Abstract
Plants produce p-aminobenzoate (pABA) in chloroplasts and use it for folate synthesis in mitochondria. In plant tissues, however, pABA is known to occur predominantly as its glucose ester (pABA-Glc), and the role of this metabolite in folate synthesis has not been defined. In this study, the UDP-glucose:pABA acyl-glucosyltransferase (pAGT) activity in Arabidopsis extracts was found to reside principally (95%) in one isoform with an apparent Km for pABA of 0.12 mm. Screening of recombinant Arabidopsis UDP-glycosyltransferases identified only three that recognized pABA. One of these (UGT75B1) exhibited a far higher kcat/Km value than the others and a far lower apparent Km for pABA (0.12 mm), suggesting its identity with the principal enzyme in vivo. Supporting this possibility, ablation of UGT75B1 reduced extractable pAGT activity by 95%, in vivo [14C]pABA glucosylation by 77%, and the endogenous pABA-Glc/pABA ratio by 9-fold. The Keq for the pABA esterification reaction was found to be 3 × 10-3. Taken with literature data on the cytosolic location of pAGT activity and on cytosolic UDP-glucose/UDP ratios, this Keq value allowed estimation that only 4% of cytosolic pABA is esterified. That pABA-Glc predominates in planta therefore implies that it is sequestered away from the cytosol and, consistent with this possibility, vacuoles isolated from [14C]pABA-fed pea leaves were estimated to contain≥88% of the [14C]pABA-Glc formed. In total, these data and the fact that isolated mitochondria did not take up [3H]pABA-Glc, suggest that the glucose ester represents a storage form of pABA that does not contribute directly to folate synthesis.
Tetrahydrofolate and its derivatives (folates) are essential cofactors for one-carbon reactions in almost all organisms, but are made only by plants and microbes (1, 2). Folates are tripartite molecules comprising pterin, p-aminobenzoate (pABA),3 and glutamate moieties (Fig. 1A), and folate synthesis in plants involves three subcellular compartments (Fig. 1B). The pterin moiety is synthesized in the cytosol, pABA is synthesized in chloroplasts, and the two are coupled together, reduced, and glutamylated in mitochondria (3). Although folate synthesis itself is fairly well understood, the metabolism and transport of the pathway intermediates, pABA and pterins, are not. Such ignorance impedes rational engineering to enhance folate levels in food crops for improved human health (4).
FIGURE 1.
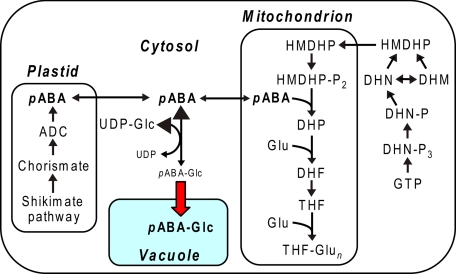
Folate structure and compartmentation of the plant folate synthesis pathway. A, tetrahydrofolate, with the pABA moiety colored red. Other natural folates have one-carbon units attached to the N5 and/or N10 positions. A γ-linked polyglutamyl tail of up to about six residues is usually attached to the first glutamate. B, the currently accepted scheme of folate biosynthesis in plants, showing subcellular locations of biosynthetic steps and of pABA esterification (highlighted in rose) and hypothetical transport steps for pABA-Glc into the mitochondrion and vacuole tested in this study (dashed arrows). Note that polyglutamylation of folates also occurs in the cytosol and plastids. ADC, aminodeoxychorismate; DHF, dihydrofolate; DHM, dihydromonapterin; DHN, dihydroneopterin; DHP, dihydropteroate; HMDHP, hydroxymethyldihydropterin; -P, phosphate; -P2, pyrophosphate, -P3, triphosphate; THF, tetrahydrofolate; UDP-Glc, UDP-glucose.
After its formation in chloroplasts, pABA must transit the cytosol before entering mitochondria for folate synthesis (Fig. 1B). As a hydrophobic weak acid, pABA can cross membranes by simple diffusion according to pH and concentration gradients (5). However, pABA has been shown to occur in plants mainly as its glucose ester (pABA-Glc), which is hydrophilic and almost certainly cannot readily diffuse across membranes (6). Because free pABA is present only at low concentrations it is entirely conceivable that mitochondria import pABA-Glc and then hydrolyze it to pABA for folate synthesis (Fig. 1B). Nothing is known about this issue or, more generally, about transport of aromatic conjugates by plant mitochondria (7).
Nor is it clear whether pABA-Glc is restricted to the cytosol. Vacuoles are well known to import glucose conjugates of aromatic compounds (8), and experiments with tissue cultures given megadoses of pABA suggest that some of the pABA-Glc formed is probably in vacuoles. Specifically, cell suspension cultures of Solanum laciniatum supplied with 2.5 mm pABA accumulate levels of pABA-Glc that, from osmotic considerations, appear too high to be confined to the cytosol (6, 9). These data invite the questions of whether pABA-Glc import into vacuoles indeed occurs (Fig. 1B), and, if so, whether it is a normal process or a pharmacological aberration.
pABA-Glc formation is known to be a reversible reaction mediated by cytosolic UDP-glucose: pABA acyl-glucosyltransferase (pAGT) activity (6) but no protein with this activity has yet been identified. However, a prediction about pAGT can be made from bioinformatics. The Arabidopsis thaliana (hereafter, Arabidopsis) genome encodes >100 UDP-glucosyltransferases (UGTs) that fall into 14 phylogenetically distinct groups (A to N) (10). Prior work on recombinant UGTs has shown that enzymes that form glucose esters with various benzoates belong to group L (11), making it likely that enzymes with pAGT activity are in this group.
In this study, we partially characterized pAGT activity extracted from Arabidopsis, screened Arabidopsis group L UGTs for pAGT activity, and biochemically characterized the three most active enzymes. This work implicated UGT75B1 as the main source of pAGT activity in planta, for which supporting evidence came from reverse-genetics. Radiolabeling approaches were used to investigate vacuolar and mitochondrial uptake of pABA-Glc, and to estimate Keq for the esterification reaction. The data strongly suggest that pABA-Glc is not taken up by mitochondria but rather represents a vacuolar storage form of pABA that does not contribute directly to folate synthesis.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—[Ring-14C]pABA (55 mCi mmol-1), [3,5-3H]pABA (26.2 Ci mmol-1), [3′,5′,7,9-3H]folic acid diammonium salt (43.2 Ci mmol-1), and [2,3,4,5-3H]proline (72.5 Ci mmol-1) were from Moravek Biochemicals (Brea, CA). [3H]Proline and [14C]pABA (when used for Keq estimation) were purified as follows. [3H]Proline (108 μCi) was dried in vacuo, redissolved in 5 μl of water and applied to a 1-cm origin on a 10-cm cellulose TLC plate, which was developed in butanol/acetic acid/water (60:15:25, v/v/v). The proline zone (Rf = 0.3) was scraped from the plate and eluted with 3 × 200 μl H2O. [14C]pABA (2.2 μCi) was dried in vacuo, redissolved in 50 μl of water, and separated by HPLC using a Discovery 5-μm C18 (250 × 4.6 mm, Supelco) column; detection was by fluorescence (270 nm excitation, 350 nm emission). The column was eluted (1 ml min-1) with 0.5% acetic acid/methanol (80:20, v/v). Unlabeled pABA-Glc was prepared as described (6). [3H]pABA-Glc was synthesized enzymatically from [3H]pABA (10 μCi, 0.38 nmol), which was dried in vacuo and redissolved in 8 μl of 25 mm Tris-HCl, pH 7.5. The reaction (10-μl final volume) contained [3H]pABA, 0.4 μg of UGT75B1, 4 mm UDP-glucose, and 14 mm 2-mercaptoethanol, and was incubated at 30 °C for 90 min. The reaction was stopped by freezing in liquid N2. Upon thawing, the [3H]pABA-Glc product was isolated by HPLC as described below. The radiochemical yield was 18%, and radiochemical purity was >99%. UDP-glucose was purchased from Sigma and checked for purity by HPLC (12); purity was determined to be >99%. Dowex (AG) resins were from Bio-Rad. TLC plates were from Merck (Darmstadt, Germany). Protein chromatography columns were from GE Healthcare (Piscataway, NJ).
Plant Materials—Arabidopsis (ecotypes Columbia-0 and Landsberg erecta) plants were grown at 23-28 °C in 12-h days (80 μmol m-2 s-1) in potting soil irrigated with water. For root production, plants were grown hydroponically (13). Pea (Pisum sativum cv. Laxton's Improved Progress 9) plants were grown as described (14). Cauliflower was obtained from a local market. The UGT75B1 mutant (line MGT.6017) was identified in the Arabidopsis (ecotype Landsberg erecta) Cold Spring Harbor Genetrap transposon insertion collection. Wild type or homozygous mutant segregants were identified by PCR using gene-specific primers located 5′ or 3′ of the Ds insertion (5′-GGCTAGACTCGAAAACAGAGT-3′ and 5′-CCTCCTCTGCTTCACACAAG-3′, respectively) and a Ds-specific primer (5′-CGCTGGCCTGCCCAACC-3′). Genomic DNA was extracted as described (15). The insertion site was confirmed by sequencing the amplicon from mutant homozygotes. Homozygous mutant and wild-type segregants were selfed, and their progeny were used for experiments.
Production and Purification of Recombinant UGTs—Procedures for Escherichia coli expression and protein isolation used to screen glutathione S-transferase-UGT fusions for pAGT activity were as described (16). For further analysis of UGT75B1, UGT74F1, and UGT74F2, fusion constructs were introduced into E. coli BL21-CodonPlus (DE3)-RIPL cells (Stratagene), which were grown at 37 °C in LB medium until the A600 reached 0.6. The temperature was then lowered to 25 °C, and isopropyl-d-thiogalactopyranoside was added (final concentration, 0.5 mm). Incubation was continued for 4 h at 25 °C. Subsequent procedures were at 0-4 °C. Cells from 100-ml cultures were pelleted, resuspended in 1 ml of phosphate-buffered saline, and broken in a Mini-Bead Beater (Biospec, Bartlesville, OK). Lysates were cleared by centrifugation (10,000 × g, 10 min). To remove co-purifying contaminant proteins, lysates were incubated with 8 m urea-denatured E. coli proteins (17). The mixture was centrifuged at 1,500 × g for 10 min, and the supernatant was mixed with glutathione-Sepharose resin and purified recombinant UGT recovered as described (16). The purified UGTs were desalted (100 mm Tris-HCl, pH 8.0) on Sephadex G-25 mini-spin columns, flash-frozen, and stored at -80 °C. Protein was estimated by the Bradford method (18) using bovine serum albumin as the standard. Recombinant proteins were analyzed by SDS-PAGE (19).
Extraction and Separation of Arabidopsis pAGT Activity—Tissues were pulverized in liquid N2. Protein extraction and purification steps were at 0-4 °C. For tests of pAGT activity in various tissues, protein was extracted in 50 mm Tris-HCl, pH 8.0, containing 10 mm 2-mercaptoethanol and 30 mg/ml polyvinylpolypyrrolidone (2 ml/g fresh weight). The extract was centrifuged (10,000 × g, 20 min), and the supernatant was filtered through a layer of Miracloth, then desalted on a PD-10 column equilibrated in 50 mm Tris-HCl, pH 7.5, containing 10% (v/v) glycerol, and 10 mm 2-mercaptoethanol. For partial purification of pAGT activity, the clarified extract from leaves (13 g) of 5-week-old Columbia-0 plants was brought to 80% saturation with (NH4)2SO4. After stirring for 30 min and centrifuging (13,000 × g, 20 min), the pellet was redissolved in 4.8 ml of 25 mm Tris-HCl, pH 7.5, containing 5 mm 2-mercaptoethanol (Buffer A) and desalted on PD-10 columns equilibrated with Buffer A. The solution was applied (1 ml min-1) to a Mono Q HR 5/5 column equilibrated with Buffer A, and the column was washed with this buffer until the A280 of the effluent fell to zero. pAGT activity was eluted (1 ml min-1) with a 15-ml gradient of 0-500 mm KCl in Buffer A, collecting 0.3-ml fractions. Active fractions were pooled, brought to 10% (v/v) glycerol, frozen in liquid N2, and stored at -80 °C until use.
pAGT Assays—Activities were measured at 30 °C; product formation was linear with time and amount of protein. Initial screening of recombinant UGTs for pAGT activity was in assay mixtures (200 μl) containing 1 μg of protein (not previously frozen and thawed), 50 mm Tris-HCl, pH 7.0, 14 mm 2-mercaptoethanol, 2.5 mm UDP-glucose, and 1 mm pABA. Reactions were incubated for 30 min, stopped by adding 20 μl of trichloroacetic acid (240 mg ml-1), flash-frozen, and stored at -20 °C before reverse-phase HPLC analysis as described (11). Kinetic constants of UGT75B1, UGT74F1, and UGT74F2 were measured on purified proteins that had been frozen once, using assay mixtures (20 μl) containing 25 mm Tris-HCl, pH 7.5, 100 ng of protein, 4 mm UDP-glucose, and the indicated amounts of [14C]pABA and glycerol. Assays were stopped in liquid N2, then applied to AG 4-X4 (OH-) columns to quantify [14C]pABA-Glc formation as described (6). A freeze-thaw cycle reduced pAGT specific activities by up to 10-fold, but use of frozen enzymes was deemed acceptable as it allowed analysis of all three UGTs at once. For Arabidopsis pAGT activity, standard reaction mixtures (final volume 20 μl) contained 25 mm Tris-HCl, 4-18 μg of protein, 45 μm [14C]pABA, and 4 mm UDP-Glc; [14C]pABA-Glc formation was measured as above. For fractions from Mono Q chromatography, assay mixtures were applied to 1-ml columns of AG 4-X4 (C2H5COO-), from which [14C]pABA-Glc was eluted with 20 ml of water and counted.
Estimation of Keq—The Keq for the pABA esterification reaction was estimated by measuring [14C]pABA-Glc formation. Assay mixtures (20 μl final volume) contained 1.5 μg of purified glutathione S-transferase-fused UGT75B1, 10 nmol of UDP-glucose, 9.25 nmol of unlabeled pABA, and 41.3 nCi (0.75 nmol) of purified [14C]pABA in 50 mm MOPS-NaOH, pH 7.5, or 50 mm Tris-HCl, pH 7.5. Reactions were incubated at 30 °C for various times, after which the [14C]pABA-Glc product was quantified by TLC as described (6).
[14C]pABA Metabolism in Arabidopsis in Vivo—The surface of the midrib was shaved from the abaxial surface of Arabidopsis leaves. A 3-μl droplet containing 68 nCi (1.23 nmol) of [14C]pABA and 939 nmol of unlabeled pABA dissolved in water was applied to the cut surface of each leaf. Leaves were incubated in Petri dishes for 3 h at 24 °C in the light (150 μmol m-2 s-1) and then shaken (100 rpm) for 15 min in 5 ml of 0.1 mm pABA to remove non-absorbed label. Tissues were extracted by grinding in 2 ml of semifrozen methanol. Aliquots (120 μl) of the methanol extract were separated by TLC as described (6); radioactive zones were detected by autoradiography and scraped from the plate for scintillation counting.
Determination of Free and Total pABA—Arabidopsis wild-type and UGT75B1 mutant leaf tissue samples (0.4 g fresh weight) were extracted and analyzed by HPLC for free and total pABA as described (6). Esterified pABA was calculated as (total pABA - free pABA). Data were corrected for recovery using free pABA spikes.
Pea Leaf Vacuole Experiments—The surface of the midrib was shaved from the abaxial surface of pea leaflets (1 g, 20-22 leaflets). A solution of [14C]pABA (0.30-0.33 μCi, 5.4-6.0 nmol) dissolved in 3 μl of water was applied to the cut surfaces. Leaflets were incubated in Petri dishes for 3 h at 24 °C in light (150 μmol m-2 s-1). After the incubation, protoplasts were prepared from the [14C]pABA-fed leaflets together with unlabeled pea leaflets (24 g) following a 5-h digestion as described (20) except that the centrifugation step used to harvest protoplasts included a cushion of 50% (v/v) Percoll in 10 mm MES-NaOH, pH 7.0, 0.5 m sorbitol, 5 mm CaCl2, 0.5 mg/ml polyvinylpyrrolidone-40. The protoplast layers were adjusted to 13% Percoll in the same buffer and purified on a three-step sucrose-sorbitol gradient as described (21). Vacuoles were prepared from lysed protoplasts (20) after adding to the lysate unlabeled folic acid to a final concentration of 50 μm and 2-25 μCi (0.05-0.58 nmol) of [3H]folic acid. Marker enzymes were extracted and assayed as described (20). Vacuolar yield was quantified by counting protoplast and vacuole preparations using a hemocytometer, and by measuring α-mannosidase activity (22). The purity of the vacuole fraction was attested by the absence of chlorophyll and by the very low recovery (<0.5%) of mitochondrial (fumarase) and stromal (NADP-linked glyceraldehyde-3-phosphate dehydrogenase) marker enzyme activities (23). The 3H and 14C contents of protoplast lysate and vacuole fractions were determined by scintillation counting. Before HPLC analysis of the vacuolar 14C label, ultrafiltration using a Centricon-10 (Millipore, Billerica, MA) was used to remove Ficoll. The ultrafiltrate was concentrated 2-fold in vacuo before HPLC analysis. HPLC separation was carried out on a Discovery 5-μm C18 (250 × 4.6 mm, Supelco); detection was by absorbance at 295 nm or fluorescence (270 nm excitation, 350 nm emission). The column was eluted (1 ml min-1) with 10% acetonitrile/0.1% trifluoroacetic acid in water; fractions (0.33 ml) were collected, and the radioactivity counted.
Mitochondrial [3H]pABA-Glc Transport Tests—Mitochondria were isolated from 500 g of cauliflower florets and purified by Percoll density gradient centrifugation (24). The purified mitochondria were resuspended in transport buffer (25 mm MOPS, pH 7.4, 0.33 m sucrose, 1 mm EDTA, and 1 mm dithiothreitol). Mitochondrial extracts were obtained by freezing in liquid N2 and thawing at 30 °C (5 cycles), followed by centrifugation at 16,000 × g for 10 min at 4 °C. As a quality control, fumarase was assayed in the freshly prepared extract (20); typical activities were 5 μmol min-1 mg-1 protein. [3H]pABA-Glc transport assays (final volume 500 μl) contained 92 nCi (3.5 pmol) of [3H]pABA-Glc and 1 mm NADH in transport buffer (25). Assays were started by adding mitochondria (330 μg protein) and incubated at 30 °C with magnetic stirring. At intervals, 75-μl aliquots were withdrawn, and mitochondria were harvested on a Whatman cellulose nitrate membrane filter (0.45 μm) using a vacuum filtration device. Filters were washed twice with 2 ml of ice-cold transport buffer, added to 3 ml of Beckman Ready Gel scintillation fluid (Beckman Coulter, Fullerton, CA), equilibrated, and counted. [3H]Proline uptake was measured by the same method using 254 nCi (3.5 pmol) of [3H]proline.
Estimation of Endogenous pABA Production Rate in Arabidopsis—The endogenous pABA production rate in Arabidopsis leaves (∼2.5 pmol mg-1 protein h-1) was estimated from published folate and total pABA contents (6, 14) by assuming a growth rate of 10% per day (26) and a protein content of 10 mg g-1 fresh weight.
RESULTS
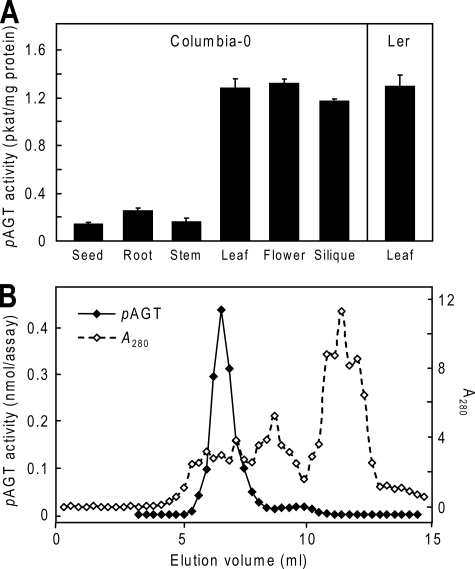
Organ Distribution and Isoform Complexity of pAGT Activity in Arabidopsis—Because pAGT activity has not been studied ex vivo in Arabidopsis, we first assayed pAGT activities in extracts from various organs of ecotype Columbia-0 to determine where pAGT is most strongly expressed (Fig. 2A). A relatively low pABA concentration (45 μm) was used to mimic the cytosolic free pABA concentration, which can be estimated as ≤45 μm from published data on plant pABA contents (6) and cytosol volume (27). pAGT activity was highest in leaves, flowers, and siliques, and was equally high in ecotype Landsberg erecta leaves, which were analyzed for comparison (Fig. 2A). To investigate pAGT isoforms, Columbia leaf activity was fractionated by anion exchange chromatography (Fig. 2B). A single major peak of activity eluted at ∼220 mm NaCl, and a minor peak (representing ≤5% of the total activity) eluted at ∼320 mm. The pAGT activity in leaves thus comes mainly from one isoform. Triplicate determinations of the Km for pABA of the major peak activity gave a value of 0.12 ± 0.02 mm (mean ± S.E.).
FIGURE 2.
pAGT activity in Arabidopsis. A, pAGT activities in extracts of organs of ecotype Columbia-0. The activity in leaves of Landsberg erecta (Ler) is shown for comparison. Enzyme assays contained 45 μm pABA and 4 mm UDP-glucose. Values for leaves, flowers, roots, and seeds are means and S.E. for two to four replicates from two or three independent extracts; stem and silique values are for single extracts. B, anion exchange chromatography of Columbia-0 leaf proteins, showing one major peak of pAGT activity. Ammonium sulfate-precipitated proteins (42 mg) were applied to a 1-ml Mono Q column and eluted with a 0-0.5 m NaCl gradient. Enzyme assays (20 μl final volume) contained 40 μm [14C]pABA (0.8 nmol) and 4 mm UDP-glucose, and were run for 30 min. Enzyme activity is in nmol of pABA-Glc formed per assay.
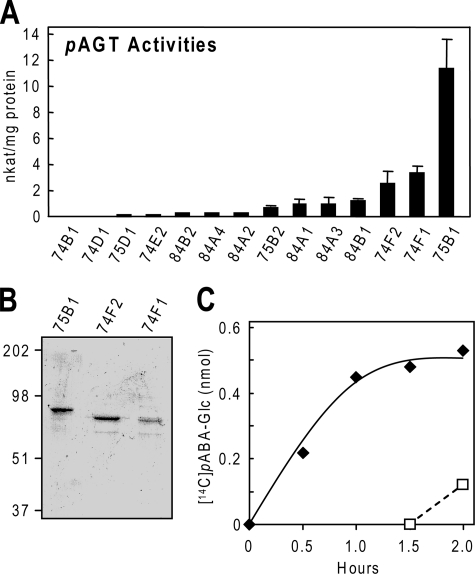
Identification and Characterization of Recombinant UGTs with pAGT Activity—Sequences from Group L of the Arabidopsis UGT family were expressed as glutathione S-transferase fusion proteins in E. coli, affinity-purified, and screened for pAGT activity by an HPLC assay (11). UGT75B1 had high activity, and UGT74F1 and UGT74F2 had moderate activity; the others had lower activity or none (Fig. 3A). The three enzymes with the most activity were kinetically characterized using a sensitive radioassay; the protein preparations used were near homogeneous (Fig. 3B). UGT75B1 was the most active enzyme in terms of its kcat/Km value and had by far the lowest apparent Km (Table 1). Significantly, the Km value of recombinant UGT75B1 was identical (0.12 mm) to that found for the enzyme extracted from Arabidopsis. These results strongly imply that UGT75B1 is the major contributor to pAGT activity in planta.
FIGURE 3.
Characterization of Arabidopsis group L UGTs and of the esterification reaction. A, pAGT activities of 16 members of group L measured using 1 mm pABA and 2.5 mm UDP-glucose. Data are means of three replicates and S.E. For UGT75B1, the activity with UDP-galactose or UDP-glucuronate as sugar donor was found to be negligible (<1% of that with UDP-glucose). B, SDS-PAGE analysis of affinity-purified preparations of the three UGTs with the highest pAGT activities. Tracks contained 1 μg (UGT75B1, UGT74F2) or 0.5 μg (UGT74F1) of protein. The gel was stained with Coomassie Blue. Running positions of molecular mass markers (kDa) are indicated on the left. C, progress curve of [14C]pABA-Glc formation in a reaction mixture containing 1.5 μg of purified UGT75B1, 10 nmol of UDP-glucose, 9.25 nmol of unlabeled pABA, and 41.3 nCi (0.75 nmol) of [14C]pABA (solid symbols and line), in 50 mm MOPS-NaOH, pH 7.5. A second reaction mixture without radiolabel was incubated in parallel with the first for 1.5 h, at which point 41.3 nCi of [14C]pABA was added (open symbols and dotted line). The formation of [14C]pABA-Glc in this reaction shows that UGT75B1 remained enzymatically active throughout the experiment.
TABLE 1.
Kinetic characteristics of Arabidopsis UGTs with pAGT activity
Enzyme activities were measured radiometrically at pH 7.5 using affinity-purified glutathione S-transferase fusion proteins. Km and kcat values are the means of three replicates ± S.E.
| Protein | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | mm-1 s-1 | |
| UGT75B1 | 0.12 ± 0.01 | 13.8 ± 0.6 | 115 |
| UGT74F2 | 0.92 ± 0.14 | 8.8 ± 1.1 | 9.6 |
| UGT74F1 | 0.85 ± 0.12 | 10.5 ± 0.2 | 12.4 |
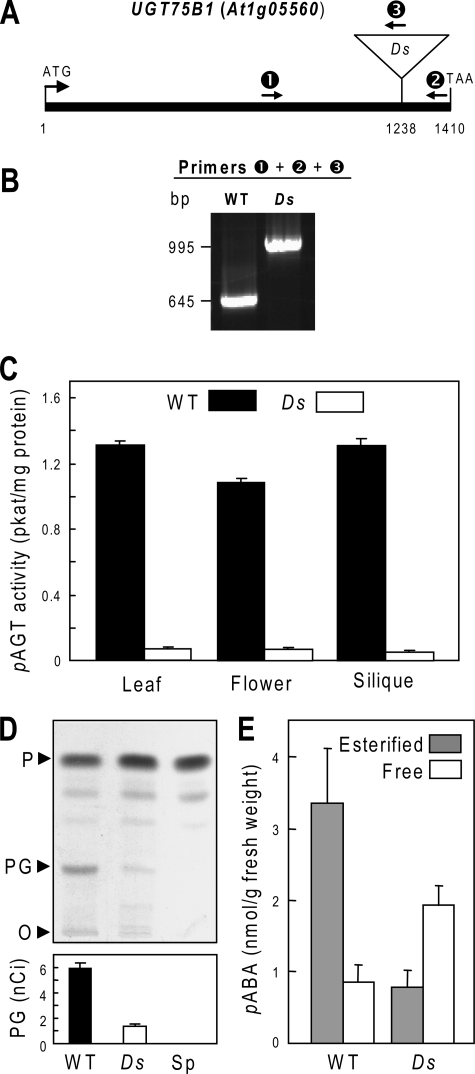
Insertional Inactivation of UGT75B1—To corroborate the involvement of UGT75B1 in pABA glucosylation in planta, the consequence of knocking out expression of the gene was investigated. A Ds mutant of the UGT75B1 gene (At1g05560), which is intronless, was identified in the Cold Spring Harbor Genetrap collection, in the Landsberg erecta background. The Ds insertion is at nucleotide 1238, corresponding to amino acid 412 of the 469-residue protein (Fig. 4A). Plants homozygous for this insertion were identified by PCR screening (Fig. 4B), and PCR tests of wild-type segregants from the same population confirmed that other Ds elements were absent (data not shown). RT-PCR analysis of mutant homozygotes detected a UGT75B1 message with an in-frame stop codon 21 nucleotides after the Ds junction. The putative translation product is most unlikely a priori to have UGT activity because the missing C-terminal region contains two conserved α-helical domains that subtend the UDP-glucose binding site (28, 29). The Ds mutation did not visibly affect germination, growth, or development.
FIGURE 4.
Insertional inactivation of the UGT75B1 gene (At1g05560) in the Landsberg erecta background. A, the UGT75B1 gene (which is intronless), showing the position of the Ds insert and of the three PCR primers (1, 2, 3) used for analysis of genomic DNA. Primer abbreviations: 1, UGT75B1-specific forward primer; 2, UGT75B1-specific reverse primer; 3, Ds-specific reverse primer. B, gel separation of PCR products amplified from genomic DNA of homozygous wild type and mutant plants using a mixture of primers 1, 2, and 3. C, pAGT activities in extracts of leaves, flowers, and siliques of wild type and mutant plants. Values are means and S.E. for two replicates from two independent extracts. D, autoradiograph of a TLC separation of extracts of wild type (WT) and mutant (Ds) leaves (0.14 g) supplied with 52 nCi (2.82 μmol) of [14C]pABA for 3 h. The origin (O) and running positions of pABA (P) and pABA-Glc (PG) standards are indicated. A control sample of unlabeled leaves (Sp) spiked with [14C]pABA just before extraction is included. Bars below the autoradiograph show the amounts of [14C]pABA-Glc present in the samples; these values are means of four replicates and S.E. E, estimation of free and esterified pABA in wild type and mutant leaves. Values are means and S.E. for three samples. The free pABA data are not corrected for the contribution of ester hydrolysis during sample processing.
Total pAGT activity was assayed in leaf, flower, and silique extracts from mutant plants and wild-type segregants using 45 μm pABA as above (Fig. 4C). Extracts from mutant plants displayed on average a 95% reduction in activity. This strongly indicates that UGT75B1 is the main pAGT isoform in all three organs. These results were extended by comparing the capacity of mutant and wild-type leaves to convert [14C]pABA to [14C]pABA-Glc in vivo, using a [14C]pABA dose (∼20 μmol/g fresh weight) sufficient to ensure that it would not all be esterified during the experiment (6). TLC analysis of labeled metabolites showed that [14C]pABA-Glc formation was drastically (77%) reduced in the mutant (Fig. 4D). Consistent with their lower pAGT activity, mutant leaves showed significantly (p < 0.05) decreased endogenous pABA-Glc levels and increased endogenous free pABA levels, so that the pABA-Glc/pABA ratio fell to 0.4 compared with 3.5 in the wild type (Fig. 4E). The total (esterified plus free) endogenous pABA levels did not differ significantly between mutant and wild type.
Measurement of Keq for the Esterification Reaction—The Keq for pABA esterification was estimated by following [14C]pABA-Glc formation in a reaction mixture containing purified recombinant UGT75B1, 10 nmol of unlabeled UDP-glucose, and 10 nmol of pABA containing a tracer amount of 14C (Fig. 3C). The reaction appeared to have reached equilibrium at 1.5 h, by which time [14C]pABA-Glc formation had plateaued. To verify that this plateau was not due to enzyme inactivation, a parallel reaction was run with unlabeled substrates, and tracer [14C]pABA was added at 1.5 h. The subsequent formation of [14C]pABA-Glc (Fig. 3C, dotted line) showed that UGT75B1 remained active throughout the experiment. The quantity of [14C]pABA-Glc present at equilibrium (0.5 nmol) corresponds to a Keq value of 3 × 10-3. Three similar experiments confirmed this value and showed that the nature of the buffer used (Tris-HCl versus MOPS-NaOH) had little if any effect (not shown).
Evidence for Vacuolar Storage of pABA-Glc—To determine whether pABA-Glc enters vacuoles, we used an in vivo dual labeling approach based on the ability of leaves to rapidly convert tracer amounts of [14C]pABA to pABA-Glc (6). A small sample of pea leaflets was supplied with a tracer dose of [14C]pABA for 3 h, then mixed with unlabeled leaves and used to prepare mesophyll protoplasts. Vacuoles were then isolated from the protoplasts. Just after protoplasts were lysed to release vacuoles, low specific activity [3H]folic acid was added to the lysate as a marker for contamination of vacuoles by extravacuolar solutes. (The low specific activity of the [3H]folic acid ensured that, if any vacuolar uptake occurred, its 3H content would be negligible. Moreover, tests with vacuolar vesicles showed no detectable [3H]folic acid uptake (not shown).) The 14C and 3H contents of the isolated vacuoles were measured and compared with those of the protoplast lysate, and with the yield of vacuoles assessed by counting and by recovery of the vacuole marker α-mannosidase (30).
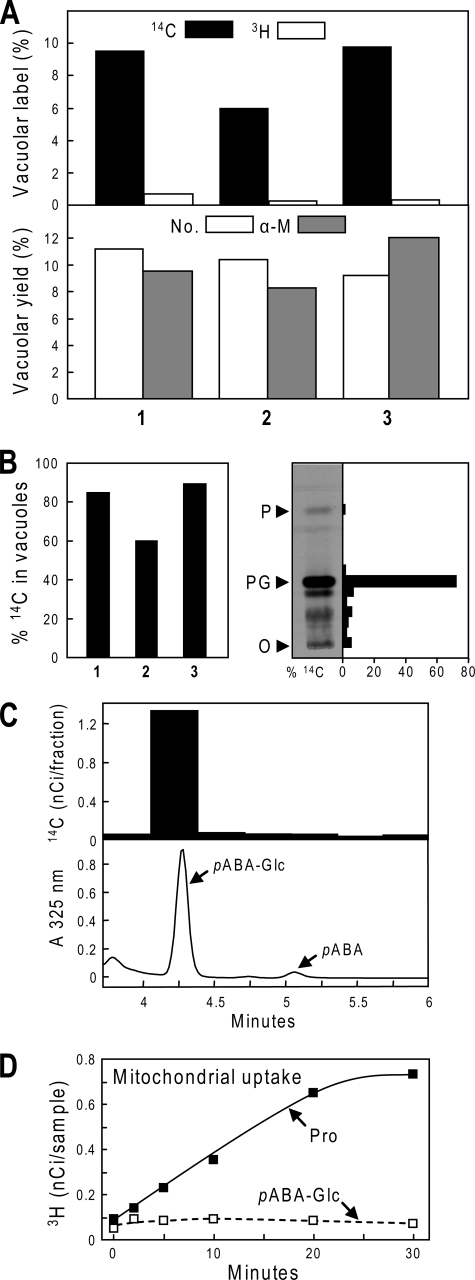
In three separate experiments, an average of 8.4% of the 14C present in the protoplast lysate was recovered in purified vacuoles versus 0.4% of the 3H (Fig. 5A), indicating that 8.0% of the 14C was intravacuolar. Because the mean yield of vacuoles was 10.1% as judged from vacuole numbers and α-mannosidase recovery (Fig. 5A), we can calculate that, on average, 79% (range 60-90%) of the 14C was located in vacuoles in vivo (Fig. 5B), this being a minimum estimate because 14C may have leaked out during isolation. Of the 14C label recovered from vacuoles, HPLC analysis demonstrated that essentially none was in the form of free pABA and that 80% was in the form of pABA-Glc (Fig. 5C) so that the [14C]pABA-Glc present in vacuoles was (0.79 × 0.8 × 100) = 63% of the 14C in the leaf tissue. Of the 14C label present in the leaf tissue whence the vacuoles came, TLC analysis showed that some 72% was in pABA-Glc (Fig. 5B, right panel). The average proportion of [14C]pABA-Glc in vacuoles may thus be estimated as (100 × 63/72) = 88%.
FIGURE 5.
Tests for pABA-Glc uptake by vacuoles and mitochondria. A, quantification of in vivo vacuolar accumulation of [14C]pABA-Glc. Pea leaflets (1 g) were supplied with [14C]pABA (118 nmol, 6.5 μCi) for 3 h, then mixed with 24 g of unlabeled leaves for protoplast preparation. Purified mesophyll protoplasts were osmotically lysed to liberate vacuoles, at which point low specific activity [3H]folic acid was added; vacuoles were then isolated by Ficoll density gradient centrifugation. The 14C and 3H contents of the purified vacuole fraction were quantified and expressed as percentages of those of the protoplast lysate (upper panel). Vacuolar yields were estimated by counting vacuole numbers (No.) and from α-mannosidase activities (α-M) (lower panel). Results are from three independent experiments (1-3). B, left panel: The estimated percentage of 14C label that was in vacuoles in vivo, in experiments 1-3. Values were calculated from the data of A using the formula: 100 × (% vacuolar 14C-% vacuolar 3H)/mean % vacuolar yield. Mean % vacuolar yield was based on the estimates from counting and α-mannosidase activity. Right panel, TLC separation of 14C-labeled metabolites in pea leaflets supplied for 8 h with the same [14C]pABA dose given to leaflets used to prepare vacuoles. After the autoradiograph on the left was taken, the 14C contents of TLC zones were determined, and expressed as percentages of the total 14C label on the plate. The pABA-Glc zone (PG) contained 72% of the total label. O, TLC origin; P, pABA zone. C, co-chromatography of the 14C label recovered from vacuoles with pABA-Glc. A sample of the vacuole fraction was freed of Ficoll by ultrafiltration, mixed with authentic pABA-Glc and pABA standards, and separated by HPLC. The pABA-Glc zone contained 80% of the total 14C in the sample. D, lack of uptake of [3H]pABA-Glc by transport-competent cauliflower floret mitochondria. Purified mitochondria (0.33 mg protein) were incubated at 30 °C in a 0.5-ml reaction mixture containing 1 mm NADH and 7 nm [3H]pABA-Glc (92 nCi) or [3H]proline (254 nCi). Samples (75 μl) were removed at the intervals shown; mitochondria were recovered by filtration on nitrocellulose membranes, which were assayed for 3H. Data points are means of duplicate samples. The experiment was repeated, with similar results.
Assay of pABA-Glc Uptake by Mitochondria—[3H]pABA-Glc was prepared from [3H]pABA and UDP-glucose using purified UGT75B1 to catalyze the reaction. The procedure (see “Experimental Procedures”) gave an overall radiochemical yield of 18%. The [3H]pABA-Glc was supplied to purified cauliflower mitochondria energized with NADH; no uptake was detectable (Fig. 5D). The mitochondrial preparations were confirmed to be transport-competent by supplying [3H]proline in place of [3H]pABA-Glc (Fig. 5D); proline is known to be readily taken up by energized plant mitochondria (31).
DISCUSSION
Humans and other higher animals do not synthesize folates de novo and therefore need a dietary supply that is mainly provided by plants. For this reason, there is considerable interest in engineering food plants for enhanced folate content (2, 4). To achieve this goal, the biosynthesis and metabolism of folates and their precursors need to be more fully understood (4).
In this study, we focused on the pABA moiety of the folate molecule. It is well established that pABA is made in the chloroplast, but is incorporated into folate in the mitochondrion (Fig. 1B). Until recently, there was a tacit assumption that pABA principally existed as the free acid in plant cells. However, the major form of pABA was found to be the d-glucopyranosyl ester, and plant extracts contained an enzyme activity that readily glucosylated pABA (6). These findings raised the possibility pABA-Glc could be the metabolite that is imported into the mitochondrion during folate synthesis. Alternatively, formation of the glucose ester could represent a mechanism of metabolite homeostasis, decreasing free pABA levels in the cytosol through vacuolar sequestration of the glucosylated molecule.
The data presented here strongly favor the second possibility, for they demonstrate that the majority of pABA-Glc in leaves is located in the vacuole, and that mitochondria do not import pABA-Glc.
The evidence for a vacuolar location was both indirect and direct. Our Keq value for pABA glucose ester formation (3 × 10-3) shows that the equilibrium disfavors product formation. Combining this Keq value with literature data on UDP-glucose/UDP ratios in plant cytosol allows calculation of the cytosolic pABA-Glc/pABA-Glc ratio. Cytosolic UDP-glucose/UDP ratios in plant tissues are generally 10-20, the value reported for leaves being 13 (32-34). Using the value of 13 and a Keq value of 3 × 10-3 gives a pABA-Glc/pABA ratio of 0.04, so that 96% of the total pABA in the cytosol is predicted to be free and only 4% esterified. This calculated situation in the cytosol is opposite to results of whole tissue analysis, which show pABA-Glc to be far more abundant than free pABA in wild-type Arabidopsis and pea leaves (Fig. 4E and Refs. 6 and 14). This discrepancy implies that the bulk of the pABA-Glc cannot be in the cytosol where pAGT activity resides (6) but must be sequestered elsewhere, most probably in the vacuole.
Direct experimental evidence for a predominantly vacuolar location for pABA-Glc came from our dual-labeling tracer studies of pea leaf protoplasts and vacuoles, which showed that at least 88% of recently synthesized [14C]pABA-Glc is compartmented in vacuoles in vivo. This result accords with the deduction from tissue culture data (6, 9) that pABA-Glc must be at least partly vacuolar. It is also in agreement with reports of other aromatic Glc esters in plant vacuoles (35, 36) and, more generally, with the observation that many UGT reaction products accumulate in vacuoles (37). Thus, the fact that pABA-Glc is rapidly cleared from the cytosol makes it an unlikely candidate for transport into mitochondria, and indeed our experiments failed to show mitochondrial pABA-Glc uptake.
Interestingly, in leaves of Arabidopsis plants 95% of pAGT activity was associated with a single peak separated by anion exchange chromatography. The Km value (0.12 mm) for this enzyme was found to be identical to that of a recombinant protein, UGT75B1, which we identified from a screen of 16 UGTs from a phylogenetic group of Arabidopsis enzymes previously shown to form glucose esters with benzoate and phenylpropanoid scaffolds. Knocking out the expression of UGT75B1, led to loss of ∼95% of pAGT activity in the mutant, suggesting UGT75B1 encodes the activity observed in Arabidopsis extracts.
In an earlier study, UGT75B1 was found associated with the cell plate at cytokinesis but was distributed throughout cells that were not dividing (38). Because pABA-Glc is in effect an activated form of glucose (6), our biochemical findings suggest the possibility that UGT75B1 functions at the cell plate by providing activated glucose units for the synthesis of a glucose polymer. Alternatively, pABA-Glc could serve as a primer for such a polymer, analogous to the hydrophobic glycosides that function as primers in the synthesis of cellulose and other polysaccharides (39). In our present study and a previous one on pea (6) pAGT enzyme activity was located in the soluble fraction. This contrasts with the data from the cell plate study in which most of the UGT75B1 protein (detected immunologically) was recovered in a 100,000 × g pellet (38). However, the extraction buffer differed from ours in having less 2-mercaptoethanol (2 mm versus 10 mm) and in lacking polyvinylpolypyrrolidone (38). Both differences generally favor cross-linking and precipitation of proteins by phenols (40) and for polyvinylpolypyrrolidone this has been demonstrated in Arabidopsis (41). Moreover, tests showed that omitting polyvinylpolypyrrolidone from our extraction buffer and cutting the 2-mercaptoethanol concentration to 2 mm reduced the pAGT activity extracted from leaves by 78%. Dissimilar extraction conditions may thus account for the dissimilar results on localization.
In conclusion, our data clearly indicate the existence of a large vacuolar pool of pABA-Glc. It is probable that this represents a readily reclaimable storage depot for pABA moieties (6). The central vacuole in plant cells contains pools of many primary and secondary metabolites, frequently in conjugated forms, for which there are transport systems in the vacuolar membrane (35). Thus the occurrence of a vacuolar pool of a glucose conjugate and a cytosolic pool of free pABA conforms to a very common pattern: a large storage pool of a metabolite in the vacuole, and a much smaller active metabolic pool in another subcellular compartment (42). Another possible function of the vacuolar pool of pABA-Glc is as a pivotal part of a system to limit the concentration of free pABA. As a hydrophobic weak acid, pABA is able to diffuse back and forth across membranes, and hence to act as an uncoupler by collapsing electrochemical proton gradients (6, 43). High concentrations of pABA could therefore in some circumstances have deleterious consequences, which are avoided by esterification with glucose and transporting the ester into the vacuole. A further reason to esterify and sequester pABA may be that, at μm levels, it is a substrate inhibitor of dihydropteroate synthase, the enzyme that couples pABA to pterin during folate synthesis (44, 45).
Finally, our data indicate that the current map of the folate synthesis pathway (e.g. 4) needs to be modestly redrawn, in its pABA branch (Fig. 6). Specifically: in the pABA ↔ pABA-Glc equilibrium in the cytosol, pABA predominates; and pABA-Glc is transported into (and stored in) the vacuole. Because pABA-Glc is located in the vacuole, it is evident that mobilizing it would involve either export from the vacuole coupled with reversal of the synthesis reaction, or intravacuolar hydrolysis of pABA-Glc. Such hydrolysis would presumably require a vacuolar esterase (6).
FIGURE 6.
An updated scheme of the plant folate synthesis pathway. Large arrowheads in the pABA ↔ pABA-Glc reaction indicate that the equilibrium strongly favors pABA. The bold red arrow denotes pABA-Glc import into the vacuole, presumably via a transporter. Mobilization of vacuolar pABA-Glc would require reversal of this transport step or intravacuolar hydrolysis of pABA-Glc.
Acknowledgments
We thank Dr. Yair Shachar-Hill for helpful discussions about cytosolic concentrations of UDP-glucose and UDP and Michael J. Ziemak for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM071382. This work was also supported by an endowment from the C. V. Griffin, Sr. Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: pABA, p-aminobenzoic acid; pABA-Glc, p-aminobenzoate β-d-glucose ester; pAGT, UDP-glucose:pABA acyl-glucosyltransferase; HPLC, high performance liquid chromatography; UGT, UDP-glucosyltransferase; MOPS, 4-morpholinepropanesulfonic acid; MES, 4-morpholineethanesulfonic acid.
References
- 1.Hanson, A. D., and Roje, S. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 119-137 [DOI] [PubMed] [Google Scholar]
- 2.Scott, J., Rébeillé, F., and Fletcher, J. (2000) J. Sci. Food Agric. 80 795-824 [Google Scholar]
- 3.Hanson, A. D., and Gregory, J. F., 3rd (2002) Curr. Opin. Plant Biol. 5 244-249 [DOI] [PubMed] [Google Scholar]
- 4.Bekaert, S., Storozhenko, S., Mehrshahi, P., Bennett, M. J., Lambert, W., Gregory, J. F., 3rd, Schubert, K., Hugenholtz, J., Van Der Straeten, D., and Hanson, A. D. (2008) Trends Plant Sci. 13 28-35 [DOI] [PubMed] [Google Scholar]
- 5.Sterling, T. M., Balke, N. E., and Silverman, D. S. (1990) Plant Physiol. 92 1121-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinlivan, E. P., Roje, S., Basset, G., Shachar-Hill, Y., Gregory, J. F., 3rd, and Hanson, A. D. (2003) J. Biol. Chem. 278 20731-20737 [DOI] [PubMed] [Google Scholar]
- 7.Haferkamp, I. (2007) FEBS Lett. 581 2375-2379 [DOI] [PubMed] [Google Scholar]
- 8.Rea, P. A. (2007) Annu. Rev. Plant Biol. 58 347-375 [DOI] [PubMed] [Google Scholar]
- 9.Panjaitan, T. S., Syahrani, A., and Indrayanto, G. (2000) J. Planar Chromat.-Modern TLC 13 114-118 [Google Scholar]
- 10.Ross, J., Li, Y., Lim, E., and Bowles, D. J. (2001) Genome Biol. 2 reviews 3004. 1-3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim, E. K., Doucet, C. J., Li, Y., Elias, L., Worrall, D., Spencer, S. P., Ross, J., and Bowles, D. J. (2002) J. Biol. Chem. 277 586-592 [DOI] [PubMed] [Google Scholar]
- 12.Goulard, F., Diouris, M., Deslandes, E., and Floc'h, J. Y. (1999) Eur. J. Phycol. 34 21-25 [Google Scholar]
- 13.Gibeaut, D. M., Hulett, J., Cramer, G. R., and Seemann, J. R. (1997) Plant Physiol. 115 317-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orsomando, G., Bozzo, G. G., de la Garza, R. D., Basset, G. J., Quinlivan, E. P., Naponelli, V., Rébeillé, F., Ravanel, S., Gregory, J. F., 3rd, and Hanson, A. D. (2006) Plant J. 46 426-435 [DOI] [PubMed] [Google Scholar]
- 15.Edwards, K., Johnstone, C., and Thompson, C. (1991) Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, E. K., Roberts, M. R., and Bowles, D. J. (1998) J. Biol. Chem. 273 34920-34925 [DOI] [PubMed] [Google Scholar]
- 17.Rohman, M., and Harrison-Lavoie, K. J. (2000) Protein Expr. Purif. 20 45-47 [DOI] [PubMed] [Google Scholar]
- 18.Bradford, M. M. (1976) Anal. Biochem. 72 248-524 [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 20.Orsomando, G., de la Garza, R. D., Green, B. J., Peng, M., Rea, P. A., Ryan, T. J., Gregory, J. F., 3rd, and Hanson, A. D. (2005) J. Biol. Chem. 280 28877-28884 [DOI] [PubMed] [Google Scholar]
- 21.Bozzo, G. G., Basset, G. J., Naponelli, V., Noiriel, A., Gregory, J. F., 3rd, and Hanson, A. D. (2008) Phytochemistry 69 29-37 [DOI] [PubMed] [Google Scholar]
- 22.Nok, A. J., Shuaibu, M. N., Kanbara, H., and Yanagi, T. (2000) Parasitol Res. 86 923-928 [DOI] [PubMed] [Google Scholar]
- 23.Trossat, C., Nolte, K. D., and Hanson, A. D. (1996) Plant Physiol. 111 965-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douce, R., Bourguignon, J., Brouquisse, R., and Neuburger, M. (1987) Methods Enzymol. 148 403-415 [Google Scholar]
- 25.Goldraij, A., and Polacco, J. C. (2000) Planta 210 652-658 [DOI] [PubMed] [Google Scholar]
- 26.Kocsis, M. G., Ranocha, P., Gage, D. A., Simon, E. S., Rhodes, D., Peel, G. J., Mellema, S., Saito, K., Awazuhara, M., Li, C., Meeley, R. B., Tarczynski, M. C., Wagner, C., and Hanson, A. D. (2003) Plant Physiol. 131 1808-1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter, H., Robinson, D. G., and Heldt, H. W. (1994) Planta 193 530-535 [Google Scholar]
- 28.Offen, W., Martinez-Fleites, C., Yang, M., Kiat-Lim, E., Davis, B. G., Tarling, C. A., Ford, C. M., Bowles, D. J., and Davies, G. J. (2006) EMBO J. 25 1396-1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, L., Modolo, L. V., Escamilla-Trevino, L. L., Achnine, L., Dixon, R. A., and Wang, X. (2007) J. Mol. Biol. 370 951-963 [DOI] [PubMed] [Google Scholar]
- 30.Boller, T., and Kende, H. (1979) Plant Physiol. 63 1123-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elthon, T. E., Stewart, C. R., and Bonner, W. D. (1984) Plant Physiol. 75 951-955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dancer, J., Neuhaus, H. E., and Stitt, M. (1990) Plant Physiol. 92 637-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dancer, J., Veith, R., Feil, R., Komore, E., and Stitt, M. (1990) Plant Sci. 66 59-63 [Google Scholar]
- 34.Farré, E. M., Tiessen, A., Roessner, U., Geigenberger, P., Trethewey, R. N., and Willmitzer, L. (2001) Plant Physiol. 127 685-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wink, M. (1997) in The Plant Vacuole: Advances in Botanical Research, Vol. 25 (Leigh, R. A., Sanders, D., and Callow, J. A., eds), pp. 141-170, Academic Press, London, UK [Google Scholar]
- 36.Martinoia, E., Klein, M., Gessler, M., Sánchez-Fernández, R., and Rea, P. A. (2001) in Vacuolar Compartments, CRC Series (Robinson, D., and Rogers, J. eds), pp. 221-253, Sheffield Academic Press, Sheffield, UK
- 37.Bowles, D., Lim, E.-K., Poppenberger, B., and Vaistij, F. V. (2006) Annu. Rev. Plant Biol. 57 567-597 [DOI] [PubMed] [Google Scholar]
- 38.Hong, Z., Zhang, Z., Olson, J. M., and Verma, D. P. (2001) Plant Cell 13 769-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, L., Kawagoe, Y., Hogan, P., and Delmer, D. (2002) Science 295 147-150 [DOI] [PubMed] [Google Scholar]
- 40.Loomis, W. D., and Bataille, J. (1966) Phytochemistry 5 423-438 [Google Scholar]
- 41.Charmont, S., Jamet, E., Pont-Lezica, R., and Canut, H. (2005) Phytochemistry 66 453-461 [DOI] [PubMed] [Google Scholar]
- 42.Lips, S. H., and Beevers, H. (1966) Plant Physiol. 41 709-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen, F. S., Eisenberg, M., and McLaughlin, S. (1977) J. Membr. Biol. 37 361-396 [DOI] [PubMed] [Google Scholar]
- 44.Hampele, I. C., D'Arcy, A., Dale, G. E., Kostrewa, D., Nielsen, J., Oefner, C., Page, M. G., Schonfeld, H. J., Stuber, D., and Then, R. L. (1997) J. Mol. Biol. 268 21-30 [DOI] [PubMed] [Google Scholar]
- 45.Rébeillé, F., Macherel, D., Mouillon, J. M., Garin, J., and Douce, R. (1997) EMBO J. 16 947-957 [DOI] [PMC free article] [PubMed] [Google Scholar]