Abstract
Choanoflagellates, unicellular organisms that are closely related to metazoans, possess cell adhesion and signaling proteins previously thought to be unique to animals, suggesting that these components may have played roles in the evolution of metazoan multicellularity. We have cloned, expressed, and purified the nonreceptor tyrosine kinase MbSrc1 from the choanoflagellate Monosiga brevicollis. The kinase has the same domain arrangement as mammalian Src kinases, and we find that the individual Src homology 3 (SH3), SH2, and catalytic domains have similar functions to their mammalian counterparts. In contrast to mammalian c-Src, the SH2 and catalytic domains of MbSrc1 do not appear to be functionally coupled. We cloned and expressed the M. brevicollis homolog of c-Src C-terminal kinase (MbCsk) and showed that it phosphorylates the C terminus of MbSrc1, yet this phosphorylation does not inhibit MbSrc to the same degree seen in the mammalian Src/Csk pair. Thus, Src autoinhibition likely evolved more recently within the metazoan lineage, and it may have played a role in the establishment of intercellular signaling in metazoans.
A defining feature of multicellular organisms is the cellular machinery that enables cell-cell interaction and communication. Recent analyses show that an unexpected diversity of cell signaling and adhesion components are present in choanoflagellates, a group of unicellular organisms that are closely related to metazoans (1, 2). The presence of these proteins in choanoflagellates demonstrates that they evolved before the origin of animals and suggests that they may have served as preadaptations for the evolution of multicellularity (3).
The genome of the choanoflagellate Monosiga brevicollis contains 128 tyrosine kinase genes.3 Expressed sequence tags (ESTs) generated from an M. brevicollis cDNA library demonstrated the expression of members of the receptor-tyrosine kinase and nonreceptor-tyrosine kinase families. Anti-phosphotyrosine Western blotting of M. brevicollis cell lysates confirmed the phosphorylation of cellular proteins in response to treatment with nutrients (1). M. brevicollis contains four Src kinase homologs (designated MbSrc1-4) (1), each of which contains the diagnostic SH34 and SH2 domains of mammalian Src kinases. Src family nonreceptor-tyrosine kinases are widely expressed in animals and are involved in the regulation of many processes including cell growth and differentiation, adhesion, and motility (4-6). Mutation of Src or elevated Src expression or activity can lead to cell transformation and metastasis (7, 8) (Src was initially identified as the cellular homolog of the transforming gene from Rous sarcoma virus). For this reason the normal activity of Src in animal cells is tightly regulated. Src kinases possess a conserved domain architecture that is critical for enzymatic regulation; they have an N-terminal myristoylation sequence followed by SH3, SH2, and kinase catalytic domains. The C terminus possesses a tyrosine (Tyr-530) that is phosphorylated by the tyrosine kinase Csk (4, 5) (human c-Src numbering is used throughout this paper). Phosphorylation at Tyr-530 by Csk produces an intramolecular interaction between the C-terminal tail and the enzyme SH2 domain that inhibits enzyme activity (9). Src is also maintained in a low activity state by intramolecular interactions involving the SH3 domain. The need to keep Src activity in check may have evolved in parallel with Src expression itself; the Csk-mediated regulatory system is present along with Src in the primitive sponge Ephydatia fluviatilis (10) and in hydra (11).
Recent functional studies of Src kinases from the choanoflagellate Monosiga ovata showed that this organism possesses orthologs of Csk and Src (10). The activity of M. ovata Src, however, was not completely inhibited by co-expression with Csk. Here, we report a biochemical characterization of a choanoflagellate Src, MbSrc1, from M. brevicollis. Despite their shared genus name, M. ovata and M. brevicollis are only distantly related within the choanoflagellates and are derived from distinct clades.5 Comparison of Src kinases from M. ovata and M. brevicollis, two evolutionarily remote choanoflagellates, provides a deeper understanding of the conservation of Src function within choanoflagellates and provides insight into the state of Src regulation before the origin of metazoans. Furthermore, unlike M. ovata, the genome sequence of M. brevicollis is now available (12), opening the door for genomic and proteomic approaches for studying choanoflagellate Src signaling. Our mechanistic studies show that the individual domains of MbSrc1 share many functional properties with Src kinases from multicellular animals and yet lack the interdomain communication in MbSrc1 seen in mammalian Src. In particular, direct phosphorylation of the C-terminal tyrosine of MbSrc1 by a Csk homolog from M. brevicollis (MbCsk) fails to shut down the activity of MbSrc1. The tight degree of Csk regulation observed in mammalian Src kinases apparently arose more recently in the evolution of metazoans and may have contributed to the establishment of metazoan cell-cell communication.
MATERIALS AND METHODS
cDNA Cloning—MbSrc1 was originally identified in an EST study (1). A BLASTp search within the JGI M. brevicollis v1 genome browser was used to identify and verify the sequence, protein ID 44220, within the M. brevicollis genome (12). MbSrc1 was amplified by PCR from an M. brevicollis cDNA library (2). For baculovirus expression, MbSrc1 DNA (encoding residues 36-495) was cloned into the BamHI and XbaI sites of pFastBac-Htb (Invitrogen). FLAG-tagged full-length MbSrc1 was expressed in mammalian cells by cloning into the XbaI and BamHI sites of p3XFLAG-CMV (Sigma). The MbSrc1 SH2 domain (residues 107-207) and SH3 domain (residues 45-105) were expressed in Escherichia coli by cloning into the BamHI and EcoRI sites of pGEX-4T-1 (GE Healthcare). Full-length MbCsk cDNA was amplified by PCR from the M. brevicollis cDNA library and cloned into the BamHI and HindIII sites of plasmid pXJ-HA for mammalian expression. MbCsk was also cloned into the BamHI and EcoRI sites of pGEX-4T-1 for bacterial expression. All constructs were confirmed by DNA sequencing.
Alignments and Identity Calculations—Amino acid sequences retrieved from the NCBI Entrez protein data base were aligned and formatted using the ClustalW (13) and BOXSHADE (Boxshade Version 3.3.1 by Kay Hofmann and Michael D. Baron) programs, respectively, within the SDSC Biology Workbench 3.2. The accession numbers for the sequences used in the Src alignment are: Src Hsap, NP_005408.1; Src64B Dmel, P00528; STK Hvul, AAA29217.1; Src6 Evlu, BAB83688.1; SrcFv Mova, BAF02920.1. The accession numbers for the sequences used in the Csk alignment are: Csk Hsap. NP_004374.1; Csk Dmel, NP_731607.1; Csk Hvul, AAC35011.1; Csk Eflu, BAA81712.3; Csk Mova, BAF02917.1. Percent amino acid identities were calculated using WU-blast (14).
Protein Expression and Purification—GST-tagged proteins (MbSrc1 SH2 and SH3 domains and MbCsk) were expressed in 1-liter E. coli cultures and purified using glutathione-agarose (Molecular Probes) as described (16). For experiments requiring soluble GST-tagged proteins, the proteins were eluted from glutathione-agarose by incubation with 20 mm glutathione in 50 mm Tris (pH 8.0). His-tagged MbSrc1 was expressed in Sf9 insect cells using the Bac-to-Bac system (Invitrogen). Sf9 cells were grown at 27 °C in Excel 401 medium (JRH Biosciences) with 2.5% heat-inactivated fetal bovine serum (Sigma) and 1% penicillin/streptomycin/amphotericin B (Invitrogen). Sf9 cells (600 ml) were infected with recombinant MbSrc1 baculovirus at a multiplicity of infection of 10 plaque-forming units/cell for a period of 72 h. MbSrc1 was purified using nickel-nitrilotriacetic acid resin (Qiagen) as described previously (17). Purified MbSrc1 was concentrated in an Ultrafree-10 concentrator (Millipore) and stored in 40% glycerol at -20 °C.
Tyrosine Kinase Assays—MbSrc1 assays were performed with [γ-32P]ATP using the phosphocellulose paper binding assay (16, 18). Reaction mixtures contained 20 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 0.1 mm Na2VO4, 0.5 mm dithiothreitol, 0.25 mm ATP, varying concentrations of peptide substrate, and [γ-32P]ATP (200-400 cpm/pmol). Peptide substrates were synthesized on an Applied Biosystems automated 431A peptide synthesizer. The peptides were purified by reverse-phase high pressure liquid chromatography and characterized by matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry. Screens of MbSrc1 specificity were carried out with the following peptides: protein kinase A, LRRASLG; Src, AEEEIYGEFEAKKKKG; Abl, EAIYAAPFAKKKG; epidermal growth factor receptor, AEEEEYFELVAKKKG; insulin receptor, KKEEEEYMMMMG; Src autophosphorylation site, RRLIEDAEYAARG; SH3 binding substrate, AEEEIYGEFGGRGAAPPPPPVPRGRG; SH3 control substrate, AEEEIYGEFGGRGAAAAAAAVARGRG; SH2 binding substrate, RRLEDAIYAAGGGGGEPPQpYEEIG (pY is phosphotyrosine); SH2 control substrate, RRLEDAIYAAGGGGGEPPQFEEIG. Steady-state kinetic measurements were performed using a continuous spectrophotometric kinase assay (19, 20) with varying concentrations of ATP and the insulin receptor substrate. For assays of Csk-treated MbSrc, RCM-lysozyme was used as the substrate to minimize Csk activity (21). The reactions were stopped by spotting onto Whatman No. 3MM paper and washing with 5% trichloroacetic acid at 55 °C. Phosphorylated RCM-lysozyme was counted by liquid scintillation counting.
MbCsk was assayed with [γ-32P]ATP and poly(Glu4-Tyr) as described (22). The phosphorylated poly(Glu4-Tyr) was spotted onto Whatman No. 3MM paper, washed with 5% trichloroacetic acid at 55 °C, and counted by liquid scintillation counting. To measure the stoichiometry of MbSrc1 phosphorylation by MbCsk, reactions were carried out in the presence of [γ-32P]ATP and analyzed by SDS-PAGE as described previously (23).
Mass Spectrometry—MbSrc1 (15 μm) was dephosphorylated by treatment with immobilized GST-tagged Yersinia tyrosine phosphatase (YOP) for 45 min at room temperature. After removal of GST-YOP by centrifugation, MbSrc1 was incubated with MbCsk (13 μg) and 0.25 mm ATP in kinase buffer for 45 min at 30 °C. A control reaction was carried out without MbCsk. The reactions were subjected to SDS-PAGE, and MbSrc1 bands were excised from the gel, cut into small pieces, and transferred to siliconized microcentrifuge tubes. After washing with 50% methanol and 5% acetic acid, gel fragments were dehydrated by the addition of acetonitrile. Proteins were reduced and alkylated by the addition of dithiothreitol and iodoacetamide and digested with trypsin in 50 mm ammonium bicarbonate overnight at 37 °C. The tryptic peptides were extracted by treatment with 50% (v/v) acetonitrile and 5% (v/v) formic acid in water. MALDI-TOF mass spectrometry was carried out on a Voyager DE-STR Biospectrometry work station (ABI). Mapping of the C-terminal phosphotyrosine by liquid chromatography-tandem mass spectroscopy (LC/MS/MS) was carried out on an API QSTAR Pulsar LC/MS/MS system (Applied Biosystems/MDS SCIEX, Foster City, CA) equipped with a Protana nanospray source (Protana Engineering A/S, Starmosegardsvej, Denmark) and an UltiMate capillary high pressure liquid chromatography system (LC Packings, San Francisco, CA).
Isothermal Titration Calorimetry (ITC)—Calorimetry experiments were performed in a VP-ITC MicroCalorimeter (MicroCal, Inc.) in buffer containing 25 mm MOPS (pH 6.8), 1 mm β-mercaptoethanol, 1 mm EDTA, and 50 mm NaCl (24). Protein and peptide solutions were dialyzed against ITC buffer and degassed before use. ITC experiments were performed with 30-60 μm MbSrc1 SH2 or SH3 domain in the ITC cell and 0.3-1.8 mm peptide in the injection syringe. Data for each titration were collected and analyzed by the Origin software package (MicroCal). The peptides used to measure SH2 binding were the pYEEI peptide (EPQpYEEIPIKQ) (19) and the MbSrc1 tail peptide (GSSpYNNPDQM). The peptide used for SH3 binding was a Pro-rich peptide (RGAAPPPPPVPRGRG) derived from the sequence of Sam68 (25).
Cell Transfection and Western Blotting—SYF cells were cultured in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum at 37 °C in 5% CO2. Cells were transfected at 50% confluency using TransIT polyamine transfection reagent (Mirus) at a ratio of 1:3.5 (DNA:TransIT) according to the manufacturer's instructions. Western blot analysis was performed after 48 h of transfection. Cells were lysed in 10 mm Tris-HCl (pH 7.4), 50 mm NaCl, 5 mm EDTA, 1% Triton X-100, 50 mm NaF, 2 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 1 mg/ml aprotinin, and 1 mg/ml leupeptin at 4 °C for 1 h. The cell lysates were centrifuged at 14,000 × g for 10 min at 4 °C. Protein concentrations were determined using a Bio-Rad protein assay. Lysates (25 μg) were separated on 7.5% SDS-PAGE and transferred onto polyvinylidene difluoride membrane. Proteins in the membranes were detected with antiphosphotyrosine antibody (4G10, Upstate), anti-FLAG antibody (M2, Sigma), and anti-Tyr(P)-419 antibody (BIOSOURCE) by ECL (GE Healthcare).
For SH2/SH3 pulldown experiments, immobilized GST-SH2, GST-SH3, or GST (25 μg in 50 μl) were incubated with 250 μl of SYF cell lysate (1.6 mg/ml protein) for 1 h at 4 °C. In some experiments competitor peptides (pYEEI or Pro-rich peptide) were added to the reactions. The GST proteins were collected by centrifugation, washed 3× with lysis buffer, and analyzed by SDS-PAGE. Bound proteins were visualized by Western blotting with anti-phosphotyrosine antibody. Immobilized pYEEI peptide was prepared by coupling 0.5 mg of pYEEI to Affi-Gel 15 resin (Bio-Rad) as described (25). Lysates from SYF cells were incubated with pYEEI resin for 45 min at 4 °C. Bound MbSrc1 was visualized by Western blotting with anti-FLAG antibody.
Luciferase Reporter Assay—The SYF cells were seeded in 12-well plates (5 × 104 cells/well) and transfected in triplicate with protein expression plasmids, interferon-gamma activation site luciferase reporter plasmid, or a promoter-less Renilla luciferase plasmid (pRL-null) as a transfection control (gifts from Dr. Nancy Reich, Stony Brook University). Transfected cells were harvested after 48 h. Preparation of cell lysates and measurement of light units were carried out using the Dual-Luciferase Reporter Assay system (Promega) following the manufacturer's instructions. Relative light units were calculated as (firefly luciferase activity/Renilla luciferase activity).
RESULTS
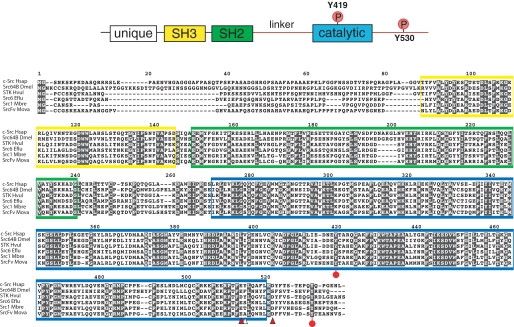
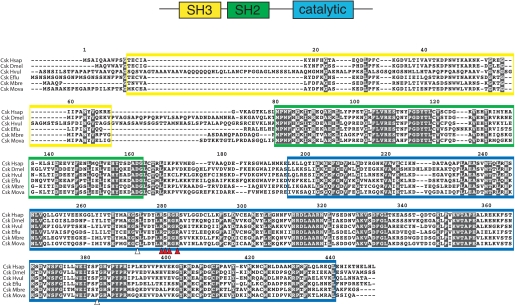
Cloning MbSrc1 from M. brevicollis—A cDNA encoding MbSrc1 was amplified by PCR from an M. brevicollis cDNA library. The domain architecture of MbSrc1 matches that of mammalian Src kinases; the enzyme possesses (from N terminus to C terminus) unique, SH3, SH2, and tyrosine kinase catalytic domains (Fig. 1). The overall amino acid identity with human c-Src is 57% (additional identities are listed in supplemental Fig. S1). The tyrosine residues involved in mammalian Src regulation are conserved in MbSrc1. Tyrosine 378 of MbSrc1 corresponds to the autophosphorylation site within the activation loop (Tyr-419) of c-Src, which is a site of enzyme activation (9, 20). The C-terminal negative regulatory site of c-Src (Tyr-530) is also conserved in MbSrc1 (Tyr-489). In mammalian Src kinases, phosphorylation of Tyr-530 by Csk represses enzymatic activity by promoting an intramolecular SH2-tail interaction (4, 5, 9). Two of the three amino acids in c-Src that are important for recognition by Csk are conserved in MbSrc1 (21); the sole exception is the position corresponding to Tyr-514 in c-Src, which is a threonine in MbSrc1 (Fig. 1). In mammalian Src kinases, polyproline sequences in the SH2 kinase linker bind intramolecularly to the SH3 domain and play an important role in enzyme regulation (9, 26-28). The sequences of the SH2 kinase linker in c-Src and MbSrc1 are divergent, with an extra four residues present in MbSrc1. Proline residues are present in the MbSrc1 linker, but it is not clear whether these sequences have the capacity to serve as SH3 ligands. Trp-263, an important regulatory site within the linker in c-Src (29), is conserved between MbSrc1 and c-Src. Thus, MbSrc1 potentially has most (if not all) of the regulatory sequences of mammalian Src kinases.
FIGURE 1.
Cloning of MbSrc1. MbSrc1 was isolated by PCR from a M. brevicollis cDNA library, as described under “Materials and Methods.” The domain structure of Src kinases is shown at the top with the regulatory tyrosines Tyr-419 and Tyr-530 (human c-Src numbering) indicated. The alignment of Src sequences shows human (c-Src Hsap), fly (Src648 Dmel), hydra (STK Hvul), sponge (Src6 Eflu), M. brevicollis (Src1 Mbre), and M. ovata (SrcFv Mova). The SH3 domain sequence is shaded yellow, the SH2 domain is green, and the catalytic domain is blue. Red circles on the sequence indicate the positions of tyrosines corresponding to Tyr-419 and Tyr-530. Arrowheads indicate residues in c-Src that are important for recognition by Csk (21); red arrowheads show residues that are conserved between c-Src and MbSrc1, and the white arrowhead shows a non-conserved residue.
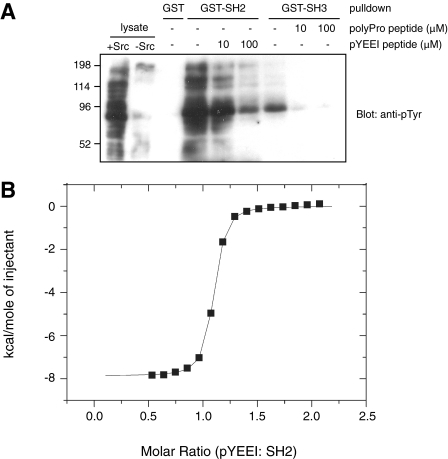
The SH2 and SH3 Domains of MbSrc1 Are Functional—The SH2 and SH3 domains of mammalian Src kinase play dual roles; they are involved in autoinhibitory interactions, and they also have a positive role in targeting the kinases to cellular substrates (4, 5, 30). It is not currently known whether these two roles are operational in earlier branching phyla, although the presence of Csk in sponges and in hydra suggests that the autoinhibitory role of the SH2 domain is intact (10, 11). Both of these roles depend on the ability of the domains to engage in protein-protein interactions. To test whether these binding properties are present in the MbSrc1 domains, we expressed the isolated domains as glutathione S-transferase fusion proteins in E. coli and purified them. We immobilized the GST-SH2 and GST-SH3 proteins and incubated them with lysates from Src-expressing mammalian cells (Fig. 2A). Western blotting experiments showed that the SH2 domain bound to tyrosine-phosphorylated proteins in the lysates. The SH3 domain also bound to at least one tyrosine-phosphorylated protein. Reprobing with a specific antibody showed that one bound protein was p130Cas, which contains C-terminal ligands for the Src SH2 and SH3 domains (31, 32) (supplemental Fig. S2). Binding to the MbSrc1 SH2 domain was inhibited by the inclusion of a phosphopeptide containing the optimal binding ligand for the mammalian c-Src SH2 domain (pYEEI) (Fig. 2A). Similarly, a proline-rich synthetic peptide (derived from the sequence of Sam68, an Src SH3 binding partner (33)) blocked the ability of the MbSrc1 SH3 domain to bind to the phosphorylated protein.
FIGURE 2.
The MbSrc1 SH2 and SH3 domains are functional. A, immobilized GST-SH2, GST-SH3, or GST were incubated with Src-transfected SYF cell lysate for 1 h at 4 °C. Competitor peptides were added to the incubations as indicated. The GST proteins were collected by centrifugation, washed with lysis buffer, and eluted with SDS-PAGE sample buffer. Bound proteins were visualized by anti-Tyr(P) Western blotting. The left two lanes show 25 μg of Src-transfected and untransfected SYF lysate, respectively, loaded directly onto the gel. B, binding of the MbSrc1 SH2 domain to pYEEI peptide measured by ITC. A representative titration is shown using 60 μm SH2 domain and 5-μl injections of 1.8 mm pYEEI peptide. The dissociation constant determined from this titration was 140 nm.
We used isothermal titration calorimetry to measure the binding affinities of the MbSrc1 SH2 and SH3 domains for their ligands. MbSrc1 bound to the pYEEI peptide with a dissociation constant (Kd) of 140 nm (Fig. 2B). This is similar to the value of 200 nm reported for the pYEEI peptide binding to the isolated SH2 domain of mammalian c-Src (34). ITC experiments on the MbSrc1 SH3 domain gave a Kd value of 2.1 μm for binding to the proline-rich peptide from Sam68 (supplemental Fig. S3); this is in the range of values typically observed for c-Src SH3 domain binding to polyproline peptides (1-10 μm) (35). Overall, the results of the binding experiments suggest that the MbSrc1 SH2 and SH3 domains share the basic function of their mammalian counterparts and that the ligand specificity is broadly similar.
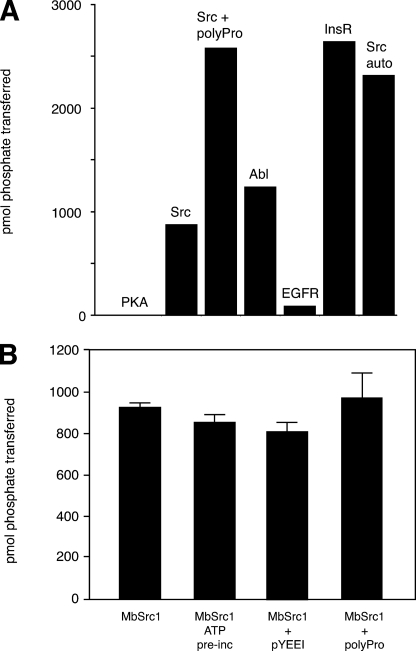
Enzymatic Activity of MbSrc1—We cloned the MbSrc1 sequence (encoding residues 36-495, from the beginning of the SH3 domain to the C terminus of MbSrc1) into a baculovirus expression vector. Similar SH3-SH2 kinase constructs of mammalian Src kinases retain the essential features of regulation and avoid potential complications due to membrane association (9). We infected Sf9 insect cells with the recombinant baculovirus and purified the protein. MbSrc1 was highly active toward synthetic peptide substrates, with a catalytic efficiency comparable with mammalian Src kinases. We screened MbSrc1 with peptide substrates containing recognition motifs for four different tyrosine kinases (36) (Src, Abl, epidermal growth factor receptor, and insulin receptor). MbSrc1 preferred the insulin receptor substrate from this group of peptides (Fig. 3A). Steady-state kinetic experiments indicated that the Km for this substrate was 609 μm, and the kcat was 50 min-1 (for comparison, the Src family kinase Hck displayed a kcat value of 60 min-1 toward a preferred substrate (29)). MbSrc had no activity toward Kemptide, a substrate for the Ser/Thr kinase protein kinase A (37) (Fig. 3A). MbSrc1 phosphorylated a peptide modeled on the Tyr-419 autophosphorylation sequence of mammalian Src, suggesting that MbSrc1 might also be capable of autophosphorylation. The enzyme also phosphorylated an Src substrate to which a polyproline sequence was attached (38) (Fig. 3A), providing evidence that the SH3 domain of MbSrc1 is capable of substrate targeting (see below).
FIGURE 3.
Activity of purified MbSrc1. A, MbSrc1 (0.7 μm) was screened with various synthetic peptides (200 μm) using the phosphocellulose paper assay. The reactions proceeded for 20 min at 30 °C and were analyzed by scintillation counting. PKA, protein kinase A; EGFR, epidermal growth factor receptor; InsR, insulin receptor. B, MbSrc1 (0.7 μm) was assayed with the Src substrate peptide (200 μm) alone or in the presence of pYEEI peptide (88 μm) or the Pro-rich peptide (200 μm) for 10 min at 30 °C. For the ATP reaction, MbSrc1 (4.6 μm) was first preincubated with ATP (0.5 mm) for 45 min at 4 °C.
In mammalian Src kinases, exogenous ligands for the SH2 and SH3 domains can disrupt the autoinhibitory interactions, promote autophosphorylation at Tyr-419, and stimulate Src kinase activity (28, 39-41). Preincubation of down-regulated Src kinases with ATP achieves the same effect because autophosphorylation weakens the intramolecular SH2 and SH3 interactions (19, 42). Incubation of purified MbSrc1 with ATP or peptide ligands for the SH2 or SH3 domain did not significantly activate the enzyme (Fig. 3B). The concentrations of ATP and ligands used were sufficient to activate the Src kinase Hck (28). These results indicate that this form of MbSrc1 was not autoinhibited.
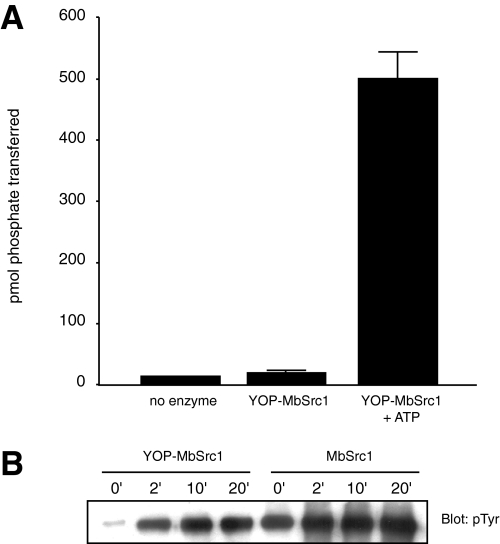
We investigated the possibility that the form of MbSrc1 purified from Sf9 cells was already phosphorylated at Tyr-419. We treated MbSrc1 with YOP tyrosine phosphatase, then assayed enzymatic activity before and after ATP incubation (Fig. 4A). YOP-treated MbSrc1 showed only background levels of activity, and ATP treatment led to a large increase in tyrosine kinase activity. These data suggested that MbSrc1, like mammalian Src kinases, is subject to regulation by autophosphorylation. Anti-phosphotyrosine Western blotting confirmed that the MbSrc1 purified from Sf9 cells was phosphorylated and that YOP treatment substantially dephosphorylated the enzyme. Incubation with ATP led to a rapid increase in phosphorylation of YOP-treated MbSrc1 (Fig. 4B).
FIGURE 4.
Autophosphorylation of MbSrc1. A, MbSrc1 (10 μl of 46 μm) was incubated with immobilized GST-YOP in 30 mm Tris (pH 7.5), 10 mm MgCl2 (200-μl final volume). The reaction was mixed at room temperature for 30 min. After removal of GST-YOP by centrifugation, MbSrc1 was assayed directly or after incubation with 0.5 mm ATP for 30 min at 30 °C. B, YOP-treated MbSrc1 and untreated MbSrc1 were incubated with 0.5 mm ATP at 30 °C. Aliquots were removed at the indicated times and analyzed by Western blotting with anti-phosphotyrosine antibody.
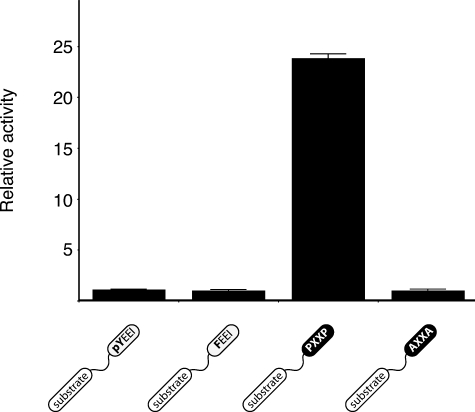
In addition to their negative regulatory role, the SH2 and SH3 domains of Src kinases play an important role in substrate recognition. This targeting role can be recapitulated in vitro; peptide substrates incorporating SH2 or SH3 ligands are phosphorylated more efficiently than control peptides (38, 43). We tested MbSrc1 with a peptide substrate linked to the pYEEI SH2 domain ligand. Phosphorylation of this peptide was similar to a control peptide in which the phosphotyrosine in the pYEEI motif was changed to Phe to prevent SH2 domain binding (Fig. 5), although MbSrc1 was capable of binding to the pYEEI substrate (supplemental Fig. S4). Peptides with longer or shorter linker lengths between the phosphorylation site and pYEEI also showed no preferential phosphorylation (supplemental Fig. S5). In contrast, when an SH3 ligand was incorporated into a substrate, phosphorylation was much higher than a control (Fig. 5). These data suggest that, whereas SH3-mediated targeting is intact in MbSrc1, the SH2 domain of MbSrc1 does not target the enzyme to substrates. The data also raise the possibility that the coupling between the tyrosine kinase and SH2 domains was not functional in the ancestral form of Src.
FIGURE 5.
Phosphorylation of peptides containing SH2 or SH3 ligands. MbSrc1 (0.7 μm) was assayed with 50 nm concentrations of the SH2-binding (pYEEI) substrate, SH2 control (FEEI) substrate, SH3 binding (PXXP) substrate, or SH3 control (AXXA) substrate. Reactions were at 30 °C for 6 min. Activities relative to control substrates are shown with S.D. Similar results were obtained at 90 nm concentrations of MbSrc1.
Cloning and Characterization of MbCsk—To test whether the regulatory properties of mammalian Src kinases were present in MbSrc1, we cloned the Csk ortholog from M. brevicollis. We identified one putative MbCsk (protein ID 33686) from the M. brevicollis genome by characteristic sequences of this tyrosine kinase family and used a PCR strategy to clone the gene from a cDNA library. MbCsk contains SH3, SH2, and kinase catalytic domains (Fig. 6), and the enzyme possesses 51% amino acid identity with human Csk (supplemental Fig. S6). Previous work on mammalian Csk has demonstrated the presence of a docking site that enables recognition of Src (44). In MbCsk, two of the six substrate-docking residues are not conserved (corresponding to Ser-273 and Phe-382 of human Csk) (Fig. 6). Ser-273 of Csk is conserved in human, fly, and hydra Csk homologs but not conserved in sponge, M. ovata, or M. brevicollis. Phe-382 is conserved in all of these species except fly and M. brevicollis (Fig. 6). This suggests that MbCsk may have low activity toward mammalian Src kinases.
FIGURE 6.
Isolation of MbCsk. MbCsk was isolated by PCR from an M. brevicollis cDNA library. The figure shows the domain arrangement of Csk and an alignment of human, fly, hydra, sponge, M. brevicollis, and M. ovata Csk sequences. The SH3 domain sequence is shaded yellow, the SH2 domain is green, and the catalytic domain is blue. Arrowheads indicate substrate docking sites in Csk that are important for Src recognition (44). Red arrowheads show residues that are conserved between human Csk and MbCsk, and the white arrowheads show non-conserved residues.
We expressed full-length MbCsk as a GST fusion protein in E. coli and purified the enzyme. We assayed enzyme activity using the synthetic substrate poly(Glu4-Tyr). MbCsk possessed tyrosine kinase activity, although the specific activity was lower than a comparable preparation of mammalian Csk (Fig. 7A).
FIGURE 7.
Activity of MbCsk. A, GST-tagged human Csk or MbCsk (175 nm) were assayed with 1 mg/ml poly(Glu4-Tyr) for 30 min at 30 °C. Reactions were analyzed by scintillation counting. B, phosphorylation of the kinase-dead K296M mutant of MbSrc1 (kd-MbSrc1) by MbCsk. The reaction mixture contained 20 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 2 mm MnCl2, 0.1 mm sodium orthovanadate, 0.55 mm [γ-32P]ATP, 11 μm MbCsk, and 8.7 μm kd-MbSrc1. At the indicated time points, aliquots were removed and quenched by the addition of Laemmli sample buffer and boiling. Reactions were analyzed by 9% SDS-PAGE and autoradiography.
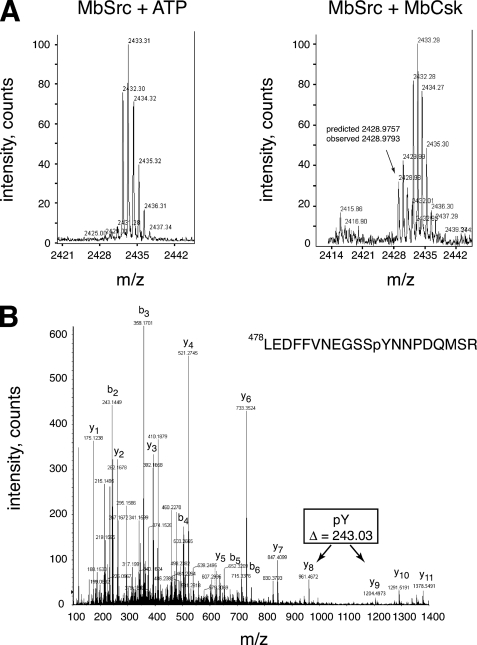
To determine whether MbSrc1 and MbCsk function as a regulatory pair, we first carried out phosphorylation experiments in vitro. To eliminate the possibility of MbSrc1 autophosphorylation, we produced a kinase-dead form of MbSrc1 (K256M) and purified it from Sf9 cells. Incubation of kinase-dead MbSrc1 with MbCsk in the presence of γ-32P-labeled ATP led to a time-dependent phosphorylation of MbSrc1, as demonstrated by autoradiography (Fig. 7B). We measured the stoichiometry of MbSrc1 phosphorylation to be 0.91 ± 0.12 under these conditions. We carried out mass spectrometry experiments to confirm that this phosphorylation occurred at the C-terminal tyrosine of MbSrc1. YOP-treated MbSrc1 was incubated together with ATP or with ATP plus MbCsk. After separation by SDS-PAGE and in-gel trypsin digestion, MALDI-TOF mass spectrometry showed that only the MbCsk-treated sample contained a peak at 2428.97 daltons, the molecular mass expected for the C-terminal MbSrc1 tryptic peptide (Leu-478—Met-495) plus the mass of a phosphate (80 daltons) (Fig. 8A). Peptide mapping by liquid chromatography/tandem mass spectrometry showed that this peptide was phosphorylated at Tyr-489 (Fig. 8B).
FIGURE 8.
Phosphorylation of the MbSrc1 C terminus by MbCsk. A, YOP-treated MbSrc1 was incubated in the presence or absence of MbCsk as described above for 45 min at 30 °C. Reactions were separated by SDS-PAGE, and gel bands corresponding to MbSrc1 were excised and subjected to in-gel tryptic digestion. The tryptic peptides for the control reaction (left) or MbCsk reaction (right) were analyzed by MALDI-TOF mass spectrometry. The position of the peak arising from the MbSrc1 C-terminal tail is indicated, with the predicted mass based on a phosphorylated sequence. This sequence is derived from residues Leu-478—Met-495 of MbSrc1, plus a C-terminal SR dipeptide derived from the baculovirus vector. Supplemental Fig. S7 presents MALDI-TOF data with an expanded m/z range. B, tandem mass spectrometric analysis of the C-terminal tryptic phosphopeptide from MbSrc1 shown in panel B. The b- and y-series of fragmentation ions are indicated. The difference in mass between the y8 and y9 ions indicate that the peptide is phosphorylated at the position corresponding to Tyr-489.
To test whether MbCsk-mediated tail phosphorylation affects the enzymatic activity of MbSrc1, we incubated YOP-treated MbSrc1 with MbCsk or mammalian Csk. We assayed the activity of MbSrc1 toward RCM-lysozyme under conditions in which Csk does not phosphorylate this substrate (21). Neither MbCsk nor mammalian Csk gave any inhibition of MbSrc1 (Fig. 9). In contrast, phosphorylation of a mammalian Src kinase (Hck) by mammalian Csk led to 65% inhibition under the same conditions, and phosphorylation of Hck by MbCsk led to 18% inhibition (Fig. 9). Thus, despite the ability of MbCsk to phosphorylate MbSrc1 at the C-terminal tyrosine, the pair of enzymes does not display the regulatory features characteristic of the mammalian Src/Csk system.
FIGURE 9.
Activity of MbCsk-treated MbSrc1. YOP-treated Hck or MbSrc1 (0.8 μm) were incubated in MbCsk assay buffer alone or in the presence of GST-Csk or GST-MbCsk (2 μm) for 15 min at 30 °C. The enzymatic activities of Hck and MbSrc1 were then measured using [γ-32P]ATP and RCM-lysozyme (0.4 mg/ml). Reactions proceeded for 1 min at 30 °C and were analyzed by spotting on Whatman No. 3MM paper, washing in warm 5% trichloroacetic acid, and scintillation counting.
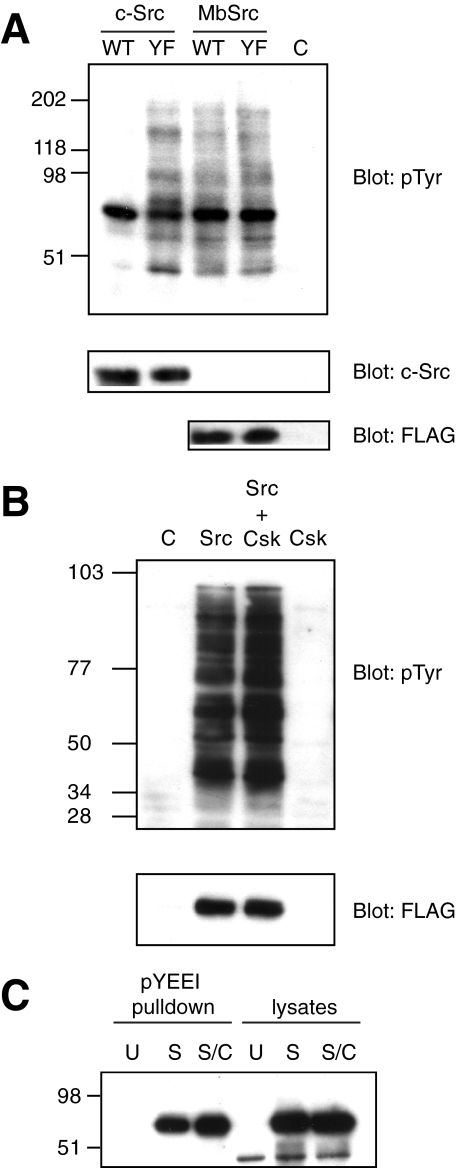
Functional Studies of MbSrc1 in Mammalian Cells—We produced a FLAG-tagged version of full-length MbSrc1 and expressed the kinase in Src/Yes/Fyn triple knock-out (SYF) cells, which lack all Src family kinases (45). MbSrc1 displayed tyrosine kinase activity toward protein substrates in these cells, as demonstrated by anti-phosphotyrosine Western blotting of whole cell lysates (Fig. 10A). A Y489F mutation of the C-terminal tyrosine did not give any significant increase in kinase activity (Fig. 10A). In contrast, mutation of Tyr-530, the C-terminal tyrosine of mammalian Src, led to a substantial activation of the kinase, consistent with previous reports (4, 5) (Fig. 10A). The levels of wild-type and Y489F MbSrc1 activity in these experiments were similar to the activity of Y530F Src. We note that MbSrc1 contains an MGC sequence at the N terminus, which in mammalian Src kinases promotes fatty acylation and membrane attachment. Introduction of the FLAG tag at the N terminus presumably disrupts membrane attachment, which could affect the activity of MbSrc1 toward cellular substrates.
FIGURE 10.
Functional studies of MbSrc1 in mammalian cells. A, SYF cells were transfected with wild-type (WT) or Y530F (YF) forms of human c-Src, WT, or Y489F forms of MbSrc1 or empty vector (C). Lysates were analyzed by SDS-PAGE and anti-Tyr(P) Western blotting. The blots were stripped and analyzed with anti c-Src antibody and with anti-FLAG antibody. B, SYF cells were transfected with MbSrc1 and MbCsk alone or in combination. Lysates were analyzed by Western blotting using anti-Tyr(P) and anti-FLAG (for MbSrc1) antibodies. C, immobilized pYEEI peptide was incubated with lysates from untransfected SYF cells (U), cells transfected with MbSrc alone (S), or cells transfected with MbSrc plus MbCsk (S/C) in buffer containing 50 mm Tris (pH 7.5), 250 mm NaCl, 0.1% Triton X-100, 5 mm EDTA, 0.5 mm sodium vanadate, and 1 mm dithiothreitol. After washing, bound MbSrc was visualized by anti-FLAG immunoblotting. Lysates (25 μg) were loaded directly onto the gel in the right-hand lanes.
Co-expression of hemagglutinin-tagged MbCsk in SYF cells did not inhibit MbSrc1 activity (Fig. 10B). To test for the presence of an intramolecular SH2-tail interaction in MbSrc1, we incubated lysates of MbSrc1- or MbSrc1/MbCsk-expressing SYF lysates with immobilized pYEEI peptide. An intramolecular SH2-tail interaction would be expected to decrease the amount of MbSrc1 in the SH2-open conformation available to bind to immobilized pYEEI, as we showed previously for Hck (3, 29). We observed no reduction in pYEEI binding upon co-expression with MbCsk (Fig. 10C). One possibility is that the affinity of the MbSrc1 C-terminal tail peptide is too low to interact stably with the SH2 domain. To test this possibility, we synthesized a phosphotyrosine-containing peptide with the sequence of the MbSrc1 C terminus. We measured the dissociation constant for this peptide to the isolated SH2 domain as 9.7 ± 0.9μm by isothermal titration calorimetry. This value is substantially weaker than the 0.2 μm measured for the pYEEI-containing peptide.
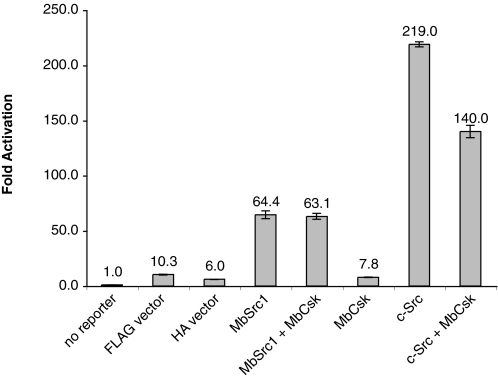
We tested the ability of MbCsk to regulate MbSrc1 using a Src-responsive reporter gene assay. MbSrc1 can functionally substitute for mammalian Src in this assay, which depends on Src phosphorylation of STAT (signal transducers and activators of transcription) molecules (12). Co-expression of MbCsk did not reduce the ability of MbSrc1 to activate the reporter construct (Fig. 11). In contrast, MbCsk partially inhibited the in vivo activity of mammalian c-Src, consistent with the results shown above in Fig. 9. Taken together, the data suggest that MbCsk phosphorylation of the MbSrc1 C terminus does not lead to MbSrc1 inhibition.
FIGURE 11.
Src reporter assay. Luciferase activity was measured in lysates from SYF cells transfected with the indicated plasmids and the interferon-gamma activation site luciferase reporter plasmid. Experiments were performed in triplicate, and error bars show S.D.
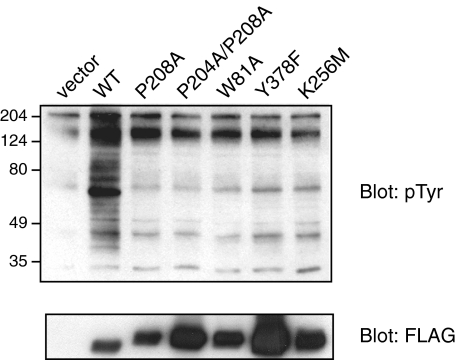
We carried out experiments to probe the molecular basis for the lack of inhibition of MbSrc1 by MbCsk. Conformational constraints play an important role in the binding of c-Src Tyr(P)-530 to the SH2 domain, and it is possible that these constraints are not present in MbSrc1. In particular, the divergent linker sequence between the SH2 and kinase domains (Fig. 1) may be unable to adopt a polyproline type II helical conformation that can bind to the SH3 domain. Targeted molecular dynamics simulations of the closed form of c-Src show that the SH3 and SH2 domains act in a concerted fashion to inhibit activity (46). Thus, inefficient SH3-linker binding would destabilize the intramolecular SH2-tail interaction in MbSrc1. Based on the structure of c-Src, we produced a series of MbSrc1 mutants in which SH3-linker interactions would be disrupted: (i) W81A, at a key residue within the SH3 domain for ligand recognition; (ii) P208A, at a residue within the linker that binds the SH3 domain of c-Src; (iii) P204A/P208A, at both proline residues within the linker. Each one of these mutations has been shown to strongly activate c-Src and related kinases (29, 47-49). In contrast, expression of these MbSrc1 mutants in SYF cells showed them to be less active than wild-type MbSrc1 (Fig. 12). Their activity was comparable with the autophosphorylation site (Y378F) and kinase-dead (K256M) mutants. In c-Src, Trp-263 is a key residue that links SH3-linker binding to kinase activation; a W263A mutation gives constitutively high kinase activity (29). We carried out a comparable mutation (W221A) in MbSrc1 (supplemental Fig. S8). In contrast to results for c-Src, the W221A mutation virtually eliminated kinase activity. Similar results were obtained for the c-Raf kinase, which showed that the corresponding tryptophan plays a structural role and that the kinase does not assume an autoinhibited conformation (50). Taken together, the data suggest that SH3-linker interactions are inefficient (or altogether absent) in MbSrc1, providing a potential explanation for the lack of regulation by C-tail phosphorylation.
FIGURE 12.
SH3-linker mutations of MbSrc1. SYF cells were transfected with the indicated wild-type (WT) or mutant forms of MbSrc1. Lysates were analyzed by SDS-PAGE and Western blotting with anti-Tyr(P) and anti-FLAG antibodies. Activities were compared with that of a Y378F mutant (at the equivalent of c-Src Tyr-419) and of the kinase-dead form (K256M).
DISCUSSION
To identify the gene products that may have enabled the transition from unicellular to multicellular organisms, metazoan genomes may be compared with genomes from their nearest relatives (3). These analyses have the potential to identify shared genes that allowed the evolution of cell-cell communication and signal transduction. Comparisons of choanoflagellate and metazoan nuclear and mitochondrial genes suggest a close phylogenetic relationship between the groups (51, 52). The presence of tyrosine kinases and associated signaling components (e.g. SH2 and SH3 domains) in M. brevicollis (1) indicates that these proteins evolved before the origin of animals. Analyses of the phosphotyrosine signaling machinery in M. brevicollis reveal highly divergent domain architectures and combinations when compared with metazoans (12).6 This suggests that the components evolved before the split between choanoflagellates and metazoans. To study an early stage in the evolution of phosphotyrosine signaling, we have investigated the common and distinct signaling properties of choanoflagellate and mammalian tyrosine kinases. We have taken a biochemical approach to compare Src kinases from M. brevicollis and mammals.
The isolated signaling domains of MbSrc1 (SH3, SH2, and kinase) are functionally very similar to the corresponding domains in mammalian Src kinases (Figs. 2 and 3A). One exception is that the substrate specificity of MbSrc1 is somewhat different from that of mammalian Src kinases, which strongly prefer a hydrophobic residue (e.g. Ile) at the P-1 position in substrates (15, 36), whereas MbSrc1 apparently does not (Fig. 3A). The potential significance of this difference in kinase specificity will be clearer when the substrates and effectors of MbSrc1 in M. brevicollis cells are identified. Despite this difference in specificity, MbSrc1 can replace mammalian Src kinases in functional assays (Fig. 8 and Ref. 12). These data indicate that MbSrc1 possesses the essential functional features of metazoan Src family kinases.
The mechanisms for interdomain communication and regulation in c-Src and MbSrc1 are divergent. Substrate targeting via the SH3 domain of MbSrc1 is similar to c-Src as measured by synthetic peptides (Fig. 5), although it is possible that more subtle differences between the enzymes would be apparent if their activities toward polyproline-containing cellular proteins were compared. In contrast to c-Src, the SH2 domain of MbSrc1 does not target the enzyme to phosphorylate synthetic peptides (Fig. 5). For mammalian Src kinases, SH2-directed targeting was not observed when the connector between the substrate sequence and the pYEEI sequence was too short to allow simultaneous binding to the kinase and SH2 domains (43). Thus, the spatial orientation of the kinase and SH2 domains in MbSrc1 may not be suited for recognition of such peptides and proteins.
Intramolecular interactions are also critical for maintaining the down-regulated state of mammalian Src kinases. It is clear that the form of MbSrc1 studied here is not autoinhibited (Fig. 3B). Although a Csk ortholog exists in M. brevicollis (Fig. 6) and the enzyme is capable of phosphorylating the C terminus of MbSrc1 (Figs. 7B and 8), no inhibition was observed (Figs. 9, 10 and 11). The lack of inhibition might be due in part to inefficient recognition of the phosphorylated C-tail by the SH2 domain (Kd = 9.7 μm); on the other hand, our data in Fig. 12 suggest that inefficient SH3-linker binding destabilizes the intramolecular SH2-tail interaction in MbSrc1.
Autoregulation is a hallmark of nonreceptor-tyrosine kinases in animal cells. The role of nonreceptor-tyrosine kinases in choanoflagellates is currently unknown, but they may play a role in detecting changes to the outside environment, such as the availability of nutrients (1). The regulatory properties of MbSrc1 are presumably tuned to the functional role of the enzyme in M. brevicollis. A definitive study of MbSrc1 function in vivo awaits the development of methods to introduce genes or to block their function in choanoflagellates. Our data suggest that the tight autoinhibition observed in mammalian Src kinases was absent in the ancestral form of Src. These results are in agreement with studies of Src kinases from M. ovata (10), and they are consistent with the proposal that stable negative regulation of Src kinases may correlate with the transition to animal multicellularity.
Supplementary Material
Acknowledgments
We thank Wendell Lim (University of California at San Francisco) for helpful discussions and a critical reading of the manuscript. We thank Nancy Reich and Yan Song (Stony Brook University) for reporter constructs and assistance with luciferase experiments and Hyejin Cho for mutagenesis of MbSrc1 W221.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EDR48627 and EDQ86681.
This work was supported, in whole or in part, by National Institutes of Health Grant CA58530 (to W. T. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S8.
Footnotes
G. Manning, personal communication.
The abbreviations used are: SH2 and SH3, Src homology 2 and 3, respectively; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight; GST, glutathione S-transferase; YOP, Yersinia tyrosine phosphatase; ITC, isothermal titration calorimetry; MOPS, 4-morpholinepropanesulfonic acid; SYF, Src/Yes/Fyn triple knock-out.
S. Baldauf, personal communication.
G. Manning, S. L. Young, W. T. Miller, and Y. Zhai, submitted for publication.
References
- 1.King, N., Hittinger, C. T., and Carroll, S. B. (2003) Science 301 361-363 [DOI] [PubMed] [Google Scholar]
- 2.King, N., and Carroll, S. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 15032-15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King, N. (2004) Dev. Cell 7 313-325 [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. T., and Cooper, J. A. (1996) Biochim. Biophys. Acta. 1287 121-149 [DOI] [PubMed] [Google Scholar]
- 5.Bjorge, J. D., Jakymiw, A., and Fujita, D. J. (2000) Oncogene 19 5620-5635 [DOI] [PubMed] [Google Scholar]
- 6.Parsons, S. J., and Parsons, J. T. (2004) Oncogene 23 7906-7909 [DOI] [PubMed] [Google Scholar]
- 7.Irby, R. B., and Yeatman, T. J. (2000) Oncogene 19 5636-5642 [DOI] [PubMed] [Google Scholar]
- 8.Frame, M. C. (2002) Biochim. Biophys. Acta 1602 114-130 [DOI] [PubMed] [Google Scholar]
- 9.Sicheri, F., and Kuriyan, J. (1997) Curr. Opin. Struct. Biol. 7 777-785 [DOI] [PubMed] [Google Scholar]
- 10.Segawa, Y., Suga, H., Iwabe, N., Oneyama, C., Akagi, T., Miyata, T., and Okada, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 12021-12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, M. A., Malik, I. A., Shenk, M. A., and Steele, R. E. (2000) Oncogene 19 3925-3930 [DOI] [PubMed] [Google Scholar]
- 12.King, N., Westbrook, M. J., Young, S. L., Kuo, A., Abedin, M., Chapman, J., Fairclough, S., Hellsten, U., Isogai, Y., Letunic, I., Marr, M., Pincus, D., Putnam, N., Rokas, A., Wright, K. J., Zuzow, R., Dirks, W., Good, M., Goodstein, D., Lemons, D., Li, W., Lyons, J. B., Morris, A., Nichols, S., Richter, D. J., Salamov, A., Sequencing, J. G., Bork, P., Lim, W. A., Manning, G., Miller, W. T., McGinnis, W., Shapiro, H., Tjian, R., Grigoriev, I. V., and Rokhsar, D. (2008) Nature 451 783-788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990) J. Mol. Biol. 215 403-410 [DOI] [PubMed] [Google Scholar]
- 15.Till, J. H., Annan, R. S., Carr, S. A., and Miller, W. T. (1994) J. Biol. Chem. 269 7423-7428 [PubMed] [Google Scholar]
- 16.Garcia, P., Shoelson, S. E., George, S. T., Hinds, D. A., Goldberg, A. R., and Miller, W. T. (1993) J. Biol. Chem. 268 25146-25151 [PubMed] [Google Scholar]
- 17.Qiu, H., and Miller, W. T. (2002) J. Biol. Chem. 277 34634-34641 [DOI] [PubMed] [Google Scholar]
- 18.Casnellie, J. E. (1991) Methods Enzymol. 200 115-120 [DOI] [PubMed] [Google Scholar]
- 19.Porter, M., Schindler, T., Kuriyan, J., and Miller, W. T. (2000) J. Biol. Chem. 275 2721-2726 [DOI] [PubMed] [Google Scholar]
- 20.Barker, S. C., Kassel, D. B., Weigl, D., Huang, X., Luther, M. A., and Knight, W. B. (1995) Biochemistry 34 14843-14851 [DOI] [PubMed] [Google Scholar]
- 21.Lee, S., Ayrapetov, M. K., Kemble, D. J., Parang, K., and Sun, G. (2006) J. Biol. Chem. 281 8183-8189 [DOI] [PubMed] [Google Scholar]
- 22.Sun, G., and Budde, R. J. (1997) Biochemistry 36 2139-2146 [DOI] [PubMed] [Google Scholar]
- 23.Patwardhan, P., Shen, Y., Goldberg, G. S., and Miller, W. T. (2006) J. Biol. Chem. 281 20689-20697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ladbury, J. E., Lemmon, M. A., Zhou, M., Green, J., Botfield, M. C., and Schlessinger, J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3199-3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu, H., and Miller, W. T. (2004) Oncogene 23 2216-2223 [DOI] [PubMed] [Google Scholar]
- 26.Sicheri, F., Moarefi, I., and Kuriyan, J. (1997) Nature 385 602-609 [DOI] [PubMed] [Google Scholar]
- 27.Xu, W., Harrison, S. C., and Eck, M. J. (1997) Nature 385 595-602 [DOI] [PubMed] [Google Scholar]
- 28.Moarefi, I., LaFevre-Bernt, M., Sicheri, F., Huse, M., Lee, C. H., Kuriyan, J., and Miller, W. T. (1997) Nature 385 650-653 [DOI] [PubMed] [Google Scholar]
- 29.LaFevre-Bernt, M., Sicheri, F., Pico, A., Porter, M., Kuriyan, J., and Miller, W. T. (1998) J. Biol. Chem. 273 32129-32134 [DOI] [PubMed] [Google Scholar]
- 30.Miller, W. T. (2003) Acc. Chem. Res. 36 393-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamoto, T., Sakai, R., Ozawa, K., Yazaki, Y., and Hirai, H. (1996) J. Biol. Chem. 271 8959-8965 [DOI] [PubMed] [Google Scholar]
- 32.Burnham, M. R., Harte, M. T., Richardson, A., Parsons, J. T., and Bouton, A. H. (1996) Oncogene 12 2467-2472 [PubMed] [Google Scholar]
- 33.Lukong, K. E., and Richard, S. (2003) Biochim. Biophys. Acta. 1653 73-86 [DOI] [PubMed] [Google Scholar]
- 34.Bradshaw, J. M., Grucza, R. A., Ladbury, J. E., and Waksman, G. (1998) Biochemistry 37 9083-9090 [DOI] [PubMed] [Google Scholar]
- 35.Kuriyan, J., and Cowburn, D. (1997) Annu. Rev. Biophys. Biomol. Struct. 26 259-288 [DOI] [PubMed] [Google Scholar]
- 36.Songyang, Z., Carraway, K. L., III, Eck, M. J., Harrison, S. C., Feldman, R. A., Mohammadi, M., Schlessinger, J., Hubbard, S. R., Smith, D. P., Eng, C., Lorenzo, M. J., Poner, B. A. J., Mayer, B. J., and Cantley, L. C. (1995) Nature 373 536-539 [DOI] [PubMed] [Google Scholar]
- 37.Kemp, B. E., Graves, D. J., Benjamini, E., and Krebs, E. G. (1977) J. Biol. Chem. 252 4888-4894 [PubMed] [Google Scholar]
- 38.Scott, M. P., and Miller, W. T. (2000) Biochemistry 39 14531-14537 [DOI] [PubMed] [Google Scholar]
- 39.Liu, X., Brodeur, S. R., Gish, G., Songyang, Z., Cantley, L. C., Laudano, A. P., and Pawson, T. (1993) Oncogene 8 1119-1126 [PubMed] [Google Scholar]
- 40.Briggs, S. D., Sharkey, M., Stevenson, M., and Smithgall, T. E. (1997) J. Biol. Chem. 272 17899-17902 [DOI] [PubMed] [Google Scholar]
- 41.Alexandropoulos, K., and Baltimore, D. (1996) Genes Dev. 10 1341-1355 [DOI] [PubMed] [Google Scholar]
- 42.Gonfloni, S., Weijland, A., Kretzschmar, J., and Superti-Furga, G. (2000) Nat. Struct. Biol. 7 281-286 [DOI] [PubMed] [Google Scholar]
- 43.Pellicena, P., Stowell, K. R., and Miller, W. T. (1998) J. Biol. Chem. 273 15325-15328 [DOI] [PubMed] [Google Scholar]
- 44.Lee, S., Lin, X., Nam, N. H., Parang, K., and Sun, G. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14707-14712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinghoffer, R. A., Sachsenmaier, C., Cooper, J. A., and Soriano, P. (1999) EMBO J. 18 2459-2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, M. A., Gonfloni, S., Superti-Furga, G., Roux, B., and Kuriyan, J. (2001) Cell 105 115-126 [DOI] [PubMed] [Google Scholar]
- 47.Erpel, T., Superti-Furga, G., and Courtneidge, S. A. (1995) EMBO J. 14 963-975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerner, E. C., and Smithgall, T. E. (2002) Nat. Struct. Biol. 9 365-369 [DOI] [PubMed] [Google Scholar]
- 49.Briggs, S. D., and Smithgall, T. E. (1999) J. Biol. Chem. 274 26579-26583 [DOI] [PubMed] [Google Scholar]
- 50.McPherson, R. A., Taylor, M. M., Hershey, E. D., and Sturgill, T. W. (2000) Oncogene 19 3616-3622 [DOI] [PubMed] [Google Scholar]
- 51.Lang, B. F., O'Kelly, C., Nerad, T., Gray, M. W., and Burger, G. (2002) Curr. Biol. 12 1773-1778 [DOI] [PubMed] [Google Scholar]
- 52.Steenkamp, E. T., Wright, J., and Baldauf, S. L. (2006) Mol. Biol. Evol. 23 93-106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.