Abstract
Soluble guanylyl cyclase (sGC), a key protein in the NO/cGMP signaling pathway, is an obligatory heterodimeric protein composed of one α- and one β-subunit. The α1/β1 sGC heterodimer is the predominant form expressed in various tissues and is regarded as the major isoform mediating NO-dependent effects such as vasodilation. We have identified three new α1 sGC protein variants generated by alternative splicing. The 363 residue N1-α1 sGC splice variant contains the regulatory domain, but lacks the catalytic domain. The shorter N2-α1 sGC maintains 126 N-terminal residues and gains an additional 17 unique residues. The C-α1 sGC variant lacks 240 N-terminal amino acids, but maintains a part of the regulatory domain and the entire catalytic domain. Q-PCR of N1-α1, N2-α1 sGC mRNA levels together with RT-PCR analysis for C-α1 sGC demonstrated that the expression of the α1 sGC splice forms vary in different human tissues indicative of tissue-specific regulation. Functional analysis of the N1-α1 sGC demonstrated that this protein has a dominant-negative effect on the activity of sGC when coexpressed with the α1/β1 heterodimer. The C-α1 sGC variant heterodimerizes with the β1 subunit and produces a fully functional NO- and BAY41-2272-sensitive enzyme. We also found that despite identical susceptibility to inhibition by ODQ, intracellular levels of the 54-kDa C-α1 band did not change in response to ODQ treatments, while the level of 83 kDa α1 band was significantly affected by ODQ. These studies suggest that modulation of the level and diversity of splice forms may represent novel mechanisms modulating the function of sGC in different human tissues.
Since the studies of the late 1970s and early 1980s, which underlined the obligatory role of the endothelium in mediating acetylcholine-induced vasodilatation, nitric oxide (NO) has been recognized as an endogenous nitrovasodilator that mediates the local regulation of basal arterial tone (1–4). Many of the physiological functions of NO in the cardiovascular, neuronal, gastrointestinal, and other systems are mediated through its primary receptor, soluble guanylyl cyclase (sGC).3 The heme-containing sGC heterodimer converts guanosine triphosphate into the secondary messenger guanosine 3′:5′-cyclic monophosphate (cGMP). The sGC activity increases more than 200-fold in response to NO (5, 6). High concentrations of cGMP produced by activated sGC modulate functions of numerous enzymes, such as cyclic nucleotide phosphodiesterases, cGMP-gated ion channels, and cGMP-dependent protein kinases (PGKs). Recently, the vital importance of sGC for mammalian physiology was directly confirmed by generation of sGC knock-out mice (7–9). The absence of sGC protein resulted in a significant increase in blood pressure, complete loss of NO-dependent aortic relaxation, and inhibition of platelet aggregation in knock-out animals, which died prematurely at the age of 4 weeks because of severe gastrointestinal disorders (7).
Four sGC isoforms, products of four genes, have been identified so far: α1, α2, β1, and β2. Only α1/β1 and α2/β1 heterodimers are activated by NO (10). The α1/β1 sGC is the most abundant isoform and is distributed ubiquitously in mammalian tissues with the highest levels of mRNA in brain, lung, heart, kidney, spleen, and muscle (11). Vascular smooth muscle and endothelial cells express predominantly α1- and β1-subunits (12). The functional importance of α1/β1 sGC was demonstrated by the significantly decreased relaxing effects of major vasodilators (acetylcholine, NO, YC-1, and BAY41-2272) in the α1 sGC knock-out mice of both genders (9).
sGC function is affected not only by NO, but also by regulation of the expression of sGC subunits at transcriptional and post-transcriptional levels. The steady state mRNA levels of α1- and β1-subunits decrease with hypertension, aging and vary during embryonic development (13). The expression of sGC subunits is regulated by estrogen (14), cAMP-elevating compounds (15, 16), cytokines (NGF, LPS, IL-1β) (17) and NO donors (18). Subcellular localization of sGC, and its activity can also be affected in proliferating tissue (19) by protein interactions and phosphorylation (13). In mammals, the alternative splicing for the α2-subunit generates a dominant-negative variant (20). Splice forms for β1- and β2-subunits have been also demonstrated (21–23). Recently, a shortened α1 sGC transcript, which lacks the predicted translation site in exon 4, has been found, and its expression was correlated with lower sGC activity in several cell lines (24). However, splice variants of α1 sGC have not been described previously.
Here we report the isolation and characterization of three new α1 sGC splice forms encoding N- and C-terminal-truncated proteins. We demonstrate that the N-terminal-truncated C-α1 splice form heterodimerizes with the β1 sGC subunit to create an active NO-sensitive enzyme both in Sf9 and human neuroblastoma BE2 cells. Moreover, this splice variant is more resistant to ODQ-induced protein degradation than the wild-type sGC. N1-α1 sGC splice form lacking the C-terminal catalytic domain has a dominant-negative effect when co-expressed with α1/β1 sGC in Sf9 or BE2 cells. The functional role of N2-α1 sGC splice variant is yet to be determined. Q-PCR and semiquantitative RT-PCR analyses of different human tissues demonstrate tissue-specific expression of the identified splice forms. Together our data suggest that alternative splicing of the α1 sGC subunit may be a novel mechanism that regulates sGC function and activation in some human tissues.
MATERIALS AND METHODS
Reagents—Dulbecco's modified Eagle's /F12 medium was from Invitrogen; Grace medium and fetal bovine serum were from Sigma. NO donor DEA-NO was from Calbiochem. The 5-cyclopropyl-2-[1-(2-fluoro-benzyl)-1H-pyrazolo[3,4-b]pyridin-3-yl]-pyrimidin-4-ylamine (BAY41-2272) activator was a generous gift from J. P. Stasch (Bayer, Wuppertal, Germany). [α-32P]GTP was from PerkinElmer Life Sciences. ODQ (1H-[1,2,4]oxadiazolo[4,3,-a]quinoxalin-1-one) was from Sigma-Aldrich. Polyclonal anti-α1 sGC antibodies were raised against the peptide FTPRSREELPPNFP of the human α1-subunit, while anti-β1 antibodies were raised against the SRKNTGTEETKQDDD peptide of the human β1-subunit.
Primers, RNA, and RT-PCR—All primers were custom synthesized by Integrated DNA Technologies (Coralville, IA). To subclone N1-α1 sGC we used the upstream primer PR1, 5′-318CAACACCATGTTCTGCACGAAGC-3′ and the downstream primer PR2, 5′-1411GCTTTCATATTCAAGATAGTATTATG-3′ (numbering according to sequence with GenBank™ no. CR618242). To subclone the N2-α1 sGC, we used the upstream primer PR3, 5′-192CAACACCATGTTCTGCACGAAGC-3′ and the downstream primer PR4, 5′-611GTATCACTCTCTTTGTGTAATCC-3′ (GenBank™ no. BC012627). To detect the deletion of exon 4 in C-α1 sGC we used the upstream primer PR5, 5′-120GCTAGAGATCCGGAAGCACA-3′ and the downstream primer 5′-317TTGCAAATACTCTCTGCCAAA-3′ (GenBank™ no. AK226125). To detect the deletion of in the exon 7 of C*-α1 sGC we used the upstream primer PR7, 5′-561GAA CGG CTG AAT GTT GCA CTT GAG-3′ and the downstream primer PR8, 5′-922GTA GGG CTG ATT CAC AAA CTC G-3′ (GenBank™ no. BX649180). Total RNA from BE2 cells was isolated using the RiboPure kit (Ambion, TX). The panel of total RNA from human tissues was purchased from Ambion (FirstChoice Human Total RNA Survey Panel, Lot 08608142). 5 μg of total RNA was used for the RT reactions, which was performed with a mixture of oligo(dT) and Random Hexamer primers using SuperScript III RT (Invitrogen) according to the manufacturer's protocol. PCR reactions with PfuUltra DNA polymerase (Stratagene) were performed for 35 cycles at the Ta of 55 °C. PCR products were separated on an agarose gel, purified using the QIAEX II gel extraction kit (Qiagen), and sequenced using PCR primers. All sequencing was performed by the Nucleic Acid Core Facility at the Medical School of University of Texas in Houston.
Real-Time Quantitative RT-PCR—Real-time quantitative RT-PCR (RT-qPCR) was performed utilizing the 7700 or 7900 Sequence Detector instrument (Applied Biosystems, Foster City, CA) (25, 26). Specific quantitative assays for α1 sGC, N1-α1 sGC, and N2-α1 sGC were developed using the Primer Express software version 1.0 for Macintosh (Applied Biosystems), Beacon Designer (Premier Biosoft), or RealTimeDesign (Biosearch Technologies) based on sequences from GenBank™. The assays are listed in supplemental Table S3. cDNA was synthesized in 10 μl (96-well plate) or 5 μl (348-well plate) total volume by the addition of 6 μl or 3 μl/well of RT master mix consisting of: 400 nm assay-specific reverse primer, 500 μm deoxynucleotides, Superscript II buffer, and 10 units of Superscript II reverse transcriptase (Invitrogen) to a 96-(ISC Bioexpress, Kaysville, UT) or 384-well plate (Applied Biosystems) and followed by a 4-μl or 2-μl volume of sample (25 ng/μl), respectively. Each sample was determined in triplicate, plus a control without reverse transcriptase to access DNA contamination levels. Each plate also contained an assay-specific sDNA (synthetic amplicon oligo) standard spanning a 5-log template concentration range and a no template control. Each plate was covered with Biofilm A (Bio-Rad) and incubated in a PTC-100 (96) or DYAD (384) thermocycler (Bio-Rad) for 30 min at 50 °C, followed by 72 °C for 10 min. Subsequently, 40 μl or 20 μl of a PCR master mix (400 nm forward and reverse primers (IDT, Coralville, IA), 100 nm fluorogenic probe (Biosearch Technologies, Novato, CA), 5 mm MgCl2, 200 μm deoxynucleotides, PCR buffer, 150 nm SuperROX dye (Biosearch Technologies, Novato, CA), and 1.25 units of Taq polymerase (Invitrogen) were added directly to each well of the cDNA plate. RT master mixes and all RNA samples were pipetted by a Tecan Genesis RSP 100 robotic work station (Tecan US, Research Triangle Park, NC); PCR master mixes were pipetted utilizing a Biomek 2000 robotic work station (Beckman, Fullerton, CA). Each assembled plate was then covered with optically clear film (Applied Biosystems) and run in the 7700 or 7900 real-time instrument using the following cycling conditions: 95 °C, 1 min; followed by 40 cycles of 95 °C, 12 s, and 60 °C, 30 s. The resulting data were analyzed using SDS 1.9.1 (7700) or SDS 2.3 (7900) software (Applied Biosystems) with ROX as the reference dye.
Synthetic DNA oligos used as standards (sDNA) encompassed the entire 5′-3′ amplicon for the assay (Invitrogen). Each oligo standard was diluted in 100 ng/μl yeast or Escherichia coli tRNA-H2O (Invitrogen or Roche Applied Sciences) and span a 5-log range in 10-fold decrements starting at 0.8 pg/reaction. It has been shown for several assays that in vitro transcribed RNA amplicon standards (sRNA) and sDNA standards have the same PCR efficiency when the reactions are performed as described above.4
Because of the inherent inaccuracies in quantifying total RNA by absorbance, the amount of RNA added to an RT-PCR from each sample was more accurately determined by measuring a housekeeping transcript level in each sample. The final data were normalized to 36B4 (500-fold dilution of sample in tRNA-H2O).
Cell Culture—BE2 human neuroblastoma cell line (American Type Culture Collection) was cultured in 1:1 mixture of Dulbecco's modified Eagle's medium/F12K media supplemented with 10% fetal bovine serum, 0.1 mm MEM nonessential amino acids, penicillin-streptomycin mixture (50 units/ml and 50 μg/ml), 10 mm Hepes (pH 7.4), 1 mm sodium pyruvate, 2 mm l-glutamine (all from Invitrogen), and maintained at 37 °C and 5% CO2. For in vivo ODQ treatments 80% confluent neuroblastoma cell cultures were treated with 20 μm ODQ for up to 24 h. To prepare lysates, the cells were collected by trypsinolysis, washed twice with phosphate-buffered saline, resuspended in 40 mm TEA (pH 7.4) containing protease inhibitor mixture (Roche Applied Science), and disrupted by sonication. The lysates were centrifuged at 15,000 × g for 30 min to prepare the cleared supernatant fractions, which were used for Western blotting, immunoprecipitation, or activity measurements.
Generation of BE2 Stable Transfectant Lines—Coding sequences of N1-, N2-, and C-type α1 sGC variants obtained from BE2 total RNA as described above were first cloned into pCR-Blunt vector (Invitrogen). For N1- and N2-type α1 forms, the coding sequence of the FLAG peptide was inserted by PCR in front of the stop codon and recloned into pCR-Blunt. The coding regions of N1-, N2-, and C-type α1 sGC variants were then isolated by restriction with NsiI/XbaI enzymes and subcloned into PstI/XbaI sites under the control of the CMV promoter of the mammalian expression vector pMGH2 (Invitrogen). These pMG-CαF, pMG-N1αF, and pMG-N2αF plasmids were transfected into BE2 cells by the Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. 48-h post-transfection, the cells were plated on 96-well plates at 2500 cells/ml density and selected by 380 μg/ml hygromycin (Sigma). Two weeks later, individual hygromycin-resistant colonies were collected and expanded on 100-mm2 tissue culture dishes. The clones stably transfected by N1- or N2-type α1 sGC were identified by anti-FLAG Western blotting of lysates prepared from the hygromycin-resistant cultures. The clones stably expressing C-type α1 sGC were identified using anti-α1 sGC Western blotting.
Western Blot Analysis—Protein samples were resolved on 8 or 12% SDS-polyacrylamide gels and transferred to methanol-activated polyvinylidene difluoride membranes. After blocking the membranes, the following antibodies were used for detection: anti-FLAG M2 monoclonal antibody (Sigma) at 1:500 dilution; anti-α1 sGC polyclonal antibody generated in our laboratory at 1:2000 dilution; anti-β1 sGC polyclonal antibodies generated in our laboratory at 1:3000 dilution; anti β-actin antibody (Santa Cruz Biotechnology) at 1:1000 dilution. Secondary horseradish peroxidase-conjugated antibodies (Sigma) were used at 1:5000 (anti-rabbit) and 1:10000 (anti-mouse) dilutions. Protein bands were visualized by enhanced chemiluminescence (ECL Plus, Amersham Biosciences).
Co-immunoprecipitation—BE2, BE-CαF, and BE-N1αF cells collected from confluent 10-cm culture dishes, were washed twice with phosphate-buffered saline, resuspended in 500 μl of phosphate-buffered saline containing protease inhibitor mixture, disrupted by sonication, spun down at 15,000 rpm for 30 min at 4 °C, and supernatants were collected. Then polyclonal anti-β1-sGC antibodies or prewashed anti-FLAG M2 affinity resin (Sigma) were added and tumbled for 1.5 h or overnight, respectively, at 4 °C. The lysate mixture with anti-β1-sGC antibodies was then combined with 100 μl of prewashed protein A-agarose beads (Upstate) and further incubated for 1.5 h. The protein A-agarose beads were washed three times with 40 mm TEA, 200 mm NaCl, 1% Nonidet P-40, pH 7.4, and bound proteins eluted by boiling in 100 μl of Laemmli buffer. The anti-FLAG M2 affinity resin was washed three times with ice-cold TBS buffer, and bound proteins were eluted with high salt buffer or with buffer containing the FLAG peptide, according to the manufacturer's instructions. The Western blot was probed for the α1- and β1-subunits of sGC using polyclonal anti-α1-sGC and monoclonal anti-β1-sGC antibodies.
Expression in Sf9 Cells—To express the α1 splice forms in Sf9 cells, the coding regions of N1, N2, or C-α1 sGC were subcloned into the pVL1392 transfer vector. The baculoviruses producing the α1 splice forms were generated by recombination with BaculoGold DNA using the manufacturer's protocol (BD Biosciences). To obtain sGC enzyme containing the splice form α1-subunits, Sf9 cells at 1.8 × 106 cells/ml were infected with the baculoviruses expressing full-length β1 sGC subunit and the corresponding splice form at a multiplicity of infection of 2.
Assay of sGC Activity—Soluble guanylyl cyclase activity in lysates of BE2 or Sf9 cells were assayed by formation of [32P]cGMP from [α-32P]GTP at 37 °C as described previously (27). The concentration of DMSO used as a vehicle for BAY41-2272 did not exceed 0.1% and alone had no effect on sGC activity.
Statistical Analysis—All data are presented as means ± S.E. or S.D. Statistical comparisons between groups were performed by Student's t test. Nonlinear regression and calculations of EC50 and IC50 were performed using Graph Pad Prism 3.0 software (GraphPad Software).
RESULTS
Identification of α1 sGC Alternative Splice Variants in the NCBI Data Base—A previous report demonstrated the existence of alternative splicing for the α1 sGC in human tissues (24). We used the α1 sGC cDNA (GenBank™ accession number Y15723) to screen a human RNA data base available on the Human Genome web page (NCBI) for additional splice variants of sGC. This in silico analysis identified twelve unique α1 sGC cDNA sequences cloned from different human tissues (supplemental Table S1). Their comparison with human genome sequences revealed that they are all generated by alternative splicing from the α1 sGC gene.
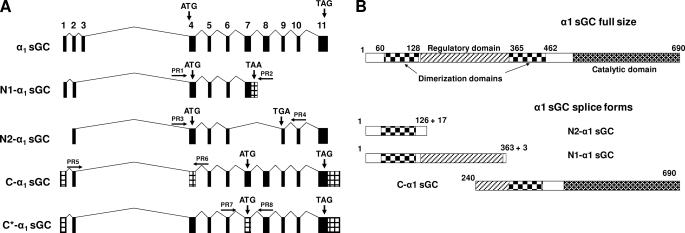
Seven of these sequences encode the full size α1 sGC protein, while five of them encode truncated α1 proteins (supplemental Table S1). With alternative splicing, one of the identified α1 sGC mRNAs (GenBank™ accession numbers CR618242, CR614534) lost the non-coding exon 2 and the coding exons 8 through 10, but acquired additional 131 bp at the end of exon 7. This mRNA encodes a protein that maintained 363 N-terminal amino acids of α1 sGC, but lost the catalytic domain because of a splice-generated frameshift (Fig. 1, A and B). Three new splice-specific amino acid residues and a premature stop codon were acquired (supplemental Table S2). We named this splice variant N1-α1 sGC (Fig. 1).
FIGURE 1.
Structure of newly identified α1 splice mRNA species and proteins. A, comparative genomic organization of human sGC mRNAs encoding full size α1-subunit and N1-, N2-, C*-, and C-type α1 sGC splice forms. The sketch is based on sequences with the following GenBank™ accession numbers: α1 sGC,Y15723; N1-α1 sGC, CR618242; N2-α1 sGC, BC012627; C-α1 sGC, AK226125, and C*-α1 sGC, BX649180. Conserved exons are marked in black, alternative sequences are in a gray pattern. Locations of start and stop codons are indicated. Positions of primers used for detection and subcloning are indicated by arrows. B, comparative domain organization of α1 sGC proteins. Schematic representation of domains encoded by various α1 sGC splices forms. The domain structure is based on previous predictions (37, 38).
In N2-α1 sGC mRNA (GenBank™ accession number BC012627), splicing eliminates exons 7 and 8 and introduces an additional 54 base pairs and a premature stop codon in exon 9. The N2-α1 protein retains only the first 126 residues of the α1 sequence, but acquires an additional 17 amino acids at the C terminus (Fig. 1, A and B and supplemental Table S2).
The third identified α1 sGC splice variant, termed C-α1 sGC, lost 240 N-terminal amino acids, but maintained part of the regulatory and the complete catalytic domains (Fig. 1B). Interestingly, the same C-α1 protein is encoded by two differentially spliced species of mRNA (Fig. 1A). In one sequence (GenBank™ accession number AK226125, C-α1 in Fig. 1A), the alternative splice acceptor in intron 3 generates a 179-bp deletion in exon 4, eliminating the translation start site. The open reading frame (ORF) starts at the alternative methionine located in exon 7. In another splice variant (GenBank™ accession number BX649180, C*-α1 in Fig. 1A), the alternative acceptor site results in a 140-bp deletion and a premature stop codon in exon 7. The alternative start codon in exon 7 restores the ORF. Thus, both C-α1 and C*-α1 splice mRNA species encode the same C-α1 sGC protein.
Alternative Splicing of α1 sGC in Human BE2 Neuroblastoma—We have previously demonstrated that the human BE2 neuroblastoma cell line expresses high levels of functional sGC enzyme (28). Using the information on the structure of α1 spliced mRNA, we designed pairs of primers to amplify the fragments specific for N1-α1, N2-α1, C-α1, and C*-α1 splice forms (Fig. 1A). As depicted in supplemental Fig. S1, RT-PCR analysis showed that BE2 cells express all found α1 splice mRNA except C*-α1. Each of the splice-specific fragments was purified, and the identity of the splice form confirmed by sequencing.
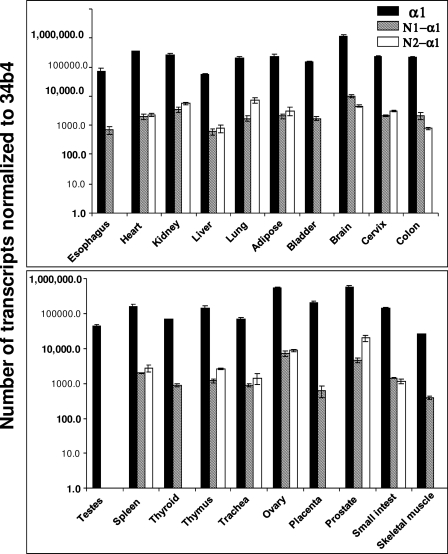
Expression of α1 sGC Alternative Splice Variants in Different Human Tissues—We next investigated the tissue-specific expression of the α1 sGC splice variants. We designed a Q-PCR assay for N1- and N2-α1 sGC splice forms based on their unique sequences inserted by alternative splicing (supplemental Table S3). We quantified the abundance of the full-length and spliced mRNA in various human tissues. As expected, full-length α1 sGC mRNA was detected in RNA of all tested tissues. N1-α1 sGC was observed at detectable levels in all human organs, except bladder, testis, thyroid, placenta, and skeletal muscle (Fig. 2). N2-α1 sGC was present in all tissues, but at significantly lower levels than α1 sGC.
FIGURE 2.
Expression of α1 sGC, N1-, and N2-α1 sGC mRNA in human tissues. Expression was quantified by qPCR analysis and normalized to the levels of ribosomal housekeeping gene 36b4. Human total RNA Survey Panel (Ambion) containing mRNA from 20 different tissues pooled from three healthy donors was used in this comparison.
The absence of sequences specific only for C-α1 or C*-α1 mRNA and the insignificant difference in size from the fulllength sGC transcript (supplemental Table S1) precluded us from using Q-PCR or Northern blotting to estimate their levels and tissue distribution. Therefore, we used a semi-quantitative RT-PCR method using primers designed to detect the deletions specific for both C- and C*-α1 sGC RNA species. The identity of amplified fragments was confirmed by sequencing. The full size α1 sGC transcript was detected in all tissues together with C-α1 sGC, albeit at different ratios (supplemental Fig. S2). However, C*-α1 sGC mRNA was not detected in esophagus, heart, kidney, liver, lung, bladder, brain, cervix, and colon (supplemental Fig. S3).
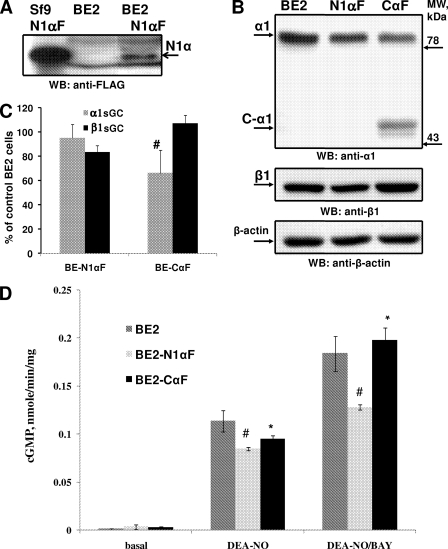
Effect of Expression of α1 sGC Alternative Splice Variants on sGC Activity—Next we tested whether α1 sGC splice variants possess any catalytic activity or whether they affect the function of full-length α1/β1 sGC in BE2 cells. We selected several stable clones expressing N1-α1 or N2-α1 sGC, which were tagged with the FLAG epitope at the C-terminal end. The stable clone expressing 41-kDa N1-α1 sGC protein was identified using anti-FLAG antibodies (BE2-N1αF in Fig. 3A). We measured the rate of cGMP production in the lysates of these cells in response to the NO donor, DEA-NO, or DEA-NO in the presence of the heme-dependent allosteric regulator BAY41-2272. As demonstrated in Fig. 3B, the BE2-N1αF clone showed a significant decrease of sGC-dependent cGMP synthesis in comparison with parental BE2 cells. This lower sGC activity cannot be attributed to a decreased expression of sGC, because Western blotting showed no significant decrease in the levels of endogenous full-length α1- and β1-subunits (Fig. 3B). The lower cGMP synthesis was observed in all tested BE-N1αF clones (results not shown), suggesting that the N1-α1 sGC splice form acts as an inhibitor of sGC function. This conclusion was confirmed when the N1-α1 splice form was co-expressed together with the full-length α1- and β1-subunits in Sf9 cells. The addition of increasing amounts of virus producing N1-α1 sGC protein correlated with the decrease of DEA-NO, BAY41-2272-, and DEA-NO/BAY41-dependent α1/β1 sGC activity (supplemental Fig. S4). The lysates from stable clones overexpressing N2-α1 sGC did not show significant changes of the NO and/or BAY41-2272-dependent cGMP synthesis. We did not observe significant changes of the NO/BAY41-dependent cGMP production in lysates of the lines overexpressing the N2-α1 sGC splice form (results not shown).
FIGURE 3.
Identification and characterization of BE2 stable clones expressing N1-α1 and C-α1 sGC splice form. A, identification of stable clone expressing N1-α1 variant by Western blotting of cell lysates with anti-FLAG antibody. The lysate from Sf9 cells infected with virus producing N1-α1F was used as a positive control. Arrows indicate the protein band of appropriate size in Sf9 cells and the BE2-N1F clone. B and C, identification of stable lines expressing C-α1 variant by Western blotting with anti-α1 sGC antibody and protein levels of endogenousα1 andβ1 sGC in stable lines and parental BE2 cells. Arrows indicate the protein bands corresponding to α1 and C-α1 sGC in BE2-CαF. B, representative Western blot; C, densitometry analysis of α1 and β1 levels normalized to β-actin and expressed as percent of expression in control BE2 cells taken as 100%. The value for BE-N1α and BE-CαF clones are presented as mean ± S.D. calculated from three independent lysates. D, rate of cGMP production in lysates of BE2 cells, BE2-N1αF and BE2-CαF in response to DEA-NO (200 μm) alone or in combination with BAY41-2272 (2 μm). Data from six (for BE2 cells) or three (for BE2-N1αF and BE2-CαF clones) independent lysates measured in triplicates are presented as mean ± S.E. #, difference with BE2 cells is statistically significant (p < 0.05); *, difference is not significant (p > 0.05).
We also selected BE2 stable lines expressing the C-α1 sGC splice variant tagged with the FLAG epitope. BE2-CαF line was selected using antibodies raised against a sequence at the C-terminal end of the human α1 sGC subunit, which recognizes both the short C-type α1 sGC (54 kDa) and the full-length (83 kDa) α1-subunit (Fig. 3A). We found that the DEA-NO- or DEA-NO/BAY41-2272-stimulated cGMP synthesis in the lysates of the BE2-CαF clone was not significantly different from parental BE2 cells. On the other hand, the amount of α1-sGC protein in BE2-CαF was lower than in BE2 cells (Fig. 3A). It appears that the expression of C-α1 variant can compensate for decreased levels of full size α1-sGC subunit through a formation of active heterodimer.
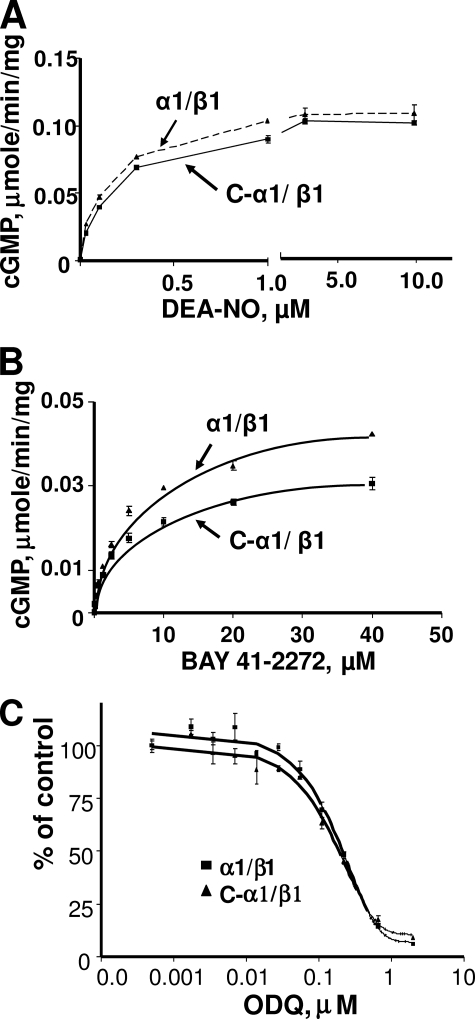
Functional Characterization of C-α1/β1 Heterodimer—Because the activity in BE2-CαF cells suggested that the C-α1 splice form can compensate for α1 function, we characterized the properties of the C-α1/β1 heterodimer. We co-expressed the C-α1 variant with β1-sGC in Sf9 cells. Sf9 lysates expressing the C-α1/β1-sGC displayed a robust guanylyl cyclase activity. We found no difference between the activation of α1/β1 and C-α1/β1 enzymes in response to various concentrations of DEA-NO, while the effect of BAY41-2272 was decreased by about 20% (Fig. 4, A and B). We also found no difference in EC50 values for DEA-NO or BAY41-2272 between C-α1/β1 and α1/β1 enzymes (125 ± 12.2 versus 108 ± 7.1 nm and 7.8 ± 0.08 versus 7 ± 0.10 μm, respectively). Moreover, both α1β1 and C-α1/β1 heterodimers demonstrated similar sensitivity to the selective sGC inhibitor ODQ, (Fig. 4C). The estimated ODQ IC50 was almost identical: 185 ± 5 nm for α1/β1 and 193 ± 11 nm for C-α1/β1.
FIGURE 4.
Functional analysis of recombinant C-α1 sGC. A, C-α1/β1 sGC and α1β1 proteins show similar response to DEA-NO. The rates of cGMP production in Sf9 cell lysates expressing C-α1/β1 or α1/β1 sGC in response to various concentrations of DEA-NO was determined as described under “Materials and Methods.” B, C-α1/β1 sGC andα1β1 proteins show similar responses to BAY41-2272. The rates of cGMP production in Sf9 cell lysates expressing C-α1/β1 or α1/β1 sGC in response to various concentrations of BAY41-2272 were determined as described under “Materials and Methods.” C, C-α1/β1 sGC and α1β1 proteins have the same sensitivity to ODQ. Sf9 lysates with α1/β1 or C-α1/β1 heterodimers were mixed with different amounts of ODQ, and the rate of cGMP production induced by 50 μm DEA-NO was measured. Data from four independent experiments are presented as mean ± S.D.
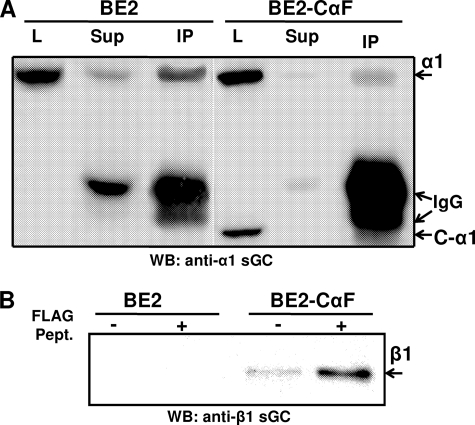
Interaction of the C-α1 Splice Variant with the β1-Subunit—To confirm the interaction of the C-α1 sGC with the β1 sGC subunit we tested for co-immunoprecipitation of the C-α1 sGC with the β1 sGC subunit from the BE2-CαF clone. As shown in Fig. 5A, polyclonal antibodies raised against the C terminus of the β1-subunit precipitated both α1 and C-α1 variants. This immunoprecipitation correlates with the depletion of the C-α1 and α1 signals from the post-immunoprecipitate lysates (Fig. 5A). Moreover, the similar intensity of the co-precipitated signals suggests that both α1 and C-α1 heterodimerize with β1 equally well. Alternatively, immunoprecipitation of C-α1 sGC with anti-FLAG M2 affinity gel precipitated the β1 sGC subunit from the lysate of BE2-CαF stable clones, but not from BE2 lysates (Fig. 5A). The amount of β1 sGC was higher in the eluates treated with the FLAG peptide, consistent with a FLAG-specific elution.
FIGURE 5.
Detection of C-α1/β1 heterodimer. A, cell homogenates of BE2 cells and the BE2-CαF stable clone were subjected to immunoprecipitation with anti β1-sGC antibodies. 0.1% SDS eluates were probed by Western blotting with anti-α1 antibodies. The arrows indicate the position of α1, C-α1, and IgG bands. L, lysate; Sup, immunodepleted lysate; IP, 0.1% SDS eluates. B, cell lysates from BE2 cells and the BE2-CαF stable line were immunoprecipitated with anti-FLAG M2 affinity gel. Proteins bound to FLAG-agarose were eluted by high-salt buffer with or without FLAG peptide and analyzed by Western blotting with anti-β1 sGC antibodies.
Despite a clear inhibitory effect of the N1-α1 splice form (Fig. 3B and supplemental Fig. S4), we were not able to observe the co-precipitation of the N1-α1 sGC with the β1-subunit (Fig. S5).
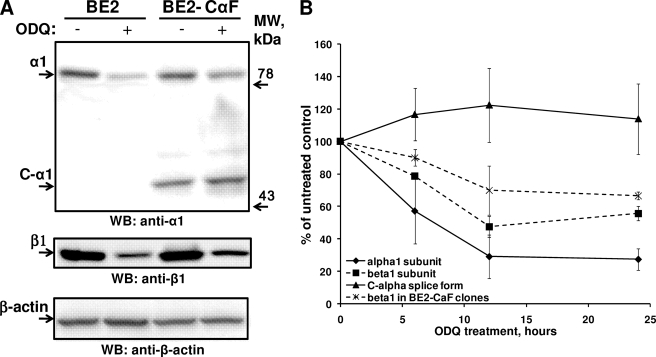
Level of C-α1 Splice Form Is Not Affected by ODQ Treatment—A recent report indicated that heme-deficient sGC or sGC with an oxidized heme prosthetic group is more prone to degradation, which may contribute to endothelial dysfunction (29). In those studies, treatment of intact cells with the sGC inhibitor ODQ decreased the level of sGC protein because of ubiquitin-dependent degradation. Thus, we tested whether the active C-α1/β1 heterodimer shows a similar response to ODQ. Indeed, as demonstrated in Fig. 6A, the amount of full-length α1 band decreased after BE2-CαF cells were treated for 24 h with 20 μm ODQ. The C-α1 splice band, however, was not affected by the same treatment. A time course study showed that the intensity of the α1 band decreased shortly after ODQ administration in all tested cells, while the level of C-α1 even slightly increased with time (Fig. 6B). These data suggest that the C-α1 subunit protein is more stable to intracellular processes occurring after the oxidation of sGC heme.
FIGURE 6.
Effect of the ODQ treatment on C-α1, α1, and β1 GC subunit. A, level of C-α1 splice form does not decline after ODQ-treatment. Amounts of α1, C-α1, β1, and β-actin were examined in BE2 and BE2-CαF cells 24 h after treatment with 20 μm ODQ. A representative blot out of four independent experiments is shown. B, time-dependent changes of α1, C-α1, and β1 bands after ODQ treatment. The intensity of α1, β1, and C-α1 bands in BE2 cells and BE2-N1αF and BE2-CαF clones was determined at different times after exposure to 20 μm ODQ. Each data point is the mean ± S.E. (n = 4 for 6 and 24 h; n = 3 for 12 h).
DISCUSSION
Alternative splicing frequently occurs in eukaryotic genes and provides an important mechanism for tissue-specific and developmental regulation of gene expression. About 15% of mammalian gene mutations associated with pathological conditions affect RNA splicing signals (30).
A number of previous reports demonstrate that alternative splicing is an essential mechanism regulating the function of several members of the cGMP signaling pathway. For example, a splice-dependent deletion of 90–100 N-terminal residues of cGMP-dependent protein kinase I (PKGI) produces two splice variant isoforms, Iα and Iβ, which have different tissue distribution and target specificity (31). Recent studies also demonstrated that PKGI splice isoforms respond differently to activation by hydrogen peroxide (32). Guanylyl cyclase B (GC-B) is another example with two truncated splice forms. It was proposed that these splice forms regulate the function of the full-length subunit and show different tissue distributions (33).
Several reports suggested that splicing may also be a method of sGC regulation. For example, the α2i sGC splice variant, carrying an insertion in the catalytic domain was detected in several human tissues (20). The α2i sGC splice variant has a dominant-negative function. In addition, the expression of two mRNA species of human α1 sGC with deletions in the exon 4 correlates with decreased sGC activity in immortalized B-lymphocyte cell lines (24). These α1 sGC splice forms, however, were never isolated or characterized.
In this report, we characterized several newly identified splice forms of the α1-subunit of human soluble guanylyl cyclase. Analyzing the abundant data available in the NCBI data base, we identified a large number of individual α1 sGC transcripts. It is notable that the diversity of uncovered α1 sGC transcripts (12 cDNAs, see supplemental Table S1) is much larger than for the β1 sGC (5 cDNAs). This diversity does not appear to be unique for humans. Analysis of the mouse genome data base also identified the N1-α1 splice variant (GenBank™ number AK031305). Conservation of this particular splice form points to a functional importance of the N1-α1 variant in mammals. Most of these transcripts are produced by sequence rearrangements in 5′- and 3′-untranslated regions, which are usually associated with altered post-transcriptional regulation of gene expression. For example, binding of the RNA-stabilizing protein HuR to the 3′-UTR of α1 sGC plays an important role in post-transcriptional regulation of sGC expression in response to cyclic nucleotides and in aging in rats (15, 34, 35). Thus, the diversity of the 5′- and 3′-untranslated region sequences most likely reflects multiple selective mechanism(s) regulating the stability of α1 sGC transcripts in response to various extracellular and intracellular stimuli in different cell types or at different developmental stages.
We concentrated our studies on four RNA splice variants which encode truncated α1 sGC, with deletions at the N or C termini of the protein (supplemental Table S1 and Fig. 1). We found that human neuroblastoma BE2 cells express three (N1-α1, N2-α1, C-α1) out of four alternatively spliced RNAs (supplemental Fig. S1). Quantitative PCR analysis of the N1-α1 and N2-α1 splice mRNAs demonstrates that the majority of human tissues express more than one splice variant, but the levels and the relative ratio of the α1 and the N1-α1 and N2-α1 splice forms is tissue-specific (Fig. 2 and supplemental Table S4). For example, N2-α1 is found in all tested tissues, while N1-α1 sGC is not detected in esophagus, bladder, testes, thyroid, placenta, and skeletal muscle (Fig. 2). Similarly, C-α1 sGC is detectable in all tested human tissues, although the relative ratio was tissue-specific (supplemental Fig. S2), while C*-α1 sGC was detected only in some tissues (supplemental Fig. S3). Taken together, these data indicate that the expression of α1 sGC splice forms is independently regulated. It should be noted, though, that the levels of N1-, N2-, C-, and C*-α1 splice mRNAs are on average lower than the full size α1 sGC. Interestingly, the semi-quantitative PCR analysis suggests that in human adipose tissue the level C-α1 and C*-α1 mRNA may be collectively comparable with the level of α1 sGC mRNA. Because the comparative analysis reported here is based on RNAs extracted from entire organs, it might not represent the true abundance of individual spliced sGC transcript at the cellular level. Cellular and subcellular localizations of individual α1 sGC splice variants in human tissues remain to be determined. Localization may significantly alter the functions of these splice forms beyond the properties reported here for BE2 neuroblastoma cells.
In this report, we identified and characterized N1-α1 and N2-α1 sGC splice proteins lacking the catalytic domain (Fig. 1). Although by themselves or in combination with the β1-subunit these truncated splice α1 proteins do not have any catalytic activity (data not shown), one of them displays dominant-negative properties. BE2 cells overexpressing N1-α1 sGC have a significantly decreased NO- and NO/BAY42-2272-induced cGMP production despite the unchanged level of endogenous α1/β1 sGC (Fig. 3). Moreover, when co-expressed in Sf9 cells with α1/β1 heterodimer, N1-α1 reduced NO- and BAY41-2272-dependent sGC activity in direct correlation with the expression level of the N1-α1 protein (supplemental Fig. S4). We were not able to detect any direct heterodimerization of the N1-α1 protein with the β1-subunit, even when both proteins were coexpressed in large quantities in Sf9 cells (supplemental Fig. S5). This suggests that the dominant-negative effect of N1-α1 is not due to a direct competition with the full-length α1 for the binding with β1-subunit, but is rather indirect. Recent deletion mapping showed that the segment spanning residues 363–372 of the α1-subunit is important for the formation of the α1/β1 heterodimer (36). Because the N1-α1 splice protein constitutes the first 363 residues of the α1-subunit, perhaps, it interferes with the heterodimerization function of the adjacent 363–372 segment of the full-length α1-subunit.
Direct regulation of guanylyl cyclase by dominant-negative splice variants has been proposed before. For example, the α2i sGC splice variant blocks the formation of a functional α1β1 sGC heterodimer (20), while the truncated GC-B splice variants hinder the formation of active full size GC-B homodimers (33). Considering the wide distribution of the N-α1 splice form in different human tissues (Fig. 2 and supplemental Table S1), modulation of N1-α1 sGC protein expression may also be a regulatory mechanism controlling the amount of active sGC heterodimer.
In contrast to N1-α1, N2-α1 sGC had no effect on sGC activity in cell lysates when overexpressed in BE2 cells or co-expressed with α1/β1 in Sf9 cells. However, it cannot be excluded that this splice variant may serve another function besides directly affecting sGC activity.
Our studies also demonstrate that the C-α1 sGC isoform clearly forms a fully functional sGC heterodimer with β1 sGC subunit. The activity of the recombinant C-α1/β1 enzyme expressed in Sf9 cells was indistinguishable from the α1/β1 sGC, at least in regard to the degree of activation by the NO donor and allosteric activator BAY41-2272 and inhibition by ODQ (Fig. 4). The preservation of the sGC activity in the BE-CαF stable line (Fig. 3) and co-immunoprecipitation of C-α1 sGC and β1 sGC subunits (Fig. 5) all support the conclusion that C-α1- and α1-subunits are interchangeable. Also, these results suggest that the lack of 240 N-terminal amino acid residues by the α1 sGC subunit does not affect the heterodimerization and enzyme activity. This observation does not support previous findings that the region between residues 61 and 128 of α1 is mandatory for heterodimerization (37). The properties of this naturally occurring splice variant, however, are in agreement with earlier reports, which show that a significant portion of the N-terminal region of the α1 sGC subunit can be deleted without affecting NO sensitivity and heterodimerization (36, 38, 39).
The existence of α1-positive bands other than the 83-kDa full-length α1 sGC subunit was mentioned in several previous reports. While screening selected human tissues with anti-α1 antibodies, Zabel et al. (40) observed in cortex, cerebellum, and lungs a band similar in size to the ∼54 kDa C-α1 described in this report. Interestingly, the antibodies used were raised against the same epitope as in current studies. Moreover, additional bands were also detected in crude lysates from human amygdale (41). Presence of several bands with electrophoretic mobility similar to C-α1 indicates that either C-α1 form may undergo additional tissue-specific processing and/or modifications, or that additional, yet unidentified, α1 sGC splice forms exist.
Recent studies suggested that some conditions of endothelial dysfunction may be associated with the accumulation of oxidized and heme-free sGC leading to a poor response to NO (29). The same report showed that in ODQ-treated cells sGC is subjected to ubiquitination and subsequent degradation. Although our current studies showed that C-α1/β1 and α1/β1 heterodimers have identical sensitivity to ODQ with a similar IC50 (Fig. 4), C-α1 and α1 sGC subunits show an opposite response to ODQ-induced degradation (Fig. 5). Western blotting confirms that the 83-kDa α1 sGC band disappears after exposure to ODQ, while the level of C-α1 protein does not decrease. These data suggest that C-α1 lacks the structural cues contributing to decreased levels of α1 after ODQ treatment. The functional studies presented here prove that the C-α1/β1 heterodimer can fully compensate for sGC activity. In light of reported previously decreased levels of α1-subunit in aged or diseased vessels (41–44), the expression of a more stable C-α1 may be a specific protective adaptation to these conditions. Thus, it is possible that the positive vasodilatory effects of BAY58-2667 in diseased blood vessels observed previously (29) are mediated by the more stable C-α1/β1 heterodimer. Future studies will show whether the C-α1 splice form may be, under certain circumstances, the main or the sole type of α1-subunit.
Although the functional studies suggest that the N-terminal fragment missing in the C-α1-subunit is not important for the activity of sGC, this region is preserved in evolution in the α1-subunits of all vertebrates. It is possible that this region, often referred to as the regulatory region (37, 38), is responsible for the integration of NO/cGMP signaling with other regulatory pathways. In this case, despite similar catalytic properties, α1/β1 and C-α1/β1 heterodimers may be modulated differently by NO-independent mechanisms, e.g. protein modifications or protein-protein interactions.
cGMP-dependent kinase is one of the major effectors of cGMP generated by sGC in response to NO. Increased cGMP levels in neuronal cells leads to a PKGI-dependent phosphorylation of the Splicing Factor 1 (SF1), which functions at early stages of pre-mRNA splicing and splice site recognition (45). Thus, it is appealing that sGC-dependent cGMP production may regulate sGC activity by affecting the spliceosome assembly and/or switching between different splice sites during pre-mRNA processing. This, in turn, may affect the diversity of expressed α1 sGC splice proteins modulating the function of the sGC heterodimer.
In summary, our present study identifies and characterizes several new splice variants of the human α1-subunit of sGC. The splicing of α1 mRNA seems to be ubiquitous and follows the domain organization of the α1 sGC subunit. Our findings point to new mechanisms of modulation of sGC function and activity that may have significant effects on nitric oxide and cyclic GMP signaling and perhaps pathophysiology.
Supplementary Material
Acknowledgments
We thank the Quantitative Genomics Core Laboratory at University of Texas Medical School (Houston, TX) for help with real time quantitative RT-PCR.
This work was supported, in whole or in part, by National Institutes of Health Grants GM061731 (to F. M.) and HL088128 (to E. M.). This work was also supported by the John S. Dunn Foundation, Robert A. Welch Foundation Grant AU-1437, and a T5 grant from the Department of Defense. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1–S4.
Footnotes
The abbreviations used are: sGC, soluble guanylyl cyclase; RT, reverse transcriptase; RT-qPCR, real-time quantitative RT-PCR; ODQ, 1H-[1,2,4]oxadiazolo[4,3-a]quinoxaline-1-one.
G. L. Shipley, personal communication.
References
- 1.Furchgott, R. F., and Zawadzki, J. V. (1980) Nature 288 373–376 [DOI] [PubMed] [Google Scholar]
- 2.Katsuki, S., Arnold, W., Mittal, C., and Murad, F. (1977) J. Cyclic Nucleotide Res. 3 23–35 [PubMed] [Google Scholar]
- 3.Murad, F. (1986) J. Clin. Investig. 78 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rapoport, R. M., Draznin, M. B., and Murad, F. (1983) Nature 306 174–176 [DOI] [PubMed] [Google Scholar]
- 5.Humbert, P., Niroomand, F., Fischer, G., Mayer, B., Koesling, D., Hinsch, K. D., Gausepohl, H., Frank, R., Schultz, G., and Bohme, E. (1990) Eur. J. Biochem./ FEBS 190 273–278 [DOI] [PubMed] [Google Scholar]
- 6.Lee, Y. C., Martin, E., and Murad, F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe, A., Mergia, E., Dangel, O., Lange, A., and Koesling, D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7699–7704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mergia, E., Friebe, A., Dangel, O., Russwurm, M., and Koesling, D. (2006) J. Clin. Investig. 116 1731–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nimmegeers, S., Sips, P., Buys, E., Brouckaert, P., and Van de Voorde, J. (2007) Cardiovasc. Res. 76 149–159 [DOI] [PubMed] [Google Scholar]
- 10.Russwurm, M., Behrends, S., Harteneck, C., and Koesling, D. (1998) Biochem. J. 335 125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budworth, J., Meillerais, S., Charles, I., and Powell, K. (1999) Biochem. Biophys. Res. Commun. 263 696–701 [DOI] [PubMed] [Google Scholar]
- 12.Mergia, E., Russwurm, M., Zoidl, G., and Koesling, D. (2003) Cell. Signal. 15 189–195 [DOI] [PubMed] [Google Scholar]
- 13.Pyriochou, A., and Papapetropoulos, A. (2005) Cell. Signal. 17 407–413 [DOI] [PubMed] [Google Scholar]
- 14.Krumenacker, J. S., Hyder, S. M., and Murad, F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kloss, S., Srivastava, R., and Mulsch, A. (2004) Mol. Pharmacol. 65 1440–1451 [DOI] [PubMed] [Google Scholar]
- 16.Papapetropoulos, A., Marczin, N., Mora, G., Milici, A., Murad, F., and Catravas, J. D. (1995) Hypertension 26 696–704 [DOI] [PubMed] [Google Scholar]
- 17.Papapetropoulos, A., Abou-Mohamed, G., Marczin, N., Murad, F., Caldwell, R. W., and Catravas, J. D. (1996) Br. J. Pharmacol. 118 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papapetropoulos, A., Go, C. Y., Murad, F., and Catravas, J. D. (1996) Br. J. Pharmacol. 117 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, H., and Murad, F. (1974) J. Biol. Chem. 249 6910–6916 [PubMed] [Google Scholar]
- 20.Behrends, S., Harteneck, C., Schultz, G., and Koesling, D. (1995) J. Biol. Chem. 270 21109–21113 [DOI] [PubMed] [Google Scholar]
- 21.Behrends, S., and Vehse, K. (2000) Biochem. Biophys. Res. Commun. 271 64–69 [DOI] [PubMed] [Google Scholar]
- 22.Chhajlani, V., Frandberg, P. A., Ahlner, J., Axelsson, K. L., and Wikberg, J. E. (1991) FEBS Lett. 290 157–158 [DOI] [PubMed] [Google Scholar]
- 23.Okamoto, H. (2004) Int. J. Biochem. Cell Biol. 36 472–480 [DOI] [PubMed] [Google Scholar]
- 24.Ritter, D., Taylor, J. F., Hoffmann, J. W., Carnaghi, L., Giddings, S. J., Zakeri, H., and Kwok, P. Y. (2000) Biochem. J. 346 811–816 [PMC free article] [PubMed] [Google Scholar]
- 25.Bustin, S. A. (2000) J. Mol. Endocrinol. 25 169–193 [DOI] [PubMed] [Google Scholar]
- 26.Heid, C. A., Stevens, J., Livak, K. J., and Williams, P. M. (1996) Genome Res. 6 986–994 [DOI] [PubMed] [Google Scholar]
- 27.Martin, E., Lee, Y. C., and Murad, F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12938–12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, E., Sharina, I., Kots, A., and Murad, F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9208–9213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stasch, J. P., Schmidt, P. M., Nedvetsky, P. I., Nedvetskaya, T. Y., H, S. A., Meurer, S., Deile, M., Taye, A., Knorr, A., Lapp, H., Muller, H., Turgay, Y., Rothkegel, C., Tersteegen, A., Kemp-Harper, B., Muller-Esterl, W., and Schmidt, H. H. (2006) J. Clin. Investig. 116 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams, M. D., Rudner, D. Z., and Rio, D. C. (1996) Curr. Opin. Cell Biol. 8 331–339 [DOI] [PubMed] [Google Scholar]
- 31.Hofmann, F., Feil, R., Kleppisch, T., and Schlossmann, J. (2006) Physiol. Rev. 86 1–23 [DOI] [PubMed] [Google Scholar]
- 32.Burgoyne, J. R., Madhani, M., Cuello, F., Charles, R. L., Brennan, J. P., Schroder, E., Browning, D. D., and Eaton, P. (2007) Science (NY) 317 1393–1397 [DOI] [PubMed] [Google Scholar]
- 33.Tamura, N., and Garbers, D. L. (2003) J. Biol. Chem. 278 48880–48889 [DOI] [PubMed] [Google Scholar]
- 34.Kloss, S., Furneaux, H., and Mulsch, A. (2003) J. Biol. Chem. 278 2377–2383 [DOI] [PubMed] [Google Scholar]
- 35.Kloss, S., Rodenbach, D., Bordel, R., and Mulsch, A. (2005) Hypertension 45 1200–1206 [DOI] [PubMed] [Google Scholar]
- 36.Rothkegel, C., Schmidt, P. M., Atkins, D. J., Hoffmann, L. S., Schmidt, H. H., Schroder, H., and Stasch, J. P. (2007) Mol. Pharmacol. 72 1181–1190 [DOI] [PubMed] [Google Scholar]
- 37.Wagner, C., Russwurm, M., Jager, R., Friebe, A., and Koesling, D. (2005) J. Biol. Chem. 280 17687–17693 [DOI] [PubMed] [Google Scholar]
- 38.Koglin, M., and Behrends, S. (2003) J. Biol. Chem. 278 12590–12597 [DOI] [PubMed] [Google Scholar]
- 39.Shiga, T., and Suzuki, N. (2005) Zoological Sci. 22 735–742 [DOI] [PubMed] [Google Scholar]
- 40.Zabel, U., Weeger, M., La, M., and Schmidt, H. H. (1998) Biochem. J. 335 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibarra, C., Nedvetsky, P. I., Gerlach, M., Riederer, P., and Schmidt, H. H. (2001) Brain Res. 907 54–60 [DOI] [PubMed] [Google Scholar]
- 42.Kagota, S., Tamashiro, A., Yamaguchi, Y., Sugiura, R., Kuno, T., Nakamura, K., and Kunitomo, M. (2001) Br. J. Pharmacol. 134 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kloss, S., Bouloumie, A., and Mulsch, A. (2000) Hypertension 35 43–47 [PubMed] [Google Scholar]
- 44.Ruetten, H., Zabel, U., Linz, W., and Schmidt, H. H. (1999) Circulation Res. 85 534–541 [DOI] [PubMed] [Google Scholar]
- 45.Wang, X., Bruderer, S., Rafi, Z., Xue, J., Milburn, P. J., Kramer, A., and Robinson, P. J. (1999) EMBO J. 18 4549–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.