Abstract
The E2F1 transcription factor activates S-phase-promoting genes, mediates apoptosis, and stimulates DNA repair through incompletely understood mechanisms. XRCC1 (x-ray repair cross-complementing group 1) protein is important for efficient single strand break/base excision repair. Although both damage and proliferative signals increase XRCC1 levels, the mechanisms regulating XRCC1 transcription remain unclear. To study these upstream mechanisms, the XRCC1 promoter was cloned into a luciferase reporter. Ectopic expression of wild-type E2F1, but not an inactive mutant E2F1(132E), activated the XRCC1 promoter-luciferase reporter, and deletion of predicted E2F1 binding sites in the promoter attenuated E2F1-induced activation. Endogenous XRCC1 expression increased in cells conditionally expressing wild-type, but not mutant E2F1, and methyl methanesulfonate-induced DNA damage stimulated XRCC1 expression in E2F1+/+ but not E2F1-/- mouse embryo fibroblasts (MEFs). Additionally, E2F1-/- MEFs displayed attenuated DNA repair after methyl methanesulfonate-induced damage compared with E2F1+/+ MEFs. Moreover, Chinese hamster ovary cells with mutant XRCC1 (EM9) were more sensitive to E2F1-induced apoptosis compared with Chinese hamster ovary cells with wild-type XRCC1 (AA8). These results provide new mechanistic insight into the role of the E2F pathway in maintaining genomic stability.
The E2F family of transcription factors plays an important role in promoting both cellular proliferation and cell death. The “activating” E2Fs (E2F1 to 3) are important for regulating S-phase specific genes as well as promoting apoptosis, particularly in the case of E2F1 (1-8). Mounting evidence implicates E2F1 as an important component of the DNA damage response as E2F1 protein is phosphorylated and stabilized via ATM/ATR and Chk2-dependent pathways (9-12). Additionally, genes that are important in DNA repair, such as homologous recombination and mismatch repair, are bona fide E2F targets (13-17). Many repair genes have also been identified as putative E2F targets during genome-wide screens (15, 18-21). Moreover, it has also been suggested that E2F1 protein may be a component of repair complexes (22, 23). Importantly, E2F1 has a functional in vivo role in promoting DNA repair and suppressing apoptosis after UVB (24). Together, these studies implicate E2F1 in DNA repair, although the actual mechanisms and specific repair pathways remain obscure.
DNA SSBs4 are one of the most common DNA lesions and pose a major threat to genetic stability and survival through accumulation of mutations or through conversion to double-stranded breaks (25). SSBs can arise by direct damage to DNA bases or sugar moieties or indirectly as intermediates during the process of BER (26). SSBR/BER requires highly coordinated overlapping enzymatic steps dependent on the nature and origin of the SSB lesion (25-27).
XRCC1 is a scaffolding protein that promotes efficient repair by interacting with many of the proteins that catalyze SSBR/BER (26, 28-30). Although XRCC1 null mice are not viable (31), mouse and Chinese hamster ovary cells with mutant or attenuated XRCC1 demonstrate decreased SSBR and hypersensitivity to many different types of DNA damage (26, 32-37). In human cells, XRCC1 is necessary for efficient SSBR, genomic stability, and survival (38-40). Furthermore, XRCC1 polymorphisms are associated with variable cancer risk, which suggests a possible role in cancer development (41). XRCC1-mediated repair includes an S-phase SSBR pathway that operates at replication forks and a rapid cell cycle independent SSBR/BER pathway (42-45). Posttranslational mechanisms, such as phosphorylation, also modulate XRCC1 function and promote genetic stability (46, 47). Additional mechanisms may also regulate XRCC1, since mRNA and protein levels increase after DNA damage in part through a mitogen-activated protein kinase signaling pathway (48, 49). However, although both cell cycle-specific and DNA damage signals are upstream of XRCC1, the mechanisms regulating XRCC1 expression remain incompletely characterized.
In this report, we demonstrate that XRCC1 is a direct E2F target gene and that E2F1 enhances SSBR/BER. These results provide new mechanistic insight into the role of E2F1 in the maintenance of genomic stability and cell survival.
EXPERIMENTAL PROCEDURES
XRCC1 Promoter and Plasmids—The 5′-region from -881 to +158, relative to the transcription start site (GI:21624595) (50), was cloned by PCR of genomic DNA (forward primer, 5′-ggacgcagaacccttctcttttgg-3′; reverse primer, 5′-accgagtcctggctgctgcaggac-3′). The PCR product was cloned into the pGL3-Basic luciferase vector (Promega) using standard techniques. Deletions of the XRCC1 promoter region were similarly engineered. All constructs were sequence-verified. The E2F expression constructs, DP1 and pRSV β-galactosidase expression vectors were gifts from Drs. William Kaelin (Dana Farber Cancer Institute, Boston, MA), W. C. Lin (University of Alabama, Birmingham, AL), and Rosalie Sears (Oregon Health and Science University, Portland, OR).
Luciferase Reporter Assays—Cells at 60% confluence in 6-well plates were transfected with the specified XRCC1 luciferase reporter plasmid, together with indicated amounts of specified E2F expression vector, pRSV β-galactosidase expression vector (0.1 μg), and an appropriate amount of empty plasmid vector, for a total of 1.0 μg of plasmid DNA per transfection. Luciferase assays were performed 36 h post-transfection with an AutoLumatB95 luminometer and relative luciferase light units were normalized to β-galactosidase activity as described previously (51).
Western Blotting—Total cellular lysates were prepared, quantitated, resolved on 10% SDS-PAGE, immunoblotted with the specified primary and horseradish peroxidase-conjugated secondary antibodies, and visualized with chemiluminescence (Pierce SuperSignal) as previously described (51). Anti-XRCC1 mouse monoclonal antibody was from NeoMarkers, anti-E2F1 antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), anti-tubulin antibody was from Sigma, and horseradish peroxidase-conjugated secondary antibodies were from Jackson Immunological.
Northern Blotting—Total RNA was prepared using the Total RNA preparation kit (Promega, Madison, MI) according to the manufacturer's instructions. Twenty μg total RNA was resolved by electrophoresis on a 1.0% formaldehyde-agarose gel, transferred, and UV-cross-linked to a Zeta-Probe®GT membrane (Bio-Rad), hybridized to a randomly primed 32P-labeled XRCC1 cDNA probe, and visualized with autoradiography using standard techniques.
Reverse Transcription-PCR—XRCC1 and glyceraldehyde-3-phosphate dehydrogenase mRNA levels were determined by semiquantitative reverse transcription-PCR using standard techniques. Briefly, equivalent amounts of total RNA were prepared from the indicated cells at the specified conditions described, and oligo(dT)-primed first strand cDNA synthesis was performed with Superscript III reverse transcriptase (Invitrogen). PCR was performed on equivalent amounts of reverse transcription product with TaqDNA polymerase for 30 cycles (95 °C for 15 s, 65 °C for 20 s, 72 °C for 40 s) (XRCC1 forward primer, 5′-gtatgcaggctccacggatgagaa-3′; XRCC1 reverse primer, 5′-aggcttgcggcaccaccccatagagc-3′; glyceraldehyde-3-phosphate dehydrogenase forward primer, 5′-acggccgcatcttcttgtgc-3′; glyceraldehyde-3-phosphate dehydrogenase reverse primer, 5′-gtgcaggatgcattgctgac-3′).
Chromatin Immunoprecipitation—Chromatin immunoprecipitation analysis on Saos2 cells was performed as described (51) with minor modifications. Briefly, cells were cross-linked with 1% formaldehyde/phosphate-buffered saline, stopped with 0.125 m glycine, lysed (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris, pH 8.0, 5 mm EDTA plus fresh protease inhibitors), and sonicated to yield 600-1000 nucleotide chromatin fragments. Extracts were centrifuged at 13,000 × g for 10 min and precleared with protein A-Sepharose/salmon sperm DNA slurry (1:1) for 30 min at 4 °C. 2 μg of anti-E2F1 antibody (Santa Cruz Biotechnology) or control antibody (normal rabbit IgG) added and incubated overnight at 4 °C, followed by protein A/G-agarose for 2 h. Beads were washed twice with ice-cold radioimmune precipitation buffer, four times with ice-cold IP wash buffer (100 mm Tris, pH 8.0, 500 mm LiCl, 1% Nonidet P-40, 1% deoxycholic acid), and twice more with ice-cold radioimmune precipitation buffer. IP elution buffer (50 mm NaHCO3, 1% SDS freshly made) was added for 15 min at room temperature, beads were spun out, and supernatant was collected. This was repeated, and supernatants were combined. Cross-links were reversed, and then DNA was purified by phenol/chloroform extraction and ethanol precipitation. PCR of XRCC1 promoter sequence was performed with 25 cycles (94 °C for 15 s, 65 °C for 45 s) and analyzed on a 2% TAE-agarose gel. XRCC1-specific primers were as follows: forward, 5′-ggacgcagaacccttctcttttgg-3′; reverse, 5′-ggctcaggcggctgcactcttctc-3′. XRCC1 control primers were 5′-ctggggagtaggacgtcagtgctg-3′ (forward) and 5′-ggcttgcggcaccaccccatagagc-3′ (reverse).
Cell Culture—Cells were grown in Dulbecco's modified Eagle's medium or McCoy's medium supplemented with 10% heat-treated fetal bovine serum and 290 μg of l-glutamine, 100 units of penicillin, and 100 μg of streptomycin per ml at 37 °C in 5% CO2. Saos2 cells stably transfected with various tetracycline-regulated E2F1 expression vectors (a gift from Dr. Karen Vousden, Beatson Institute, UK) were maintained in tetracycline-free fetal bovine serum (Clontech). E2F1+/+ and E2F1-/- mouse embryonic fibroblasts (MEFs) were gifts from Dr. Joseph Nevins (Duke University, Durham, NC). Chinese hamster ovary cell lines AA8 and EM9 were obtained from the American Type Culture Collection.
Comet Assay—Single cell agarose alkaline gel electrophoresis was performed per standard methods with minor modifications (52). Equivalent numbers of cells (1 × 105) were collected at the indicated time points, mixed with low melt temperature agarose (Sigma), and then layered onto agarose-coated glass slides. Slides were maintained at 4 °C to solidify and for all subsequent steps. Slides were submerged in lysis buffer (2.5 m NaCl, 0.1 m EDTA, 10 mm Tris-Cl (pH 7.0), 1% Triton X-100, 1% DMSO) for 1 h, washed with nanopure H2O, and incubated for 45 min in alkaline electrophoresis buffer (50 mm NaOH, 1 mm EDTA, 1% DMSO, pH 12.8). After electrophoresis (25 min, 25 V), air-dried and neutralized slides were stained with 2 μg/ml propidium iodide. Average comet tail moment was scored for duplicate slides (50 cells/slide) in randomly selected fields, and automated calculations were performed using Comet Assay II software (Perceptive Instruments, Suffolk, UK).
In Situ DNA Strand Break Detection—A DNA polymerase I-mediated labeling assay was performed as described previously with minor modifications (53). Briefly, cells grown on coverslips were fixed in 10% formalin for 10 min. Cells were rinsed in phosphate-buffered saline, permeabilized with 1% Triton X-100 for 20 min, and then incubated in a moist air chamber at 37 °C for 90 min in a labeling mixture containing 10 μm each dGTP, dATP, and dCTP and 7 μm dTTP, 3 μm FITC-dUTP, 20 units/ml Escherichia coli DNA polymerase I (Sigma) in reaction buffer containing 5 mm MgCl2, 10 mm 2-mercaptoethanol, and 20 μg/ml bovine serum albumin. The reaction was terminated by washing the slides twice in phosphate-buffered saline. Nonspecific labeling was determined by incubation in the reaction buffer without the enzyme.
Apoptosis Assay—Equivalent numbers of indicated cells were plated and allowed to settle for 24 h at 50% confluence. Adenovirus infections were performed at a multiplicity of infection of 1 × 1010 (Stratagene AdEasy system). E2F1-expressing and green fluorescent protein (GFP)-expressing adenoviruses were a gift from Dr. Rosalie Sears (Oregon Health and Science University). Forty-eight h after infection, cells were harvested and subjected to Annexin V and flow cytometry as previously described (54). Briefly, attached and floating cells were collected and resuspended in Annexin V binding buffer (BD Biosciences) at a concentration of 1 × 106 cells/ml. Allophycocyanin-conjugated Annexin V was immediately added, incubated for 15 min at room temperature, and then analyzed on a BD Biosciences FACScan. The percentage of apoptotic cells was determined by the increase in Annexin V-allophycocyanin signal on stained cells over the base-line signal (defined on unstained cells) per standard flow cytometry convention.
RESULTS
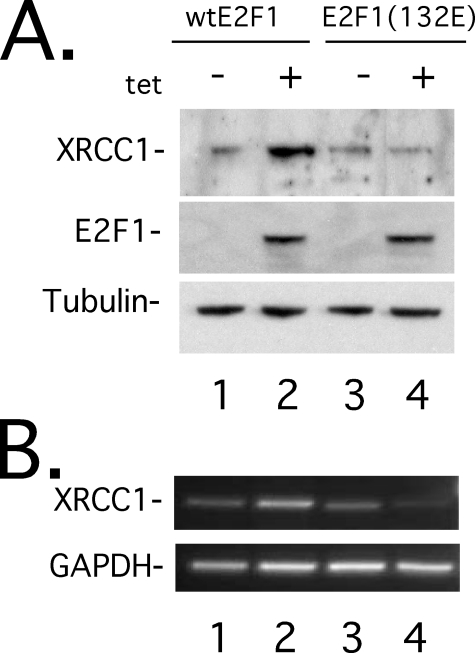
E2F1 Expression Stimulates Endogenous XRCC1 Expression—Despite the important role that XRCC1 plays in SSBR/BER (26), little is known about the upstream pathways that regulate its expression. Because DNA damage signals and proliferative signals stimulate XRCC1 expression (26, 48, 49), we wondered if E2F1 could stimulate endogenous XRCC1 expression (Fig. 1). To test this, we utilized a Saos2 stable cell line containing an E2F1 cDNA expression vector under control of a tetracycline-regulated promoter (55). We found that tetracycline-induced E2F1 expression stimulated an increase in XRCC1 protein (Fig. 1A, lanes 1 and 2) and mRNA levels (Fig. 1B, lanes 1 and 2). To demonstrate that E2F1 transcription function was required to induce endogenous XRCC1 expression, we additionally utilized a tetracycline-regulated Saos2 stable cell line containing a DNA binding-incompetent mutant E2F1(132E) (55). Conditional expression of mutant E2F1(132E) could not stimulate endogenous XRCC1 protein (Fig. 1A, lanes 3 and 4) or mRNA (Fig. 1B, lanes 3 and 4). These results suggest that E2F1 stimulates endogenous XRCC1 expression by a transcription-mediated mechanism.
FIGURE 1.
E2F1 expression stimulates endogenous XRCC1. Shown are Western blots (A) and ethidium bromide-stained agarose gel (B) of semiquantitative reverse transcription-PCR, on Saos2 cell lines conditionally expressing wild-type E2F1 (lanes 1 and 2) or DNA binding-incompetent mutant E2F1(132E) (lanes 3 and 4). Prior to induction, cell lines were maintained in 0.5% fetal bovine serum-containing medium for 30 h, induced with 2.0 μg/ml doxycycline, and then harvested 24 h later.
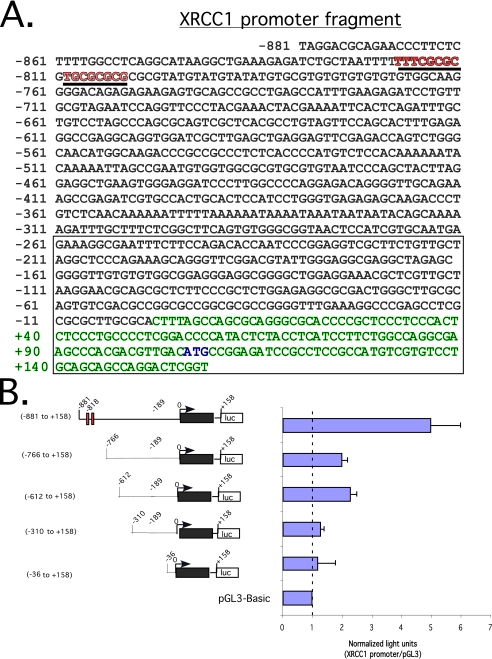
XRCC1 Promoter Has Features Suggestive of an E2F Target Gene—Since transcription-competent wild-type E2F1 was capable of stimulating XRCC1 mRNA and protein levels (Fig. 1), we examined the genomic sequence encompassing the XRCC1 promoter (Fig. 2A). Sequence analysis from -881 to +158 relative to the putative transcription start site (green sequence) (50) revealed a CpG island (boxed sequence) as well as putative E2F binding sites from -819 to -803 (red sequence). To verify that an active promoter was present in this region, we cloned this genomic fragment into the pGL3-Basic luciferase reporter vector. After transfection into Saos2 cells, we found that this genomic region stimulated the luciferase reporter nearly 6-fold relative to empty vector (Fig. 2B). Sequential deletions from the 5′-end attenuated luciferase activation relative to empty vector. This finding suggests that a functional XRCC1 promoter is contained within this region, although the possibility of other control elements outside of this region cannot be excluded.
FIGURE 2.
XRCC1 promoter fragment sequence and luciferase reporter assay define an active promoter region. A, genomic sequence from -881 to +158 relative to the predicted transcription start site/cDNA 5′-end indicated as +1. Exon 1 is indicated in green type, and the translation-initiation codon is shown in blue type. The open box indicates a CpG island. Underlined sequences in red denote putative E2F-binding sites. B (left), schematic representation of a series of promoter deletion mutants in pGL3-Basic-luciferase reporter. The red boxes denote putative E2F-binding sites. The black box represents exon 1. An open box with [luc] denotes luciferase reporter. Right, luciferase readout of the indicated promoter-reporters after transfection into Saos2 cells. Light units normalized to a β-galactosidase signal. In all, 1.0 μg of the indicated promoter-reporter and pRSV β-galactosidase plasmids was used for all transfections. S.D. of triplicate experiments is shown.
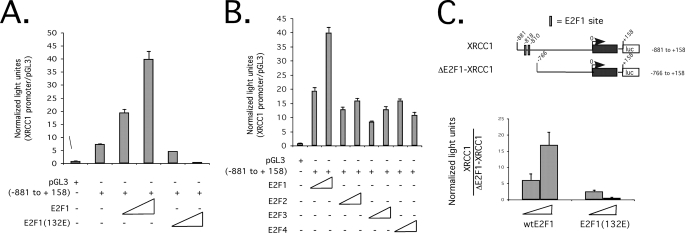
The XRCC1 Promoter-Luciferase Reporter Is Stimulated by E2F1—Since the XRCC1 promoter sequence was typical for an E2F target gene (Fig. 2), we reasoned that the XRCC1 promoter-luciferase reporter would be stimulated by E2F. Co-transfection of increasing amounts of E2F1 expression vector with the XRCC1 promoter-luciferase reporter caused a dose-dependent increase in luciferase activation (Fig. 3A). In contrast, DNA binding-incompetent E2F1(132E) could not activate the XRCC1 promoter-luciferase reporter (Fig. 3A). We also found that E2F1 more efficiently stimulated the XRCC1 promoter-reporter relative to other family members (E2F2, E2F3, and E2F4) (Fig. 3B). To determine the contribution of the predicted E2F binding sites for their ability to influence XRCC1 promoter activation, we co-transfected E2F1 expression vector with an XRCC1 promoter-luciferase reporter, which lacked the E2F binding sites (ΔE2F1-XRCC1), as shown in Fig. 3C. We found that in response to E2F1 co-transfection, stimulation of the XRCC1 promoter-luciferase reporter containing the E2F binding sites was over 15-fold higher than the ΔE2F1-XRCC1 promoter-reporter (Fig. 3C, left). As a further control for specificity, mutant E2F1(132E) did not stimulate either promoter (Fig. 3C, right). Together, these results suggest that the XRCC1 promoter is stimulated by E2F.
FIGURE 3.
E2F stimulates the XRCC1 promoter-luciferase reporter. A, relative light units, normalized to a β-galactosidase signal, of pGL3-XRCC1-(-881 to +158) (0.1 μg) co-transfected with increasing amounts of wild-type E2F1 or mutant E2F1(132E) expression plasmids (50-250 ng) into Saos2 cells. B, relative light units, normalized to a β-galactosidase signal, of pGL3-XRCC1-(-881 to +158) (0.1 μg) co-transfected with increasing amounts of the indicated E2F expression vectors (50-250 ng) into Saos2 cells. C, relative light units, normalized to a β-galactosidase signal, of the relative -fold change of the XRCC1-(-881 to +158) promoter-reporter versus an E2F binding site-deleted XRCC1 promoter-reporter (ΔE2F1-XRCC1), in response to increasing amounts of co-transfected wild-type E2F1 or mutant E2F1(132E) expression plasmids (50-250 ng). S.D. of triplicate experiments is shown.
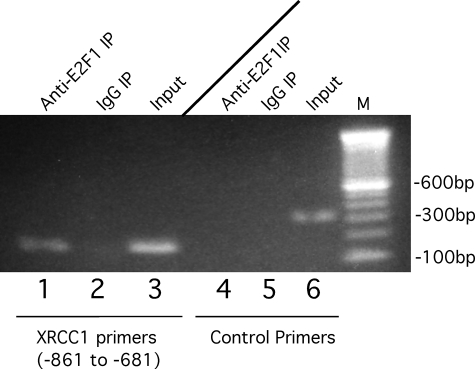
Endogenous XRCC1 Promoter Binds Endogenous E2F1—Since XRCC1 transcription is stimulated by E2F overexpression (Figs. 1 and 3), we wanted to demonstrate that endogenous E2F1 could bind the endogenous XRCC1 promoter in its native chromatin context. To examine this, we performed a chromatin immunoprecipitation assay on the proximal XRCC1 promoter region (Fig. 4). XRCC1 promoter-specific PCR primers amplified this region after chromatin IP with an anti-E2F1-specific antibody but not nonimmune IgG (Fig. 4, lanes 1-3). To further demonstrate that this was a specific interaction, XRCC1-specific primers that amplify a region that does not contain E2F-binding sites (control primers) could not generate a PCR product after chromatin IP with anti-E2F-1-specific antibody (Fig. 4, lanes 4-6). These findings demonstrate that the XRCC1 promoter, in its native chromatin context, can specifically bind E2F1 protein and that XRCC1 is a bona fide E2F target gene.
FIGURE 4.
Chromatin immunoprecipitation of E2F1 at the XRCC1 promoter. Shown is an ethidium bromide-stained agarose gel of PCR products performed after chromatin immunoprecipitation using anti-E2F1 antibody (lanes 1 and 4) or nonimmune control IgG (lanes 2 and 5). Shown is input chromatin (lanes 3 and 6) at a 1:1000 dilution of PCR products from IP reactions performed with XRCC1-specific primers (lanes 1-3) or control primers (lanes 4-6).
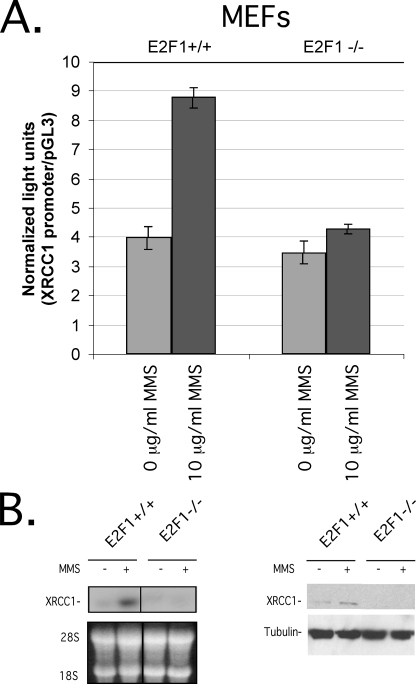
Endogenous E2F1 Stimulates the XRCC1 Promoter after DNA Damage—Since XRCC1 is an E2F target gene (Figs. 1, 2, 3 and 4) and E2F1 is a DNA damage-inducible protein (9-12), we wondered whether MMS, which causes heat-labile DNA damage repaired by an XRCC1-mediated BER pathway (56), could stimulate XRCC1 transcription in an E2F1-dependent manner (Fig. 5). To investigate this, we transfected E2F1-/- and E2F1+/+ MEFs with the XRCC1 promoter-luciferase reporter and then treated them with MMS. Compared with no treatment, we found that the XRCC1 promoter-luciferase reporter was stimulated by MMS in E2F1+/+ but not E2F1-/- cells (Fig. 5A). Consistent with transcriptional activation, an increase in XRCC1 mRNA and protein levels (Fig. 5B, left and right, respectively) was observed after MMS treatment in E2F1+/+ but not E2F1-/- cells. These results demonstrate that MMS-induced DNA damage stimulates XRCC1 transcription at least in part through an E2F1-dependent pathway.
FIGURE 5.
Endogenous E2F1 stimulates the XRCC1 promoter after DNA damage. A, relative light units, normalized to a β-galactosidase signal, of pGL3-XRCC1-(-881 to +158) (0.1 μg) luciferase reporter plasmid transfected into E2F1+/+ or E2F1-/- MEFs, followed by incubation with 10 μg/ml MMS (columns 2 and 4) or 0 μg/ml MMS (columns 1 and 3) for 12 h prior to assay. S.D. of triplicate experiments is shown. B, Northern blot (left) and Western blot (right) on equivalent amounts of lysates prepared from E2F1+/+ or E2F1-/- MEFs after incubation with 10 μg/ml MMS (lanes 2 and 4) or 0 μg/ml MMS (lanes 1 and 3).
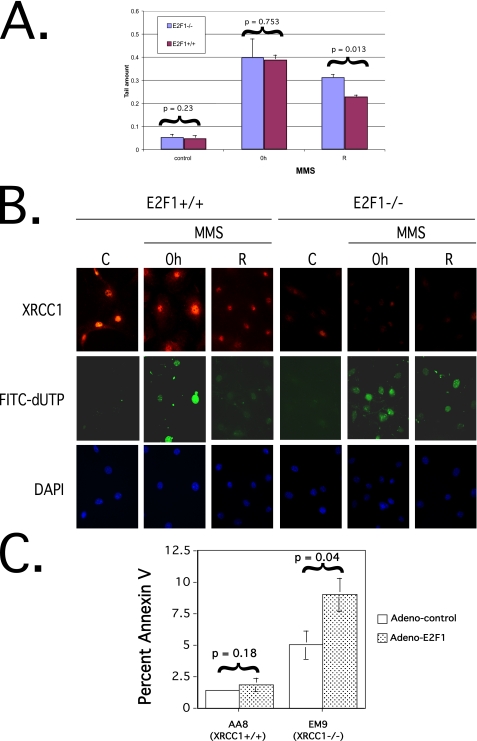
E2F1 Enhances DNA Repair after MMS-induced Damage—Since XRCC1 is a bona fide E2F1 target gene (Figs. 1, 2, 3, 4 and 5), we wished to determine if loss of E2F1 would result in attenuated DNA repair after MMS-induced DNA damage (Fig. 6). To investigate this, we treated E2F1-/- and E2F1+/+ MEFs with MMS and then measured strand break repair by single cell alkaline agarose gel electrophoresis (comet assay) (52) as shown in Fig. 6A. The average tail moment was similar in undamaged cells (left-hand bars, control). As expected after incubation in MMS, the average tail moment increased equivalently in both E2F1-/- and E2F1+/+ MEFs (middle bars, 0 h). However, if cells were allowed to recover in MMS-free medium (right-hand bars, R), E2F1-/- MEFs had a statistically significant longer average tail moment compared with E2F1+/+ MEFs (33% versus 22%, respectively; p = 0.013, unpaired two-tailed t test). To further confirm that E2F1-/- MEFs had decreased DNA repair capacity, we utilized an in situ DNA polymerase I-mediated FITC-dUTP labeling assay (53) to detect persistent DNA strand breaks after recovery from MMS treatment (Fig. 6B). As expected, immunofluorescence staining demonstrated reduced XRCC1 staining in E2F1-/- MEFs compared with E2F1+/+ MEFs (top panels). Without damage, no DNA polymerase I-mediated FITC-dUTP labeling was detected (middle panels, C). After MMS exposure, FITC-dUTP labeling was detected in both E2F1+/+ and E2F1-/- MEFs (middle panels, 0 h). However, if MMS exposure was followed by recovery with incubation in MMS-free medium, less FITC-dUTP labeling was detectable in E2F1+/+ MEFs compared with E2F1-/- MEFs (middle panels, R).
FIGURE 6.
Loss of E2F1 attenuates DNA repair. A, comet assay expressed as average tail moment on E2F1+/+ and E2F1-/- MEFs untreated (left-hand graph, control), after incubation for 1 h in medium containing 50 μg/ml MMS (middle graph, 0 h), and after incubation for 1 h in medium containing 50 μg/ml MMS followed by recovery with incubation in fresh drug-free medium for 2 h (right-hand graph, R). Experiments were performed in duplicate, and measurement of mean tail moment was from 50 cells/slide from 15-20 randomly selected fields representing the whole area of each slide. Statistical analysis was performed using unpaired two-tailed t test on comet tail moments that were determined using Comet Assay II software (Perceptive Instruments; Suffolk, UK). Error bars, S.D. from separate experiments. B, indirect immunofluorescence microscopy on XRCC1 immunostaining (top), DNA polymerase I-mediated FITC-dUTP labeling assay (middle), and 4′,6-diamidino-2-phenylindole (DAPI) staining (bottom) performed on E2F1-/- and E2F1+/+ MEFs untreated (C), after incubation for 1 h in medium containing 50 μg/ml MMS (0 h panels), and after incubation for 1 h in medium containing 50 μg/ml MMS followed by recovery with incubation in fresh drug-free medium for 6 h (R panels). C, percentage of apoptosis as determined by Annexin V staining and flow cytometry 48 h after infection of the indicated cells with adenovirus expressing wild-type E2F1 (Adeno-E2F1) or adenovirus expressing GFP (Adeno-control). Error bars, S.D. of triplicate experiments and statistical analysis performed using Student's unpaired two-tailed t test.
If XRCC1 was downstream of E2F1 and could mediate E2F1 function, we reasoned that loss of XRCC1 would result in increased E2F1-induced apoptosis due to diminished DNA repair. To examine this, we utilized Chinese hamster ovary cells containing wild-type XRCC1 (AA8 cell line) and a cell line derived from AA8 cells that is mutant for XRCC1 (EM9 cell line) (26, 32). We infected cells with an adenovirus expressing wild-type E2F1 or a control adenovirus expressing GFP and quantified the level of E2F1-induced apoptosis at 48 h using Annexin V staining and flow cytometry (Fig. 6C). Under these conditions, AA8 cells did not have a significant increase in apoptosis after infection with E2F1-expressing adenovirus compared with control GFP-expressing adenovirus (left-hand graph). In contrast, EM9 cells demonstrated a statistically significant 1.8-fold increase in apoptosis after infection with E2F1-expressing adenovirus compared with control GFP-expressing adenovirus (right-hand graph) (p = 0.04, unpaired two-tailed t test). Together, these results suggest a mechanism by which the E2F1-XRCC1 pathway can stimulate DNA repair and promote genomic stability and cell viability. However, the existence of other E2F1-mediated pathways that stimulate BER independent of XRCC1 remains likely.
DISCUSSION
The E2F1 pathway is centrally involved in the highly complex networks coupling cellular proliferation and apoptosis (1, 5, 7, 8, 12, 57). We have shown that XRCC1, which plays a critical role in SSBR/BER (26), is a direct E2F1 target gene. Because E2F1 is a damage-inducible protein (9-12), our data provide novel insight into an important DNA repair function of the E2F pathway and open new avenues for investigation.
Our finding that E2F1 is upstream of XRCC1 significantly expands on prior observations that E2F plays a role in other repair pathways, such as MMR and NER (13-17, 19-21, 24). Our data are consistent with the report that the BER protein uracil-DNA glycosylase is also E2F-regulated (58). Intriguingly, although E2F1 is best characterized as a transcription factor, E2F1 protein may have a direct role in DNA repair, as suggested by its localization to repair complexes (22, 23). Thus, it is likely that multiple E2F-regulated mechanisms function in parallel with XRCC1 to stimulate repair.
We found that enforced E2F expression stimulated XRCC1 levels (Figs. 1 and 3) and that MMS, which induces predominantly heat-labile DNA damage repaired by an XRCC1-mediated BER pathway (56), causes an E2F1-dependent increase in XRCC1 expression (Fig. 5). This is consistent with prior reports demonstrating that cellular stress (such as from γ-irradiation) increases endogenous XRCC1 levels (48, 49, 59), although this may be cell type-specific (60). How MMS-induced stress activates the E2F1-XRCC1 axis remains unknown. Cellular sensitivity to MMS may involve an ATR-dependent pathway, and genetic evidence suggests that MMS-induced damage activates the yeast Rad53 (Chk2 human homologue) pathway (61, 62). Given that the ATM/ATR and Chk2 pathways phosphorylate and activate E2F1 protein (9-12), it is possible that these kinases stimulate XRCC1 expression through E2F1 activation, although this remains to be demonstrated. Interestingly, Chk2-mediated stabilization of the FoxM1 transcription factor stimulates expression of DNA repair genes, including XRCC1 (63). Given that XRCC1 function is complex, it is likely that its control involves multiple levels. Indeed, posttranslational mechanisms modulate XRCC1 function, as evidenced by the ability of DNA-dependent protein kinase to phosphorylate XRCC1 (46) as well as the requirement of protein kinase CK2 to phosphorylate XRCC1 and enhance SSBR and genetic stability (47). Consistent with the complex control of XRCC1, we found that serum starvation followed by refeeding stimulated XRCC1 expression (data not shown). This is consistent with cell conditions of high E2F activity but also suggests that serum/mitogenic factors may be important too. This could be a cell type-specific phenomenon, since density arrest and release does not alter XRCC1 levels in human T24 cells (64). Nevertheless, our discovery of an E2F1-XRCC1 pathway provides a previously undescribed mechanism for regulating XRCC1 function.
The biological importance of E2F1 regulation of XRCC1 is suggested by the attenuated in vivo DNA repair in E2F1-/- versus E2F1+/+ MEFs (Fig. 6, A and B). Two different methods demonstrated reduced DNA repair after MMS-induced DNA damage, which correlates with the decreased XRCC1 levels observed in E2F1-/- cells. The repair of MMS-damaged DNA still occurs in E2F1-/- cells, suggesting that the E2F1-XRCC1 axis is not an absolute requirement in these systems. This is not surprising, given the complex and overlapping repair pathways involved. However, the significance of even a modestly reduced XRCC1-mediated repair function may have important implications for maintaining genomic stability and cell viability. Consistent with this notion of XRCC1 mediating E2F1 activity is the observation that loss of XRCC1 function resulted in an enhanced E2F1-induced apoptotic response in EM9 cells compared with AA8 cells (Fig. 6C).
Although E2F1 is a damage response protein, it also plays an important role in promoting the expression of a large number of genes required for replication and proliferation (15, 19-21). Given the intimate relationship between proliferation and replication/repair, the control of XRCC1 by E2F in undamaged cells further integrates SSBR with cell cycle progression as might be expected if enhanced SSBR were necessary to repair SSBs at replication forks (26, 42-45, 65). Whether and in what context the other E2F family members play a role, as well as what specific SSBR pathways are utilized (e.g. long patch BER), remains to be explored.
The p53-E2F network controls and integrates critical functions, such as proliferation, cell cycle checkpoints, apoptosis, and DNA repair (7, 8, 12, 66, 67). In particular, p53 can promote BER (68-72), and our discovery that E2F1 may also promote BER expands our understanding of the p53-E2F1 network in regulating DNA repair. Disruption of these cooperative pathways has profound implications for tumorigenesis, as evidenced by enhanced tumor formation in knock-out mouse models for both p53 and E2F1, although intriguingly, both oncogenic and tumor suppressor functions for E2F1 are suggested in compound p53-/-;E2F1-/- mice (12, 73-75). Our finding that E2F1 regulates XRCC1 implies that an intact E2F1 pathway may be important for maintaining genomic stability in response to highly dangerous SSBs. This observation opens new avenues for investigation that may ultimately allow us to develop novel preventive and therapeutic strategies for cancer patients.
This work was supported, in whole or in part, by National Institutes of Health, Grant CA104997 (to C. D. L.). This work was also supported by the Oregon Health and Science University Division of Hematology and Medical Oncology and the Oregon Health and Science University Cancer Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SSB, single strand break; SSBR, single strand break repair; BER, base excision repair; IP, immunoprecipitation; MEF, mouse embryo fibroblast; FITC, fluorescein isothiocyanate; MMS, methyl methanesulfonate; GFP, green fluorescent protein.
References
- 1.Sears, R., and Nevins, J. (2002) J. Biol. Chem. 277 11617-11620 [DOI] [PubMed] [Google Scholar]
- 2.Ginsberg, D. (2002) FEBS Lett. 529 122-125 [DOI] [PubMed] [Google Scholar]
- 3.DeGregori, J., Leone, G., Miron, A., Jakoi, L., and Nevins, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7245-7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leone, G., Sears, R., Huang, E., Rempel, R., Nuckolls, F., Park, C.-H., Giangrande, P., Wu, L., Saavedra, H., Field, S., Thompson, M., Yang, H., Fujiwara, Y., Greenberg, M., Orkin, S., Smith, C., and Nevins, J. (2001) Mol. Cell 8 105-113 [DOI] [PubMed] [Google Scholar]
- 5.Trimarchi, J., and Lees, J. (2002) Nat. Rev. Mol. Cell Biol. 3 11-20 [DOI] [PubMed] [Google Scholar]
- 6.Pediconi, N., Ianari, A., Costanzo, A., Belloni, L. R. G., Cimino, L., Porcellini, A., Screpanti, I., Balsano, C., Alesse, E., Gulino, A., and Levrero, M. (2003) Nat. Cell Biol. 5 552-558 [DOI] [PubMed] [Google Scholar]
- 7.Dimova, D., and Dyson, N. (2005) Oncogene 24 2810-2826 [DOI] [PubMed] [Google Scholar]
- 8.Bell, L., and Ryan, K. (2004) Cell Death Differ. 11 137-142 [DOI] [PubMed] [Google Scholar]
- 9.Lin, W.-C., Lin, F.-T., and Nevins, J. (2001) Genes Dev. 15 1833-1844 [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens, C., Smith, L., and La Thangue, N. (2003) Nat. Cell Biol. 5 401-409 [DOI] [PubMed] [Google Scholar]
- 11.Blattner, C., Sparks, A., and Lane, D. (1999) Mol. Cell. Biol. 19 3704-3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens, C., and La Thangue, N. (2004) DNA Repair 3 1071-1079 [DOI] [PubMed] [Google Scholar]
- 13.Iwanga, R., Komori, H., and Ohtani, K. (2004) Oncogene 23 8581-8590 [DOI] [PubMed] [Google Scholar]
- 14.Youn, C.-K., Cho, H.-J., Kim, S.-H., Kim, H.-B., Kim, M.-H., Chang, I.-Y., Lee, J.-S., Chung, M.-H., Hahm, K.-S., and You, H.-J. (2004) Nat. Cell Biol. 7 137-147 [DOI] [PubMed] [Google Scholar]
- 15.Polager, S., Kalma, Y., Berkovich, E., and Ginsberg, D. (2002) Oncogene 21 437-446 [DOI] [PubMed] [Google Scholar]
- 16.Bindra, R., Gibson, S., Meng, A., Westermark, U., Jasin, M., Pierce, A., Bristow, R., Classon, M., and Glazer, P. (2005) Cancer Res. 65 11597-11604 [DOI] [PubMed] [Google Scholar]
- 17.Prost, S., Lu, P., Caldwell, H., and Harrison, D. (2007) Oncogene 26 3572-3581 [DOI] [PubMed] [Google Scholar]
- 18.Bracken, A., Ciro, M., Cocito, A., and Helin, K. (2004) Trends Biochem. Sci. 29 409-417 [DOI] [PubMed] [Google Scholar]
- 19.Ishida, S., Huang, E., Zuzan, H., Spang, R., Leone, G., West, M., and Nevins, J. (2001) Mol. Cell. Biol. 21 4681-4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R., and Dynlacht, B. (2002) Genes Dev. 16 245-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinmann, A., Yan, P., Oberley, M., Huang, T., and Farnham, P. (2002) Genes Dev. 16 235-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maser, R., Mirzoeva, O., Wells, J., Olivares, H., Williams, B., Zinkel, R., Farnham, P., and Petrini, J. (2001) Mol. Cell. Biol. 21 6006-6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, K., Lin, F.-T., Ruppert, M., and Lin, W.-C. (2003) Mol. Cell. Biol. 23 3287-3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berton, T., Mitchell, D., Guo, R., and Johnson, D. (2005) Oncogene 24 2449-2460 [DOI] [PubMed] [Google Scholar]
- 25.Slupphaug, G., Kavli, B., and Kroken, H. (2003) Mutat. Res. 531 231-251 [DOI] [PubMed] [Google Scholar]
- 26.Caldecott, K. (2003) DNA Rep. 2 955-969 [DOI] [PubMed] [Google Scholar]
- 27.Campalans, A., Marsin, S., Nakabeppu, Y., O'Connor, T., Boiteux, S., and Radicella, J. (2005) DNA Rep. 12 826-835 [DOI] [PubMed] [Google Scholar]
- 28.Wiederhold, L., Leppard, J., Kedar, P., Karimi-Busheri, F., Rasouli-Nia, A., Weinfeld, M., Tomkinson, A., Izumi, T., Prasad, R., Wilson, S., Mitra, S., and Hazra, T. (2004) Mol. Cell 15 209-220 [DOI] [PubMed] [Google Scholar]
- 29.Marsin, S., Vidal, A., Sossou, M., Menissier-de Murcia, J., Le Page, F., Boiteux, S., de Murcia, G., and Radicella, J. (2003) J. Biol. Chem. 278 44068-44074 [DOI] [PubMed] [Google Scholar]
- 30.Vidal, A., Boiteux, S., Hickson, I., and Radicella, J. (2001) EMBO J. 20 6530-6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tebbs, R., Flannery, M., Meneses, J., Hartmann, A., Tucker, J., Thompson, L., Cleaver, J., and Pedersen, R. (1999) Dev. Biol. 208 513-529 [DOI] [PubMed] [Google Scholar]
- 32.Thompson, L., Brookman, K., Dillehay, L., Carrano, A., Mazrimas, J., Mooney, C., and Minkler, J. (1982) Mutat. Res. 95 427-440 [DOI] [PubMed] [Google Scholar]
- 33.Zdzienicka, M., Vanderschans, G., Natarajan, A., Thompson, L., Neuteboom, I., and Simons, J. (1992) Mutagenesis 7 265-269 [DOI] [PubMed] [Google Scholar]
- 34.Caldecott, K., Tucker, J., Stanker, L., and Thompson, L. (1995) Nucleic Acids Res. 23 4836-4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen, N., Zdzienicka, M., Mohrenweiser, H., Thompson, L., and Thelen, M. (1998) Nucleic Acids Res. 26 1032-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cantoni, O., Murray, D., and Meyn, R. (1987) Chem. Biol. Interact. 63 29-38 [DOI] [PubMed] [Google Scholar]
- 37.Churchill, M., Peak, J., and Peak, M. (1991) Photochem. Photobiol. 53 229-236 [DOI] [PubMed] [Google Scholar]
- 38.Brem, R., and Hall, J. (2005) Nucleic Acids Res. 33 2512-2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, H., Chan, D., Yang, T., Rodriguez, M., Chen, B., Leng, M., Mu, J.-J., Chen, D., Songyang, Z., Wang, Y., and Qin, J. (2004) Mol. Cell. Biol. 24 8356-8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitehouse, C., Taylor, R., Thistlethwaite, A., Zhang, H., Karim-Busheri, F., Lasko, D., Weinfeld, M., and Caldecott, K. (2001) Cell 104 107-117 [DOI] [PubMed] [Google Scholar]
- 41.Ladiges, W. (2006) Oncogene 25 1612-1619 [DOI] [PubMed] [Google Scholar]
- 42.Fan, J., Otterlei, M., Wong, H.-K., Tomkinson, A., and Wilson, D. (2004) Nucleic Acids Res. 32 2193-2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, D., Taylor, R., Clements, P., and Caldecott, K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13649-13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor, R., Thistlethwaite, A., and Caldecott, K. (2002) Mol. Cell. Biol. 22 2556-2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, R., Moore, D., Whitehouse, J., Johnson, P., and Caldecott, K. (2000) Mol. Cell. Biol. 20 735-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy, N., Martz, A., Bresson, A., Spenlehauer, C., de Murcia, G., and Menissier-de Murcia, J. (2006) Nucleic Acids Res. 34 32-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loizou, J., El-Khamisy, S., Ziatanou, A., Moore, D., Chan, D., Qin, J., Sarno, S., Meggio, F., Pinna, L., and Caldecott, K. (2004) Cell 117 17-28 [DOI] [PubMed] [Google Scholar]
- 48.Yacoub, A., Park, J., Qiao, L., Dent, P., and Hagan, H. (2001) Int. J. Radiat. Biol. 77 1067-1078 [DOI] [PubMed] [Google Scholar]
- 49.Yacoub, A., McKinstry, R., Hinman, D., Chung, T., Dent, P., and Hagan, H. (2003) Radiat. Res. 159 439-452 [DOI] [PubMed] [Google Scholar]
- 50.Thompson, L., Brookman, K., Jones, N., Allen, S., and Carrano, A. (1990) Mol. Cell. Biol. 10 6160-6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen, D., Padiernos, E., Ding, F., Lossos, I., and Lopez, C. D. (2005) Cell Death Differ. 12 358-368 [DOI] [PubMed] [Google Scholar]
- 52.Speit, G., and Hartmann, A. (1999) Methods Mol. Biol. 113 203-212 [DOI] [PubMed] [Google Scholar]
- 53.Chen, J., Jin, K., Chen, M., Pei, W., Kawaguchi, K., Greenberg, D., and Simons, R. (1999) J. Neurochem. 69 232-245 [DOI] [PubMed] [Google Scholar]
- 54.Zhu, S., Ramos, J., Kampa, K., Sirisawad, M., Yu, Z., Chen, D., Naumovski, L., and Lopez, C. D. (2005) J. Biol. Chem. 280 34473-34480 [DOI] [PubMed] [Google Scholar]
- 55.Bates, S., Phillips, A., Clark, P., Stott, F., Peters, G., Ludwig, R., and Vousden, K. (1998) Nature 395 124-125 [DOI] [PubMed] [Google Scholar]
- 56.Lundin, C., North, M., Erixon, K., Walters, K., Jenssen, D., Goldman, A., and Helleday, T. (2005) Nucleic Acids Res. 33 3799-3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pickering, M., and Kowalik, T. (2006) Oncogene 25 746-755 [DOI] [PubMed] [Google Scholar]
- 58.Walsh, M., Shue, G., Spidoni, K., and Kapoor, A. (1995) J. Biol. Chem. 270 5289-5298 [DOI] [PubMed] [Google Scholar]
- 59.Shung, B., Miyakoshi, J., and Takebe, H. (1994) Mutat. Res. 307 43-51 [DOI] [PubMed] [Google Scholar]
- 60.Thompson, L., and West, M. (2000) Mutat. Res. 459 1-18 [DOI] [PubMed] [Google Scholar]
- 61.Collins, S., Swartz, M., Nelson, W., and DeWeese, T. (2003) Cancer Res. 63 1550-1554 [PubMed] [Google Scholar]
- 62.Nakada, D., Shimomura, T., Matsumoto, K., and Sugimoto, k. (2003) Nucleic Acids Res. 31 1715-1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan, Y., Raychaudhuri, P., and Costa, M. (2007) Mol. Cell. Biol. 27 1007-1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong, Z., and Tomkinson, A. (2006) Nucleic Acids Res. 34 5721-5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caldecott, K. (2001) BioEssays 23 447-455 [DOI] [PubMed] [Google Scholar]
- 66.Dynlacht, B. (2005) Cell Death Differ. 12 313-314 [Google Scholar]
- 67.Harris, S., and Levine, A. (2005) Oncogene 24 2899-2908 [DOI] [PubMed] [Google Scholar]
- 68.Sengupta, S., and Harris, C. (2005) Nat. Rev. Mol. Cell Biol. 6 44-55 [DOI] [PubMed] [Google Scholar]
- 69.Adimoolam, S., and Ford, J. (2003) DNA Repair 2 947-954 [DOI] [PubMed] [Google Scholar]
- 70.Seo, Y., Fishel, M., Amundson, S., Kelley, M., and Smith, M. (2002) Oncogene 21 731-737 [DOI] [PubMed] [Google Scholar]
- 71.Offer, H., Wolfkowicz, R., Matas, D., Blumenstein, S., Livneh, Z., and Rotter, V. (1999) FEBS Lett. 450 197-204 [DOI] [PubMed] [Google Scholar]
- 72.Zhou, J., Ahn, J., Wilson, S., and Prives, C. (2001) EMBO J. 20 914-923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winkonkal, N., Remenyik, E., Knezveic, D., Zhang, W., Liu, M., Zhao, H., Berton, T., Johnson, D., and Brash, D. (2003) Nat. Cell Biol. 5 655-660 [DOI] [PubMed] [Google Scholar]
- 74.Yamasaki, L., Jacks, T., Bronson, R., Goillot, E., Harlow, E., and Dyson, N. (1996) Cell 85 537-548 [DOI] [PubMed] [Google Scholar]
- 75.Field, S., Tsai, F., Kuo, F., Zubiaga, A., Kaelin, W. G. J., Livingston, D., Orkin, S., and Greenberg, M. (1996) Cell 85 549-561 [DOI] [PubMed] [Google Scholar]