; β2K213C: MTSET+, 30 μm, 20 s; β2K215C: MTSET+, 30 μm, 20 s. The effects of co-applying GABA, SR-95531 (GABA antagonist), or PB (modulator) on reaction rates were assayed by co-applying GABA (EC80–90), 10 μm SR-95531, or PB (50 or 500 μm) with the MTS reagent. For these experiments, IGABA was stabilized as follows: EC40–60 GABA was applied for 10 s, washed for 40 s, high concentrations of GABA, SR-95531, or PB were applied for 5–20 s, and the oocyte washed for 2.5–5 min. The procedure was repeated until IGABA from EC40–60 GABA was <3% of the previous IGABA peak. This allowed complete wash out of the different drugs and ensured that any alteration in the current amplitudes following MTS treatment in the presence of drug was the result of MTS modification and not a result of inadequate wash out of drug. Concentrations of MTS reagents and times of applications in the presence of GABA (EC80–90) were as follows: α1K219C: MTSEA-biotin, 30 μm, 20 s; α1K221C: MTSEA-biotin, 10 μm, 20 s; β2K213C: MTSET+, 30 or 60 μm, 10 s; β2K215C: MTSET+, 30 μm, 10 s. In the presence of 500 μm PB: α1K219C: MTSEA-biotin, 30 μm, 20 s; α1K221C: MTSEA-biotin, 10 μm, 20 s; β2K213C: MTSET+, 30 μm, 20 s; β2K215C: MTSET+, 30 μm, 10 s. In the presence of 50 μm PB: α1K219C: MTSEA-biotin, 30 μm, 20 s; α1K221C: MTSEA-biotin, 10 μm, 20 s; β2K213C: MTSET+, 30 μm, 20 s; β2K215C: MTSET+, 50 SR-95531: α1K219C: MTSEA-biotin, 100 μm, 10 s; α1K221C: MTSEA-biotin, 100 μm, 10 s; β2K213C: MTSET+, 50 μm, 10 s; β2K215C: MTSET+, 75 μm, 20 s.
For all rate experiments, the decrease or increase in GABA-induced current was plotted versus cumulative time of MTS exposure. Peak current at each time point was normalized to the initial peak current (t = 0) and fit to a single exponential function using GraphPad Prism software to obtain a pseudo-first-order rate constant (k1). The second-order rate constant (k2) was calculated by dividing k1 by the concentration of the MTS reagent used (18).
Statistical Analysis—Log (EC50) values, changes in PB EC50 after MTS modification, and second-order (k2) rates were analyzed using a one-way analysis of variance, followed by a post-hoc Dunnett's test to determine the level of significance between wild-type and mutant receptors.
Structural Modeling—A model of the entire GABAA receptor was built as previously described (13).
RESULTS
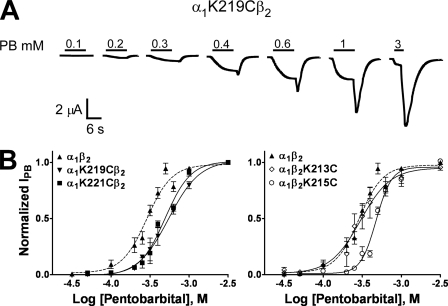
Functional Characterization of Pre-M1 Mutant Receptors—We previously showed (13) that cysteine substitutions in the pre-M1 region of the α1 (K219C, K221C) and β2 (K213C, K215C) subunits had no effects on GABA EC50 (α1β2 receptors, EC50 = 6.9 ± 0.7 μm). To determine whether the same cysteine substitutions altered PB activation, we measured PB concentration responses using two-electrode voltage clamp (Fig. 2). Cysteine substitutions of α1K219, α1K221, and β2K215 had little (2-fold) to no effect (β2K213) on PB EC50 values relative to α1β2 receptors (EC50 = 271 ± 18 μm; Fig. 2; Table 1). Mutant receptors had Hill coefficients for PB activation that were not significantly different from wild-type α1β2 receptors (Table 1). PB maximal macroscopic currents elicited from mutant receptors were similar to α1β2 receptors, ranging from 600 nA to 18 μA. The fact that these mutations had little effect on GABA and PB EC50 values, Hill coefficients, or surface expression suggests that the side chains of the introduced cysteines are in similar positions as the side chains of the native residues, making the introduced cysteines at positions α1K219, α1K221, β2K213, and β2K215 ideal candidates to probe the dynamics of the pre-M1 region induced by PB binding/gating.
FIGURE 2.
PB concentration response curves of wild-type α1β2 and mutant GABAAR. A, representative current responses from an oocyte expressing α1K219Cβ2 receptors elicited by increasing concentrations of PB (mm). B, PB concentration response curves from oocytes expressing α1β2 (▴; dashed line), α1K219Cβ2 (▾), α1K221Cβ2 (▪), α1β2K213C (⋄), and α1β2K215C (○) receptors. Peak PB-activated currents were measured after PB wash out (tail current) and used for concentration response fitting. Data points represent the mean ± S.E. from four to six independent experiments. Data were fit by nonlinear regression analysis as described under “Experimental Procedures.” PB EC50 and nH values are reported in Table 1.
TABLE 1.
PB concentration response data for α1β2 and mutant receptors Concentration response data for PB activation of wild-type and mutant receptors are tabulated. EC50 and Hill coefficient (nH) values are expressed as mean ± S.E. for n number of independent experiments from at least two batches of oocytes. *, p < 0.05, **, p < 0.01, significantly different from control.
| Receptor | EC50 | nH | n |
|---|---|---|---|
| μm | |||
| α1β2 | 271 ± 18 | 2.7 ± 0.3 | 6 |
| α1(K219C)β2 | 495 ± 32** | 2.4 ± 0.2 | 4 |
| α1(K221C)β2 | 471 ± 41* | 2.8 ± 0.4 | 5 |
| α1β2(K213C) | 224 ± 5.1 | 2.5 ± 0.4 | 5 |
| α1β2(K215C) | 577 ± 44** | 3.8 ± 0.5 | 4 |
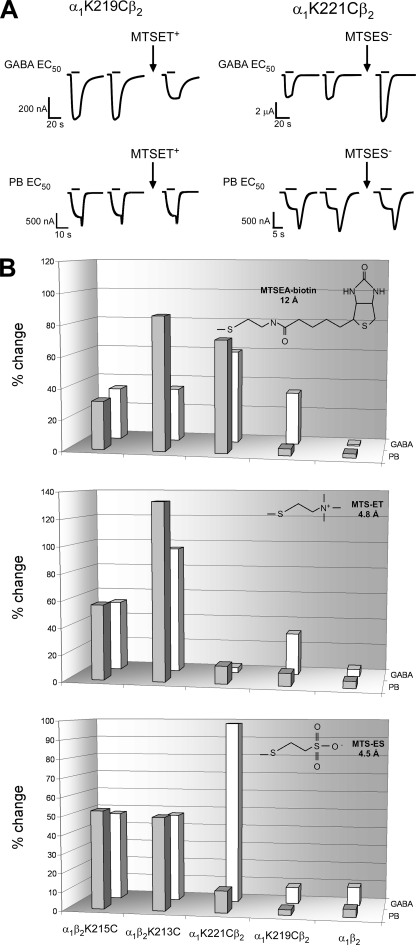
Effects of Cysteine Modification on PB- and GABA-evoked Currents—We measured current responses elicited with PB EC50 (IPB) concentrations before and after MTS reagent application (Fig. 3) to examine how covalently modifying the introduced cysteines would affect PB currents. The MTS reagents used were 1) MTSEA-biotin, which covalently adds a neutral biotinylaminoethyl group (12 Å long); 2) MTSET+, which adds a positively charged ethyl-trimethylammonium group (4.5 Å long); and 3) MTSES-, which adds a negatively charged ethyl-sulfonate group (4.8 Å long). Application of 2 mm MTSEA-biotin, MTSET+, or MTSES- for 2 min to wild-type receptors had no effect on IPB (≤5 ± 3% for all reagents), indicating that any effects observed in the mutant receptors are due to modification of the introduced cysteines (Fig. 3B). MTSEA-biotin modification of β2K213C and β2K215C increased IPB by 86 ± 12 and 31 ± 4% (n ≥3), respectively, and decreased IPB by 72 ± 3% (n = 6) in α1K221C-containing receptors (Fig. 3B). Similar functional effects on GABA EC50 current responses (IGABA) were observed after MTSEA-biotin modification of β2K213C, β2K215C, and α1K221C (Fig. 3B and Ref. 13). In contrast, MTSEA-biotin modification of α1K219C did not affect IPB whereas IGABA was increased by 34 ± 2% (Fig. 3B). This indicates that MTSEA-biotin covalently modified α1K219C but modification had no functional effect (i.e. a “silent” reaction) on PB-induced currents.
FIGURE 3.
Effects of MTS reagents on α1β2 and mutant GABAAR. A, representative current traces elicited by GABA (top traces) or PB (bottom traces) of EC50 concentrations from oocytes expressing α1K219Cβ2 and α1K221Cβ2 receptors before and after treatment with MTSET+ and MTSES- (2 min, 2 mm). MTSET+ and MTSES- treatment altered the GABA current responses but had no effect on PB current responses. B, summary of the effects of a 2-min application of 2 mm MTSEA-biotin (top), MTSET+ (middle), or MTSES- (bottom) on GABA (EC50) -activated currents (IGABA) (previously reported in Ref. 13) and PB (EC50) -activated currents (IPB) from α1β2 and mutant receptors. The absolute percent change in IPB and IGABA after MTS treatment is defined as: [((Iafter/Iinitial) - 1) × 100]. Bars represent the mean from at least three independent experiments. Values >20% are significantly different from α1β2 values (p <0.01).
Derivatization of β2K213C and β2K215C with MTSES- increased IPB by 50 ± 3 and 53 ± 16%, respectively, while MTSET+ increased IPB 133 ± 2 and 56 ± 5%, respectively (n ≥ 3) (Fig. 3B). Similar functional effects on IGABA were observed after derivatization of β2K213C and β2K215C with MTSES- or MTSET+ (13) (Fig. 3B). As we previously reported, modification of α1K219Cβ2 and α1K221Cβ2 by MTSES- and MTSET+ differentially affected IGABA (Fig. 3B). Tethering a negative charge (MTSES-) onto α1K221C enhanced IGABA (98 ± 14%; n = 4), whereas treatment with the positively charged MTSET+ had no functional effect on IGABA (Fig. 3B). In contrast, tethering a positive charge (MTSET+) onto α1K219C decreased IGABA (31 ± 7%; n = 3), whereas modification with MTSES- had no effect. These results are likely due to differences in the local electrostatic environments near α1K219C and α1K221C rather than steric effects because MTSET- and MTSES+ are similar in size (Fig. 3B, insets) and have a common reaction mechanism. Surprisingly, modification of α1K219Cβ2 and α1K221Cβ2 by MTSES- or MTSET+ had no functional effects on IPB (Fig. 3A), indicating that introducing a positive or negative charge at these positions has different effects on PB and GABA current responses, suggesting that the physicochemical structural elements in the pre-M1 region important for GABA and PB current activation are distinct.
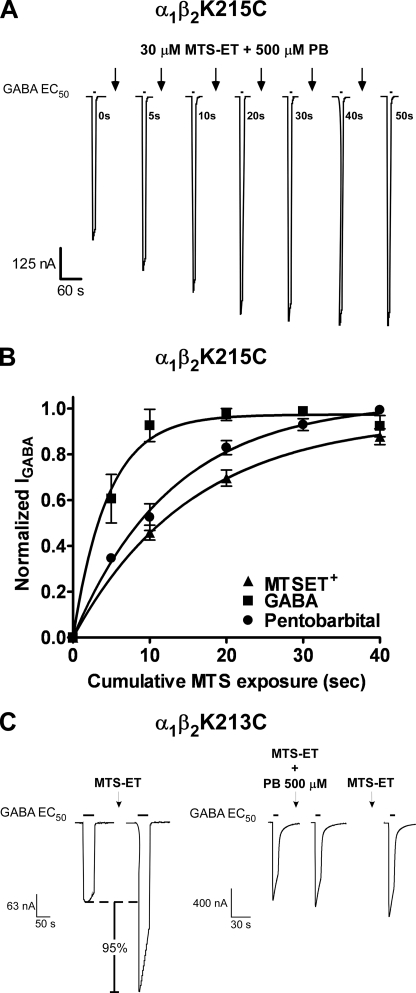
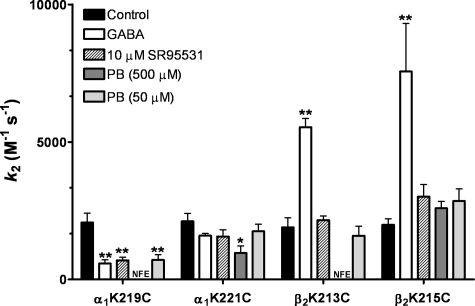
Effect of PB on MTS Reaction Rates—To determine whether PB binding/gating induces different structural rearrangements in the pre-M1 regions than GABA, we measured the rates of MTS modification of α1K219C, α1K221C, β2K213C, and β2K215C in the presence of a directly activating concentration of PB (500 μm) and compared these to rates measured in the presence of GABA (Figs. 4 and 5). The rate of modification of a cysteine by a MTS reagent mainly depends on the ionization of the thiol group and the access pathway of the reagent. Thus, changes in rates measured in the presence of PB or GABA provide a measure of structural changes in the receptor that are triggered by their binding. PB had no effect on the MTSET+ rate of modification of α1β2K215C whereas GABA increased the rate by 4-fold. For α1K221Cβ2 receptors, PB decreased the MTSEA-biotin rate by 2-fold whereas GABA had no significant effect. For α1β2K213C receptors and α1K219Cβ2 receptors, GABA increased and decreased their rates of modification by 3- and 4-fold, respectively. In contrast, when PB was present during the MTS reaction, the MTS treatment no longer altered subsequent GABA current responses for α1β2K213C and α1K219Cβ2 receptors (Fig. 4C). A subsequent application of MTS reagent in the absence of PB had little effect on GABA current, indicating that the thiol was modified in the presence of PB but modification now resulted in no detectable effect on IGABA. Although the mechanism underlying this loss of functional effect is unknown, one can conclude that cysteine modification in the presence of 500 μm PB is different from modification in the presence of GABA. A low concentration of PB that does not activate the receptor but potentiates GABA responses (50 μm) decreased the rate of modification ofα1K219C by ∼3-fold and had no effect on the rates of modification of α1K221C, β2K213C, and β2K215C (Fig. 4, Table 2), indicating that the effects of PB (500 μm) on α1K221Cβ2 and α1β2K213C receptors likely reflect gating-associated motions. Overall, these data indicate that GABA and PB induce different structural rearrangements in the pre-M1 regions of the α1 and β2 subunits.
FIGURE 4.
Rates of MTSET+ modification of α1β2K215C receptors in the presence and absence of GABA or PB. A, representative GABA current traces recorded while applying MTSET+ (30 μm) in the presence of PB (500 μm). GABA EC40–60 current responses were recorded before and after successive application (10 s) of 30 μm MTSET+ co-applied with PB (arrows). B, normalized GABA current responses were plotted versus cumulative time of MTSET+ (▴), MTSET+ co-applied with EC80–90 GABA (▪), and MTSET+ co-applied with 500 μm PB (•) and fit with single exponential functions. Data were normalized to the maximal amount of potentiation of IGABA for each experiment and represent mean ± S.E. from at least three independent experiments. C, representative GABA-mediated current traces from oocytes expressing α1β2K213C receptors. Currents elicited by an EC50 concentration of GABA were recorded before and after MTSET+ (2 mm, 2 min) in the absence or presence of 500 μm PB. MTSET+ treatment alone potentiated the subsequent current response (95%), but when MTSET+ was co-applied with 500 μm PB the treatment had no functional effect. On the subsequent GABA current and following wash out, MTSET treatment alone no longer resulted in a significant potentiation of current.
FIGURE 5.
Summary of effects of GABA, PB, and SR-95531 on MTS second-order rate constants. Second-order rate constants (k2) for MTS modification of cysteine mutants in the absence and presence of EC80–90 GABA, 500 μm PB, 50 μm PB, or 10 μm SR-95531. k2 values are reported in Table 2. Data are the mean ± S.E. from at least three independent experiments. * and ** indicate values significantly different from control at p <0.05 and p <0.001, respectively. NFE (no functional effect) MTS reagent reacts with the cysteine mutant in the presence of PB but has no functional effect on subsequent GABA responses.
TABLE 2.
Second-order rate constants (k2) for reaction of MTS reagents with mutant receptors in the absence (Control) and presence of GABA, PB, and SR-95531 NFE, no functional effect; MTS reagents react but have no functional effect on subsequent GABA responses. *, ** indicate values significantly different from control, with p < 0.05 and p < 0.001, respectively. Values are the mean ± S.E.
|
Receptor |
Controla |
GABAaEC80–90 |
SR-95531 |
Pentobarbital |
Pentobarbital |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| k2 M–1S–1 | n | k2 M–1S–1 | n | k2 M–1S–1 | n | k2 M–1S–1 | n | k2 M–1S–1 | n | |
| 10 μm | 500 μm | 50 μm | ||||||||
| α1K219Cβ2b | 2070 ± 300 | 4 | 580 ± 130** | 3 | 700 ± 110** | 4 | NFE | 3 | 710 ± 190 | 4 |
| α1K221Cβ2b | 2110 ± 290 | 6 | 1600 ± 80 | 4 | 1570 ± 250 | 4 | 970 ± 250* | 4 | 1750 ± 260 | 3 |
| α1β2K213Cc | 1900 ± 340 | 5 | 5540 ± 320** | 7 | 1610 ± 200 | 4 | NFE | 3 | 1590 ± 340 | 3 |
| α1β2K215Cc | 1990 ± 220 | 6 | 7560 ± 1750** | 3 | 3050 ± 510 | 3 | 2590 ± 250 | 3 | 2850 ± 440 | 4 |
Effect of SR-95531 on MTS Reaction Rates—To explore whether the structural rearrangements in the pre-M1 regions induced by GABA reflect conformational movements associated with gating, we measured the rates of MTS modification of α1K219C, α1K221C, β2K213C, and β2K215C in the presence of the GABA binding site competitive antagonist SR-95531 (10 μm) (Fig. 5). Because GABA and SR-95531 bind to the same site, but GABA promotes channel opening/desensitization whereas SR-99531 does not, co-application of SR-95531 and MTS should capture motions associated with stabilization of a closed state whereas co-application of GABA and MTS should capture motions associated with open/desensitized states (i.e. gating). SR-95531 had no significant effect on the rate of modification of α1K221Cβ2, α1β2K213C, and α1β2K215C receptors compared with control (Fig. 5 and Table 2), suggesting that occupancy of the GABA binding site alone does not induce structural rearrangements in or near these positions. SR-95531 slowed the MTSEA-biotin rate of modification of α1K219Cβ2 receptors 3-fold (Fig. 5 and Table 2), indicating that binding of a competitive antagonist can induce structural rearrangements in the pre-M1 region of the α1 subunit.
DISCUSSION
The Monod-Wyman-Changeux allosteric theory has been used with great success to model LGIC gating behavior (19). In this theory, channel gating is accomplished by a concerted quaternary movement of all the subunits switching from an inactive to an active conformation. GABA and PB activation both evoke the same single channel conductances (3) despite binding to different parts of the receptor (20). Thus, a key question is whether binding of an allosteric modulator such as PB triggers similar allosteric gating transitions as GABA.
For Cys-loop LGICs, it has been suggested that the pre-M1 region acts as a central hub that couples neurotransmitter-induced motions in the ligand binding site to movements in Loop 2 and the M2-M3 linker, which then ultimately triggers movements in the M2 channel region that opens the channel (Fig. 1) (10, 13). A key element in the transduction pathway coupling neurotransmitter binding to gating of the channel is believed to be a salt bridge between two highly conserved residues present in all Cys-loop LGIC subunits: an arginine in the pre-M1 region with a glutamic acid in Loop 2 (Fig. 1) (10). Previously, we showed that cysteine substitution of this highly conserved arginine (Arg-216) in the β2 pre-M1 region of the GABAAR abolished channel gating by GABA without altering binding of the GABA agonist [3H]muscimol (13), suggesting that this residue plays a key role in allosterically coupling GABA binding to gating. Interestingly, the β2R216C mutation also abolished channel gating by PB, suggesting that this residue and the pre-M1 region may also play a role in PB activation (13).
Here, we provide evidence that the α1 and β2 pre-M1 regions move in response to PB activation of the GABAAR and that PB triggers different movements in this region than GABA. Cysteine substitutions of the conserved pre-M1 lysine residues had little to no effect on PB EC50 (Fig. 2 and Table 1); thus, the positions occupied by the cysteine side chains in the mutant receptors are likely similar to the native lysine positions. The rates of modification of α1K221Cβ2, α1K219Cβ2, α1β2K213C, and α1β2K215C receptors differ depending upon whether GABA or PB is present. For α1K221Cβ2 receptors, only 500 μm PB caused a significant change in the rate of MTS modification whereas for α1β2K215C receptors only GABA caused a change (Fig. 5, Table 2). For α1K219Cβ2 and α1β2K213C receptors, GABA altered their rates of modification, whereas in the presence of PB the mutant receptors were modified but MTS modification no longer altered subsequent GABA-induced current (Fig. 5, Table 2), demonstrating that PB stabilizes the receptor in a different conformation(s) than GABA. Moreover, structurally perturbing the pre-M1 regions by tethering chemically diverse thiol-reactive groups onto these mutant cysteines had different effects on PB and GABA current responses (Figs. 2 and 3). Based on these data, we infer that the structural transitions evoked and/or stabilized by PB channel activation differ from those triggered by GABA in this region of the receptor.
Alternatively, one could argue that some of the differences measured in the effects of modifying these cysteines on GABA and PB current responses result from the pre-M1 residues being located near the PB binding site pocket. Several lines of evidence indicate that α1K219, α1K221, β2K213, and β2K215 do not form part of the PB binding site. First, mutations of these residues to cysteine had little to no effect on PB EC50. If these residues directly formed part of the PB binding site, one would expect bigger shifts in PB EC50, especially because of the non-conservative cysteine for lysine substitution. Second, modification of β2K213C and β2K215C with a variety of MTS reagents all increased PB-induced current and modification of α1K219C had no effects on PB-induced currents. If these residues were lining the PB binding site, one would expect that tethering bulky/charged groups at these positions would sterically inhibit the ability of PB to bind and would decrease PB-mediated current. Although modification of α1K221C with MTSEA-biotin caused a decrease in PB-mediated current, modification with MTSET and MTSES had no effect on PB-mediated currents, again suggesting that this residue is not part of the PB binding site. Third, PB caused an increase in the rate of MTS modification of β2K215C. If β2K215C were part of the PB binding site, one would expect that PB would decrease the rate of modification. Moreover, based on our homology model of the GABAAR and given the size of PB (∼7 Å), it seems unlikely that α1 and β2 pre-M1 region residues are forming part of the general anesthetic binding site. Residues in the α1 and β2 pre-M1 regions are separated by 20 Å or more from the residues that have been previously identified as forming the potential PB/general anesthetic binding site (Fig. 1) (8, 21–24). Mutations in M2 (Asn-265) and M3 (Met-286) in the β subunit eliminate PB activation of the receptor (8, 25) and the actions of the related general anesthetics etomidate and propofol (26–28). More recently, α1M236 (in M1) and βM286 (in M3) have been directly identified as being part of a general anesthetic binding site by photolabeling with an etomidate analog (6). Propofol blocks covalent modification of βM286C by sulfhydryl-specific reagents, indicating that this residue forms part of a general anesthetic binding site (29). Furthermore, a knock-in mouse for βN265M removes the immobilizing and hypnotic actions of PB as well as the actions of etomidate and propofol (30). Taken together, the data indicate that general anesthetics, including PB, likely share a similar binding site, which is located in a water-filled pocket ∼50 Å below the GABA binding site between M1, M2, and M3.
In the presence of GABA, the receptor undergoes transitions between an ensemble of open and desensitized states (31, 32). If PB were stabilizing similar states as GABA, one might expect similar changes in the rate of modification in the presence of PB and GABA. Because this was not the case, we infer that PB binding/gating induces structural rearrangements near the pre-M1 region that are structurally distinct from movements induced by GABA. Interestingly, a recent report using disulfide-trapping experiments demonstrated that GABA and PB induce a similar open state structure at the 6′-position in M2 (12). This is consistent with functional studies that have shown similar single-channel conductances (2–4) regardless of whether GABAAR channels are opened by GABA or by PB. We speculate that the unique movements induced by PB and GABA in the pre-M1 regions are the result of their binding to different sites and triggering different activation pathways that lead to their functional effects.
We envision that PB binding between transmembrane helices initiates a conformational change in the M2-M3 linker that propagates to the pre-M1 region via Loop 2 (Fig. 1). Mutational studies have implicated the α1 and β2 M2-M3 linker as involved in PB activation (21, 33). The movements in the pre-M1 region triggered by PB are then likely to be transmitted to various regions of the GABAAR extracellular ligand binding domain as well as channel membrane domain. The pre-M1 region may be the conduit by which the actions of PB are propagated to the GABA binding site. Binding studies have shown that PB enhances GABA apparent affinity (34), suggesting that the structure of the GABA binding site changes in the presence of PB. Moreover, we have identified 13 positions in the GABA binding site interface that change accessibility during pentobarbital binding/gating (β2T160C, β2D163C, β2G203C, β2S204C, β2R207C, β2S209C, β2D62C, α1S68C, α1E122C, α1R131C, α1V180C, α1A181C, and α1R186C) (17, 35–38), indicating that the extracellular domain undergoes conformational rearrangements during PB binding/gating.
In summary, we have shown that the α1 and β2 pre-M1 regions of the GABAARs are structural elements involved in both GABA- and PB-mediated channel activation. PB binding, however, induces different structural movements in this region than when the receptor binds GABA, suggesting that PB stabilizes a different state or ensembles of states than GABA. These differences reveal distinct molecular mechanisms of action of these two ligands.
Acknowledgments
We thank Dr. Ken Satyshur for assistance in construction of the structural model.
This work was supported, in whole or in part, by National Institutes of Health Grant NS34727 from NINDS (to C. C.). This work was also supported by the Diversity Program in Neuroscience of the American Psychological Association (to J. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: LGIC, ligand-gated ion channel; GABAAR, γ-aminobutyric acid type A receptor; PB, pentobarbital; MTS, methanethiosulfonate; MTS, MTS-ethyltrimethylammonium; MTSES, MTS-ethylsulfonate.
References
- 1.Belelli, D., Pistis, M., Peters, J. A., and Lambert, J. J. (1999) Trends Pharmacol. Sci. 20 496-502 [DOI] [PubMed] [Google Scholar]
- 2.Mathers, D. A., and Barker, J. L. (1980) Science 209 507-509 [DOI] [PubMed] [Google Scholar]
- 3.Jackson, M. B., Lecar, H., Mathers, D. A., and Barker, J. L. (1982) J. Neurosci. 2 889-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald, R. L., Rogers, C. J., and Twyman, R. E. (1989) J. Physiol. 417 483-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rho, J. M., Donevan, S. D., and Rogawski, M. A. (1996) J. Physiol. 497 Pt. 2, 509-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, G. D., Chiara, D. C., Sawyer, G. W., Husain, S. S., Olsen, R. W., and Cohen, J. B. (2006) J. Neurosci. 26 11599-11605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pistis, M., Belelli, D., McGurk, K., Peters, J. A., and Lambert, J. J. (1999) J. Physiol. 515 Pt. 1, 3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amin, J. (1999) Mol. Pharmacol. 55 411-423 [PubMed] [Google Scholar]
- 9.Grosman, C., Zhou, M., and Auerbach, A. (2000) Nature 403 773-776 [DOI] [PubMed] [Google Scholar]
- 10.Lee, W. Y., and Sine, S. M. (2005) Nature 438 243-247 [DOI] [PubMed] [Google Scholar]
- 11.Chakrapani, S., Bailey, T. D., and Auerbach, A. (2004) J. Gen. Physiol. 123 341-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen, A., Bali, M., Horenstein, J., and Akabas, M. H. (2007) Biophys. J. 92 3130-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercado, J., and Czajkowski, C. (2006) J. Neurosci. 26 2031-2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boileau, A. J., Kucken, A. M., Evers, A. R., and Czajkowski, C. (1998) Mol. Pharmacol. 53 295-303 [DOI] [PubMed] [Google Scholar]
- 15.Liman, E. R., Tytgat, J., and Hess, P. (1992) Neuron 9 861-871 [DOI] [PubMed] [Google Scholar]
- 16.Robertson, G. A., Warmke, J. M., and Ganetzky, B. (1996) Neuropharmacology 35 841-850 [DOI] [PubMed] [Google Scholar]
- 17.Holden, J. H., and Czajkowski, C. (2002) J. Biol. Chem. 277 18785-18792 [DOI] [PubMed] [Google Scholar]
- 18.Pascual, J. M., and Karlin, A. (1998) J. Gen. Physiol. 111 717-739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monod, J., Wyman, J., and Changeux, J. P. (1965) J. Mol. Biol. 12 88-118 [DOI] [PubMed] [Google Scholar]
- 20.Amin, J., and Weiss, D. S. (1993) Nature 366 565-569 [DOI] [PubMed] [Google Scholar]
- 21.Serafini, R., Bracamontes, J., and Steinbach, J. H. (2000) J. Physiol. 524 Pt. 3, 649-676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mihic, S. J., Ye, Q., Wick, M. J., Koltchine, V. V., Krasowski, M. D., Finn, S. E., Mascia, M. P., Valenzuela, C. F., Hanson, K. K., Greenblatt, E. P., Harris, R. A., and Harrison, N. L. (1997) Nature 389 385-389 [DOI] [PubMed] [Google Scholar]
- 23.Mihic, S. J., and Harris, R. A. (1996) J. Pharmacol. Exp. Ther. 277 411-416 [PubMed] [Google Scholar]
- 24.Harrison, N. L., Kugler, J. L., Jones, M. V., Greenblatt, E. P., and Pritchett, D. B. (1993) Mol. Pharmacol. 44 628-632 [PubMed] [Google Scholar]
- 25.Pistis, M., Belelli, D., McGurk, K., Peters, J. A., and Lambert, J. J. (1999) J. Physiol. 515 3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belelli, D., Lambert, J. J., Peters, J. A., Wafford, K., and Whiting, P. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 11031-11036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegwart, R., Jurd, R., and Rudolph, U. (2002) J. Neurochem. 80 140-148 [DOI] [PubMed] [Google Scholar]
- 28.Siegwart, R., Krahenbuhl, K., Lambert, S., and Rudolph, U. (2003) BMC Pharmacol. 3 13, 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bali, M., and Akabas, M. H. (2004) Mol. Pharmacol. 65 68-76 [DOI] [PubMed] [Google Scholar]
- 30.Zeller, A., Arras, M., Jurd, R., and Rudolph, U. (2007) BMC Pharmacol. 7 2, 1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macdonald, R. L., and Twyman, R. E. (1992) in Ion Channels (Narahashi, T., ed), pp. 315-343, Plenum Press, New York [DOI] [PubMed]
- 32.Lema, G. M., and Auerbach, A. (2006) J. Physiol. 572 183-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigel, E., Buhr, A., and Baur, R. (1999) J. Neurochem. 73 1758-1764 [DOI] [PubMed] [Google Scholar]
- 34.Olsen, R. W. (1982) Annu. Rev. Pharmacol. Toxicol. 22 245-277 [DOI] [PubMed] [Google Scholar]
- 35.Wagner, D. A., and Czajkowski, C. (2001) J. Neurosci. 21 67-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newell, J. G., and Czajkowski, C. (2003) J. Biol. Chem. 278 13166-13172 [DOI] [PubMed] [Google Scholar]
- 37.Newell, J. G., McDevitt, R. A., and Czajkowski, C. (2004) J. Neurosci. 24 11226-11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloda, J. H., and Czajkowski, C. (2007) Mol. Pharmacol. 71 483-493 [DOI] [PubMed] [Google Scholar]