Abstract
The Ca2+ release-activated Ca2+ (CRAC) channel is a plasma membrane (PM) channel that is uniquely activated when free Ca2+ level in the endoplasmic reticulum (ER) is substantially reduced. Several small interfering RNA screens identified two membrane proteins, Orai1 and STIM1, to be essential for the CRAC channel function. STIM1 appears to function in the PM and as the Ca2+ sensor in the ER. Orai1 is forming the pore of the CRAC channel. Despite the recent breakthroughs, a mechanistic understanding of the CRAC channel gating is still lacking. Here we reveal new insights on the structure-function relationship of STIM1 and Orai1. Our data suggest that the cytoplasmic coiled-coil region of STIM1 provides structural means for coupling of the ER membrane to the PM to activate the CRAC channel. We mutated two hydrophobic residues in this region to proline (L286P/L292P) to introduce a kink in the first α-helix of the coiled-coil domain. This STIM1 mutant caused a dramatic inhibition of the CRAC channel gating compared with the wild type. Structure-function analysis of the Orai1 protein revealed the presence of intrinsic voltage gating of the CRAC channel. A mutation of Orai1 (V102I) close to the selectivity filter modified CRAC channel voltage sensitivity. Expression of the Orai1V102I mutant resulted in slow voltage gating of the CRAC channel by negative potentials. The results revealed that the alteration of Val102 develops voltage gating in the CRAC channel. Our data strongly suggest the presence of a novel voltage gating mechanism at the selectivity filter of the CRAC channel.
The CRAC2 channel is activated by depletion of the intracellular Ca2+ stores and mediates sustained Ca2+ entry in hematopoietic cells (1–3) and some other cell types (1, 3). Until recently, the molecular identity of the machinery linking store depletion with Ca2+ entry remained elusive. However, recent studies identified two proteins, STIM1 and Orai1, as being essential for CRAC function (4–11). Although STIM1 resides predominantly in the ER and the plasma membrane (PM) (11), Orai1 is almost exclusively expressed in the PM (12). STIM1 functions as the Ca2+ sensor in the ER that triggers the CRAC channel activation (5, 6, 11). However, its translocation to the PM upon activation (11, 13) and functional role in the PM have also been demonstrated (6, 14). Even though a report by Mignen et al. (14) revealed that PM-STIM1 regulates another Ca2+ channel (arachidonate-regulated Ca2+ channels), the same group later revealed that arachidonate-regulated Ca2+ channels have the same molecular identity as CRAC channels (Ref. 41; see also Ref. 15). STIM1 aggregates into puncta and translocates toward the PM, when the stores are depleted (5, 16). Physical incorporation of STIM1 molecules into the PM after Ca2+ store depletion has been clearly demonstrated (11, 13). The time course of the ER Ca2+ depletion followed by CRAC activation correlates with the time course of STIM1 translocation that is ∼52 s (5, 17).
How STIM1 relays signals about ER Ca2+ levels from the ER to the PM has not been determined. STIM1 contains one transmembrane domain and a number of potential protein-interacting domains on its N and C termini. The luminal/extracellular N terminus contains a helix-rich sterile α-motif in addition to an EF-hand Ca2+ sensor (6, 18). The cytoplasmic C terminus contains two prominent coiled-coil regions followed by a flexible proline-rich domain (6, 18). Based on the protein structure and several studies on STIM1, we hypothesize that two distinct types of interactions are likely important in the function of STIM1. First, homotypic interactions likely mediate the aggregation of the molecule into puncta after Ca2+ store depletion (19). Recent studies indicate that an N-terminal fragment of STIM1 that contains the sterile α-motif and the EF-hand domains is able to undergo self-association upon removal of Ca2+ (19). These results suggest that the ER luminal N terminus of STIM1 mediates the punctal self-assembly of STIM1 when the ER Ca2+ is depleted. Second, the “punctal” STIM1, which lies within ∼25 nm of the PM (5, 17), likely interacts directly with components in the PM, through its cytosolic domain. The latter can be achieved through homotypic interactions between the ER-bound STIM1 (STIM1-ER) and the PM-bound STIM1 (STIM1-PM). Earlier studies revealed that the coiled-coil region of STIM1 is indeed involved in homotypic interactions between STIM1 molecules (18). Alternatively STIM1-ER might be interacting with other PM protein or directly with the pore-forming protein Orai1. We consider the later less likely, based on a recent study that showed a lack of detectable interaction between STIM1 and Orai1 (20). Furthermore, biochemical cross-linking of the ER with the PM revealed that the distance between the ER and the PM at the sites of STIM1 puncta is too large (11–14 nm) for direct interactions of STIM1-ER with the PM Orai1 (21). That, however, is not the case for putative STIM1-ER and STIM1-PM interactions; hence the first coiled-coil region of STIM1 is an approximately 8-nm-long α-helix containing 15 turns (6) and can provide the structural means for ER-PM interactions during CRAC activation. The possibility that this cytosolic segment of STIM1 is essential for bridging the ER with the PM to activate CRAC channel was explored in the present study. Our data reveal that the intact structure of the first coiled-coil domain of STIM1 protein is crucial for the CRAC channel activation.
Orai1 is an integral membrane protein that works in synergy with STIM1 to reconstitute CRAC channel function (8, 9). In resting cells, Orai1 is diffusely distributed in the PM, but following store depletion it migrates to areas in close proximity to the near-PM STIM1 puncta, where local Ca2+ entry occurs (22). Orai1 has four predicted transmembrane domains (TM), a topology similar to the KCNK two pore domains K+ channels (23). The putative TM1-TM2 extracellular loop of human Orai1 contains a number of acidic residues, and the first one, Glu106, is the most conserved across species. Several groups simultaneously identified Glu106 (Glu180 in Drosophila) as a residue that coordinates the Ca2+ binding to the selectivity filter of the CRAC channel (24, 25).3 These experiments were repeated by another group using our Orai1E106D construct (26). All these results provided unequivocal evidence that Orai1 is a pore-forming subunit of the CRAC channel. In the present study we further examined the function of this residue and adjacent residues by structure-function analysis. Uniquely, our study was conducted on cells in which endogenous STIM1 and Orai1 proteins were knocked down, with a background overexpression of the constitutively active STIM1 EF-hand double mutant D76A/E87A. Under this condition, expression of Orai1 resulted in massive constitutive CRAC channel activity allowing a precise evaluation of the function of Orai1 and mutants thereof. In addition, using the constitutively active STIM1 protein, we could examine the channel properties independent of store emptying and the effects of Ca2+ release from the ER. We examined the properties of the CRAC channel selectivity for monovalent and divalent cations and demonstrated that expression of the nonselective E106D mutant results in a CRAC channel with preserved inward rectification in divalent cation-free solution (DVF), suggesting an intrinsic voltage gating of CRAC channel. Mg2+ and polyamines are known to induce inward rectification in K+ channels by voltage-dependent block (27–29). Our hypothesis of the presence of intrinsic voltage gating is further supported by the lack of effect of intracellular Mg2+ and polyamines on the endogenous CRAC channel conductance (30, 31). Here we further demonstrate that mutation of the Val102 residue of Orai1, close to the selectivity filter-defining Glu106 residue, results in a time-resolved voltage gating of the CRAC channel. This voltage gating was not affected by the ionic composition of the extracellular solution that is further consistent with an intrinsic mechanism.
EXPERIMENTAL PROCEDURES
Cells, Cell Culture, and Reagents—HEK293 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 1% penicillin/streptomycin. RBL cells were grown in the same medium but with heat-inactivated serum. ATP and G418 were from Sigma. Thapsigargin was from EMD Biosciences (San Diego, CA). Stealth siRNA duplexes for rat STIM1 and rat Orai1 were from Invitrogen. Enhanced yellow fluorescent protein (YFP) vector was from BD Biosciences (Mountain View, Ca).
RNA Interference Design and RNA Interference/DNA Transfection—All siRNA sequences were designed using Invitrogen Block-It software. A mixture of two siRNA duplex sequences exclusively targeting rat and not human STIM1 were used with start nucleotides 935 and 970. Two rat-specific siRNA targeting different regions of rat Orai1 and not human Orai1 were used for Orai1 knockdown with start nucleotides 488 and 837. Scrambled double-stranded RNA sequences were used as negative control. Transfections were performed by electroporation using the Gene Pulser II Electroporation system (Bio-Rad) at 350 V, 960 microfarads, and infinite resistance. Transfection with siRNA was followed by co-transfection with the indicated DNA expression constructs and YFP after 24 h. After 48 h the cells were selected by YFP fluorescence for recordings as previously described (6). In some cases the cells were transfected with one of two transfection reagents (FuGENE 6, Roche Applied Science; or Lipofectamine, Invitrogen) with 2–3 μg of DNA, ∼24 h after plating in 35-mm dishes of 250,000 cells.
DNA Expression Constructs and Mutagenesis—Wild type human STIM1 (STIM1WT) was subcloned into pIRESneo (Clontech, Palo Alto, CA) as previously described (18, 32). Orai1, expressed in pCMV6-XL4, was obtained from Origene (Rockville, MD). YFP conjugated to the N terminus of STIM1 and STIM1D76A constructs were gifts from Dr. Tobias Meyer. The human STIM1 and Orai1 mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing.
Immunoblot Analysis of Protein Expression—The cells were grown in 35-mm dishes and harvested 48 h after transfection as previously described (33). Protein concentrations were determined using a Bradford assay. The samples were retained on ice, mixed with prewarmed 2× loading buffer and immediately loaded onto the gel. Standard SDS-PAGE and transfer buffers were used for running the gel and for transferring the gel to a 0.45-μm pore nitrocellulose membrane (Schleicher & Schuell) at ∼70 mA. The membrane was blocked in 5% dry milk in washing buffer containing Tween (0.05%) and Tris-buffered saline and incubated overnight with primary anti-green fluorescent protein antibody at 1:3000 dilution (Sigma-Aldrich). The membrane was washed and incubated with a horseradish peroxidase-labeled secondary antibody and visualized with the enhanced chemiluminescence kit (LumiGlo, Upstate Cell Signaling Solutions, Lake Placid, NY).
Fluorescence Localization and Immunocytochemistry—To characterize the fluorescence distribution of the YFP-STIM1, YFP-STIM1D76A, and YFP-STIM1L286P/L292P proteins, the cells grown on coverslips were fixed 48 h after transfection, with or without 20 min stimulation by thapsigargin, using 4% paraformaldehyde in phosphate-buffered saline. Cell imaging was performed using a confocal microscope (Leica TCS-NT).
Electrophysiology—We used conventional whole cell recordings as described previously (6). The cells were transferred into the recording chamber on poly-l-lysine-coated coverslips prior to experiments and allowed to settle. Immediately after establishing the whole cell configuration, linear voltage ramps of 50-ms duration spanning the voltage range of –100 to +100 mV were delivered from a holding potential of 0 mV at a rate of 0.5 Hz, in some cases preceded by a prestep to various voltages as described in the text. The currents were filtered at 6 kHz and sampled at 10–50-μs intervals. We used automatic capacitive and series resistance compensation of the EPC-10 amplifier (HEKA). PatchMaster, Pulsetools, FitMaster, and Origin software were used for acquisition and analysis. The temporal development of inward (at –80 mV) and outward (at +80 mV) currents were measured from the individual ramps. The intracellular solution contained 145 mm CsGlu, 10 mm HEPES, 10 mm BAPTA, 8 mm NaCl, 4 mm MgCl2, 2mm Mg2ATP (8 mm Mg2+ total), pH 7.2. The excess 8 mm Mg2+ and ATP were used to ensure the inhibition of the endogenous TRPM7 channels (31, 34). The extracellular solution contained 145 mm NaCl, 10 mm CsCl, 2.8 mm KCl, 10 mm HEPES, 10 mm glucose, pH 7.4, with 2 mm EDTA added to form the DVF. 2 mm Mg2+ with 1 or 10 mm Ca2+ were added as indicated. The 20 and 30 mm Ca2+ solutions were prepared with addition of the appropriate amount of 120 mm CaCl2, pH 7.4 solution and the addition of MgCl2 to 2 mm Mg2+ final concentration. Na+ was replaced by NMDG+ in some experiments as indicated. We applied a 10-mV junction potential compensation as described for solutions with similar composition (35). In some cases we used a glutamate-based external solution 130 mm CsGlu, 20 mm CaGlu2, 2mm MgCl2, 10mm HEPES, pH 7.4, and the corresponding DVF containing 160 mm NaGlu, 10 mm EDTA, 10 mm HEPES, pH 7.4. These solutions did not give different results from the Cl–-based solutions. All current-voltage (I/V) relationships were filtered off-line at 1 kHz. The I/V curves in DVF were derived from the current ramp at the time of maximum activation defined from the current time dependence at –80 mV. All I/V curves recorded in Ca2+ are shown at the time of maximum activation. The currents after channel inactivation in DVF or block by 5 μm 2-aminoethoxydiphenyl borate or Gd3+ were used for leak subtraction. In some cases control cells with STIM1 and Orai1 knockdown were used for leak subtraction. The maximal CRAC currents at –100 mV after leak subtraction were used for statistical analysis. Each statistical bar plot is an average of three or more experiments. The stars on the bar plots represent significant difference from control (*, p < 0.05; **, p < 0.01; ***, p < 0.005). When referring to CRAC channel activity that is measured in cells co-expressing STIM1D76A/E87A and Orai1X mutants on a combined knockdown background of rOrai1 and rSTIM1, we refer to it as “Orai1X-CRAC channel” activity. The cells with transient expression of STIM1D76A/E87A when co-expressed with Orai1 or its mutants were healthy for recordings within 48 h after the transfection.
Ca2+ Measurement—HEK cells grown on coverslips were placed in “cation safe” medium free of sulfate and phosphate anions (107 mm NaCl, 7.2 mm KCl, 1.2 mm MgCl2, 11.5 mm glucose, 20 mm HEPES-NaOH, pH 7.2) and loaded with fura-2/acetoxymethylester (2 μm) for 30 min at 20 °C. The cells were washed, and dye was allowed to de-esterify for a minimum of 30 min at 20 °C. The cells on coverslips were placed in cation safe medium in the absence or presence of 1 mm CaCl2. Ca2+ measurements were performed using an InCyt dual wavelength fluorescence imaging system (Intracellular Imaging Inc.). Fluorescence emission at 505 nm was monitored with excitation at 340 and 380 nm; intracellular Ca2+ measurements are shown as 340/380-nm ratios. Ca2+ entry through CRAC channel was induced by incubation with 2 μm thapsigargin. All traces are averages from multiple cells and are representative of at least three separate experiments.
RESULTS AND DISCUSSION
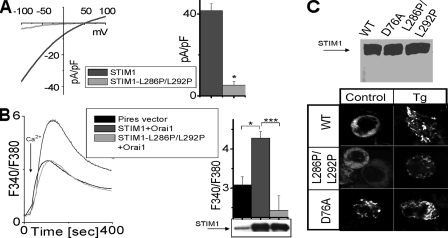
Introduction of a Kink in the First STIM1 Coiled-coil α-Helix Prevents the CRAC Channel Activation—The STIM1 protein is a Ca2+ sensor in the ER that transmits information to the PM to activate the CRAC channels when ER Ca2+ is depleted (5, 6, 11). Following store depletion, STIM1 aggregates into “punctal” regions of ER that lie within ∼25 nm of the PM (17, 22). Interaction with the PM proteins is likely through the large C-terminal cytoplasmic domain of STIM1, which includes two extended coiled-coil regions and a less structured proline-rich region (6). We previously demonstrated that deletion of the proline-rich region of STIM1 still permits CRAC channel activation (6). Here we address the role of the coiled-coil region of STIM1, in coupling of the ER with the PM to activate the CRAC channel. We expressed human STIM1WT and mutants thereof on a rat STIM1 knockdown background in RBL cells, as we undertook previously (6). We introduced a kink in the first, unusually long coiled-coil region of STIM1 by substituting two close leucine residues with prolines to perturb the α-helical structure. Expression of the single STIM1L292P mutation did not result in significant change in CRAC channel current amplitudes compared with the STIM1WT-expressing cells. However, expression of the double mutant STIM1L286P/L292P dramatically reduced the amplitude of ICRAC measured at its maximum activation in DVF (Fig. 1A). On average the current was decreased by 85% in cells expressing the mutated STIM1 compared with the STIM1WT (Fig. 1A, right panel). We also compared the ability of STIM1WT and STIM1L286P/L292P to increase CRAC-mediated Ca2+ entry in HEK293 cells using intracellular Ca2+ imaging (Fig. 1B). Co-overexpression of STIM1 and Orai1 resulted in a large increase of CRAC-mediated Ca2+ entry (Fig. 1B). However, when STIM1L286P/L292P was co-expressed with Orai1, the resulting CRAC-mediated Ca2+ entry was not higher than the background level (Fig. 1B). In fact, on average, the resulting Ca2+ elevation with co-expression of STIM1L286P/L292P and Orai1 was slightly decreased compared with the expression of the empty vector (Fig. 1B, right panel). To verify whether STIM1L286P/L292P was properly expressed, we examined the protein levels in HEK293 cells overexpressing STIM1WT and STIM1L286P/L292P. The cells expressing mutated STIM1L286P/L292P had protein levels similar to the wild type as detected by Western blot (Fig. 1B, right panel, bottom).
FIGURE 1.
The STIM1 coiled-coil domain is essential for CRAC channel activation. A, CRAC channels were measured in RBL cells expressing STIM1WT or STIM1L286P/L292P on a rSTIM1 knockdown background. The current-voltage relationship of CRAC channel was recorded at the time of maximal activation in DVF (left panel). Average currents at –100 mV reveal significant reduction of the CRAC currents in STIM1L286P/L292P expressing RBL cells (n = 5) compared with STIM1WT expressing cells (n = 4) (right panel). B, HEK cells co-expressing STIM1 or STIM1L286P/L292P together with Orai1 were pretreated with thapsigargin (Tg, 2 μm) in the absence of extracellular Ca2+ (10 min) to deplete Ca2+ stores. Control cells were transfected with empty Pires vector. 1 mm Ca2+ was added at the arrow to assess the store operated channel. A summary of the experiments is presented on the right with a Western blot of empty vector, STIM1, and STIM1L286P/L292P overexpressed in HEK cells (right panel, bottom). The STIM1-Orai1 co-expressing HEK cells had significantly elevated store operated channel, whereas STIM1L286P/L292P-Orai1 co-expressing cells had the store operated channel at the background level. C, top panel, Western blot of YFP-STIM1, YFP-STIM1L286P/L292P, and YFP-STIM1D76A overexpressed in HEK cells; bottom panel, distribution of the YFP-STIM1, YFP-STIM1L286P/L292P, and YFP-STIM1D76A proteins in HEK cells before (left column) and after (right column) store depletion by 2 μm thapsigargin.
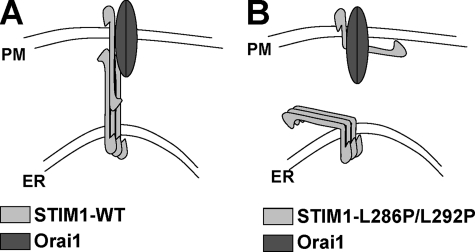
These results indicated that the STIM1L286P/L292P mutant was deficient in coupling from the ER to activate the PM channel. We considered whether the coupling defect was in the ability of the STIM1 protein to associate into puncta. After depletion of Ca 2+ stores with thapsigargin, the expressed STIM1L286P/L292P mutant in HEK293 cells was still able to enter puncta as observed by YFP-conjugated STIM1 and STIM1L286P/L292P localization (Fig. 1C, bottom right panel). Constitutively active YFP-STIM1D76A was distributed in puncta before and after store depletion as expected (Fig. 1C, bottom panel). The EF-hand of the STIM1D76A mutant has lower affinity to Ca2+ because of the reduced net negative charge (D76A) and is presumably free of Ca2+ in resting cells. Thus, the YFP-STIM1D76A distribution in resting HEK cells is similar to the STIM1WT distribution after stimulation. The YFP-STIM1D76A distribution demonstrates that the puncta formation under our experimental condition is not an artifact and depends on the Ca2+ dissociation from the EF hand of STIM1. Our results reveal that the STIM1L286P/L292P mutant is able to aggregate into puncta but is likely defective in the conformational coupling that allows successful gating of the CRAC channel (Fig. 2). Based on our data and that of others, we propose that the oligomerization of STIM1 within the ER membrane is dominated by the intermolecular interactions of the STIM1 EF-hand sterile α-motif domain region (19), whereas the interactions of ER-STIM1 with the PM proteins are structurally supported by the coiled-coil region of STIM1. Such interactions likely involve homomultimerization of ER-STIM1 with the PM-STIM1 (Fig. 2). That is consistent with an earlier study showing that the coiled-coil region of STIM1 plays a role in STIM1 homomultimerization (18) and with the lack of direct physical interactions between STIM1 and Orai1 (20, 21). The study by Varnai et al. (21) hypothesized the presence of intermediate players that connect the ER-STIM1 with the PM-Orai1 to open CRAC channel. That hypothesis is based on the finding that the distance between the ER and the PM at the sites of STIM1 puncta is ∼11–14 nm, too large for direct interaction between ER-STIM1 and PM-Orai1. However, interactions between the ER-STIM1 and the PM-STIM1 can span distance larger than 11 nm, based on the ∼8-nm length of the first coiled-coil domain (based on its predicted α-helical structure with 15 turns (6)). Overall our results reveal that the long coiled-coil region of STIM1 is necessary for successful gating of the CRAC channel.
FIGURE 2.
Model of STIM1 mediated ER-PM interactions via its coiled-coil domain. ER-STIM1 forms oligomers after store depletion that relocalize close to the PM. A, ER-STIM1 oligomers relocalized close to the PM interact with PM-STIM1 and/or Orai1 protein to activate CRAC channel. B, the interaction between the ER and the PM is abolished by the kink introduced in the coiled-coil domain of STIM1 by L286P/L292P mutation.
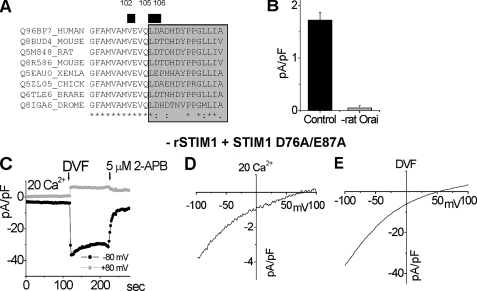
Analysis of the CRAC Channel Selectivity by Co-expression of Orai1 Mutants with a Constitutively Active STIM1 Mutant—The recently discovered Orai1 protein is a pore-forming protein of the CRAC channel (24, 25, 36).3 These results were later confirmed by Vig et al. (26) using our Orai1E106D construct. Orai1 has predicted topology similar to the KCNK class of K+ channel proteins (23), with four TMs, one extracellular loop between the TM1 and TM2, and a second one between TM3 and TM4. We examined the function of Orai1 mutants that affect the CRAC channel pore properties using RBL cells. In these cells the endogenous rat STIM1 was knocked down by rat-specific RNA interference, and the constitutively active hSTIM1D76A/E87A EF-hand mutant was expressed. The mutant causes constitutive CRAC channel activity obviating the nonspecific problems associated with the harsh conditions of cells Ca2+ store-emptying. When co-expressed, STIM1 and Orai1 are known to work in synergy to massively amplify CRAC channel function (8–10); thus we used this system to examine structure-function parameters of the Orai1 protein with precision. We initially focused on the domains within the Orai1 molecule that appear to form the functional CRAC channel pore. The first extracellular loop of Orai1 has a number of acidic residues, reminiscent of the pore-forming loop of Ca2+ channels known to have carboxylate-rich selectivity filters. Alignment of the first extracellular loop of Orai1 with Drosophila Orai and with Orai1 orthologs in other species is shown in Fig. 3A. The most conserved acidic residue, Glu106, coordinates Ca2+ binding at the selectivity filter. We introduced conserved mutations at and close to this position that were likely to influence the permeability properties of the channel. To knockdown endogenous rat Orai1 we used rat-specific siRNA, as previously described (6), and examined human Orai1 and mutants thereof on this knockdown background. Application of a mixture of two siRNAs targeted to different regions of Orai1 resulted in 98% reduction of the endogenous CRAC current (Fig. 3B). Thus, we examined the reconstitution of CRAC channel function by human STIM1D76A/E87A and Orai1 or its mutants. The cells with combined knockdown of STIM1 and Orai1 did not have measurable CRAC current. Characterization of STIM1D76A/E87A expressed alone in the rSTIM1 knockdown background cells is shown in Fig. 3 (C–E). The resulting CRAC channel was constitutively active with highly positive reversal potentials in 20 mm Ca2+ (Fig. 3D) and DVF (Fig. 3E), demonstrating preserved Ca2+ and monovalent ions selectivity. The current was rapidly inhibited by 5 μm 2-aminoethoxydiphenyl borate (Fig. 3C), similar to our previous report on STIM1E87A expression-induced CRAC channel activity (6). The selectivity for Cs+ versus Na+ was ∼0.1 calculated from the reversal potential (Fig. 3E).
FIGURE 3.
Functional characterization of the constitutively active STIM1D76A/E87A mutant used with different Orai1 mutants. A, alignment of the first extracellular loop (gray) of Orai1 generated using Clustal W. The most conserved residues are labeled with stars. The residues mutated in this study are labeled with a black bar and numbered. B, transfection with mixture of two siRNAs targeting rat Orai1 resulted in 98% reduction of the CRAC channel activity measured at –100 mV in 20 mm Ca2+ (n = 3) compared with the cells transfected with control scrambled siRNA and YFP (n = 3). C–E, characterization of the CRAC channels in cells expressing the constitutively active STIM1D76A/E87A. The STIM1D76A/E87A mutant is used further in the study to bypass store depletion. We used cells expressing STIM1D76A/E87A on rSTIM1 knockdown background (6). C, time course of constitutively active CRAC current after break-in, during DVF exchange, and after inhibition by 5 μm 2-aminoethoxydiphenyl borate (2-APB) shown at –80 mV and +80 mV. D, current-voltage relationship of the current in 20 mm Ca2+. E, current-voltage relationship in DVF measured at maximal activation.
With STIM1 and Orai1 co-expression on a knockdown background, the CRAC current levels were increased up to 60 pA/pF in 10 mm Ca2+ (Fig. 4, A–C), 900-fold higher than the knockdown background (Fig. 3B). The initial level of the constitutively active current was further increased by perfusing the cell cytosol with 10 mm BAPTA (Fig. 4, A and B). This partial initial inactivation of the current at the time of break-in, later recovered by BAPTA perfusion, is likely because of saturation of the endogenous Ca2+ buffers and CRAC inactivation by elevated cytosolic Ca2+ (37). To measure the fully activated CRAC current, we perfused the cell cytosol after break-in for at least 2 min with 10 mm BAPTA to allow diffusion of BAPTA into the cell before taking any statistical measurements. The current through Orai1WT-CRAC was decreased in proportion to the Ca2+ concentration, when extracellular Ca2+ was decreased from 10 to 1 mm (Fig. 4B). The current was highly Ca2+-selective, with a reversal potential above 50 mV, and was not affected by replacement of extracellular Na+ with nonpermeant NMDG+ (38) (Fig. 4B, see also Fig. 6C). The I/V curves in 10 mm Ca2+ and DVF (Fig. 4C) were identical to endogenous CRAC current, with a reversal potential in DVF of ∼50 mV.
FIGURE 4.
Mutation E106D in the first extracellular loop of Orai1 changes the CRAC channel selectivity. A–C, characterization of the CRAC current in cells co-expressing Orai1WT and STIM1D76A/E87A on combined rSTIM1 and rOrai1 knockdown background. A, time course of the constitutively active CRAC current after break-in. The observed activation is likely due to perfusion with 10 mm BAPTA (see text). B, reduction of the current by reduction of Ca2+ to 1 mm and inactivation in DVF (white bar); the current in 1 mm Ca2+ was not affected by replacement of Na+ with NMDG+. C, current-voltage relationship of Orai1WT-CRAC in 10 mm Ca2+ (black) and DVF (gray). D–F, characterization of the Orai1E106D-CRAC channel. D, the current at +80 mV and –80 mV in 10 Ca2+-Na+ external solution is reduced by replacement of Na+ by NMDG+, suggesting it is largely carried by Na+ ions. E, current-voltage relationship of Orai1E106D-CRAC current in DVF, 10 mm Ca2+-Na+, and 10 mm Ca2+-NMDG+. Orai1E106D-CRAC had much lower reversal potential in Ca2+ and DVF compared with Orai1WT-CRAC (C). F, average Orai1E106D-CRAC currents under the indicated extracellular ionic conditions. Note the reduction of the current in 10 Ca2+ compared with 1 Ca2+, suggesting inhibition of the channel by Ca2+.
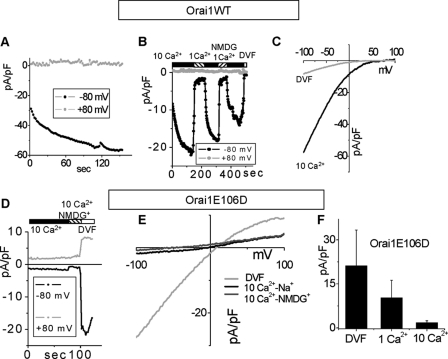
FIGURE 6.
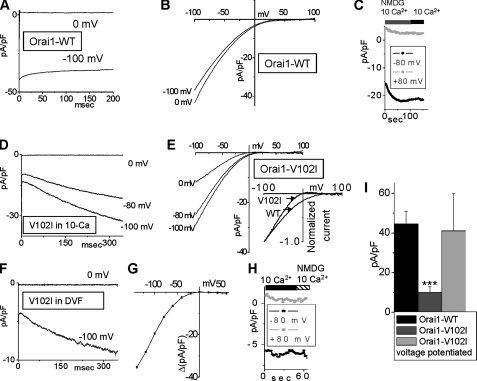
The Orai1V102I-CRAC channel is potentiated by negative voltages. A, Orai1WT-CRAC current did not increase at –100 mV, after equilibration in 10 mm Ca2+ and 0 mV holding potential. B, I/V values recorded immediately after the voltage steps shown in A. C, replacement of Na+ with NMDG+ did not affect the Orai1WT-CRAC current in 10 mm Ca2+. D, potentiation of the Orai1V102I -CRAC current during hyperpolarizing voltage steps, after equilibration in 10 mm Ca2+. E, I/V values were recorded immediately after the voltage steps in D. Inset, scaled I/Versus of Orai1V102I-CRAC and Orai1WT-CRAC for comparison. Orai1V102I-CRAC shows steeper voltage dependence then Orai1WT-CRAC. F, voltage potentiation of Orai1V102I-CRAC current in DVF. G, current difference between the beginning and the end of 350-ms voltage steps in 10 mm Ca2+ at different voltages is plotted versus voltage. The current was potentiated by potentials lower than 20 mV. H, replacement of Na+ with NMDG+ did not affect the Orai1V102I-CRAC current in 10 mm Ca2+, demonstrating preserved selectivity for Ca2+. I, maximal currents at –100 mV in 10 Ca2+ of Orai1WT-CRAC (n = 6), Orai1V102I-CRAC before (n = 9), and after voltage-dependent potentiation (n = 5).
Using the knockdown background of Orai1 and STIM1, we further re-examined the properties of the E106D mutant under different ionic conditions (Fig. 4, D–F). The Orai1E106D-CRAC current was outwardly rectifying and constitutively active in 10 mm external Ca2+ (Fig. 4, D–F). However, the inward current was reduced by more than 50% when Na+ solution was replaced by NMDG+ (Fig. 4, D and E), indicating that it was carried largely by Na+. Similar substitution did not have any effect on the Orai1WT-CRAC activity (Fig. 4C, see also Fig. 6C), demonstrating that similar to the endogenous CRAC, the constitutively active Orai1WT-CRAC current is highly selective for Ca2+, the major charge carrier in high extracellular Ca2+. The negative reversal potential of approximately –50 mV measured from the I/V relationship in 10 mm Ca2+-NMDG+ further revealed that the Orai1E106D-CRAC channel is not selective for Ca2+ versus Cs+ (Fig. 4E). Furthermore, when the external solution was switched to DVF (Fig. 4, D–F), the Orai1E106D-CRAC current exhibited a reversal potential of 5 ± 1 mV on average (n = 3), revealing low selectivity of Na+ versus Cs+ compared with Orai1WT-CRAC. Further characterization of the Orai1E106D-CRAC channel revealed that the inward current was on average lower in 10 mm Ca2+ then in 1 mm Ca2+ (Fig. 4F), demonstrating inhibition of the Na+ current by Ca2+. Our data demonstrate that Orai1E106D expression converts the CRAC channel permeability properties from a Ca2+ selective to nonselective, with a severely reduced selectivity for Cs+ versus Na+ and selectivity ratio increased from ∼0.1 for Orai1WT CRAC to ∼0.8 for the Orai1E106D-CRAC channel. The selectivity of Ca2+ versus Cs+ was also drastically reduced, suggesting that the E106D residue coordinates the binding of both monovalent and divalent ions to the selectivity filter. These results agree well with those published after completion of this work (24, 25) and further establish that the Orai1E106D-CRAC channel is nonselective for Ca2+ versus Cs+ (25, 26). We present these results here to demonstrate that the nonselective mutant Orai1E106D-CRAC has preserved inward rectification in DVF, suggesting that the channel has an intrinsic voltage gating mechanism. This conclusion is supported by the data on another nonselective mutant, Orai1E190Q-CRAC (26). Orai1E190Q-CRAC has similar to Orai1E106D-CRAC reversal potential and outward current amplitude in DVF but much smaller inward current amplitude and thus lacks the inward rectifying properties (26).
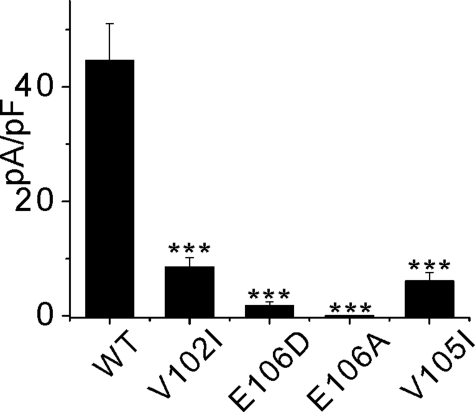
We characterized the Orai1 protein function, when residues close to the E106D residue were mutated to further examine the voltage gating properties of the CRAC channel. CRAC currents in 10 mm Ca2+ in cells expressing the V105I, V102I, E106D, and E106A mutants of Orai1 co-expressed with the STIM1D76A/E87A mutant were measured at –100 mV and summarized in Fig. 5A. Expression of E106A resulted in a complete absence of current above the background level (dead pore mutant) and expression of the E106D, V105I, and V102I mutants revealed drastic reduction of CRAC current in 10 mm Ca2+.
FIGURE 5.
Summary of the Orai1X-CRAC current amplitudes. Different mutants of Orai1 were co-expressed with the STIM1D76A/E87A mutant in RBL cells and the CRAC current amplitude was measured at the time of maximal activation at –100 mV in 10 mm Ca2+. Each bar represents average of three or more experiments.
Single Point Mutation of Orai1, V102I, Close to the Selectivity Filter Induces Slow Voltage Gating of the CRAC Channel—We examined in more detail the Orai1 function using the mutated V102I at the end of the first putative transmembrane domain (TM1) of Orai1 (Fig. 6). On average Orai1V102I-CRAC was constitutively active, with severely reduced whole cell Ca2+ conductance after equilibration in high Ca2+ (Fig. 5). However, atypical potentiation of the Orai1V102I-CRAC current by negative voltages was present, and we studied this phenomenon more thoroughly. The control Orai1WT-CRAC current was slightly inhibited after a hyperpolarizing prestep to –100 mV, likely because of fast Ca2+ inactivation. Fig. 6B compares the CRAC I/V after a prestep to –100 mV to the control CRAC I/V, where the cell was held at 0 mV before the voltage ramp. All of the recordings were done after equilibration in 10 mm extracellular Ca2+ to avoid effects of Ca2+-dependent potentiation. The control Orai1WT-CRAC I/V curves in Fig. 6B were recorded immediately after the voltage presteps shown in Fig. 6A. Replacement of Na+ with NMDG+ did not have any effect on the current (Fig. 6C). However, with Orai1V102I expression, a prestep to –100 mV increased CRAC current ∼3-fold with a time constant of >200 ms (Fig. 6, D and E). The amount of the inward rectification of Orai1V102I-CRAC correlated with the amplitude of the negative voltage prestep (Fig. 6E). Similar voltage potentiation of Orai1V102I-CRAC was observed in DVF (Fig. 6F), revealing that it is a phenomenon independent from the extracellular ion composition. Consistently, when extracellular Ca2+ was elevated to 30 mm, the voltage potentiation was preserved (not shown). Furthermore, the voltage dependence of the Orai1V102I-CRAC current was steeper compared with the control Orai1WT-CRAC current-voltage dependence after holding at 0 mV (Fig. 6E, inset). That further confirmed increased voltage dependence of the OraiV102I-CRAC channel. The potentiated portion of the current in 10 mm Ca2+ (Fig. 6G) had inwardly rectifying properties, consistent with the notion that the potentiated current has similar properties to CRAC channel current. The current amplitude of OraiV102I-CRAC in 10 mm Ca2+ was not affected by replacement of Na+ with NMDG+ (Fig. 6H), revealing that the Ca2+ selectivity of the Orai1V102I-CRAC channel is preserved and is similar to the Orai1WT-CRAC current. It is important to note that the reduced conductance of Orai1V102I-CRAC was not a result of lower expression or reduced plasma membrane localization of the Orai1V102I proteins, because the potentiated current revealed maximal whole cell currents similar to Orai1WT-CRAC (Fig. 6I). We hypothesize the presence of a gating charge at the selectivity filter of the CRAC channel. A similar phenomenon is observed in the KcsA K+ channel, where the reorientation of the Glu71 residue in the electric field induces destabilization of the inactivated conformation of the channel pore (39, 40). Our results, however, suggest that the gating charge assumes a position that stabilizes the open channel conformation (destabilizes the closed conformation) at negative voltages. We further propose that in the Orai1V102I-CRAC channel, the gating charge movement has a much slower component, compared with the WT-CRAC. The data reveal that the Val102 residue likely interacts with the gating charge, and the V102I mutation of this residue alters the gating charge movement.
Overall, we present strong evidence that the STIM1 coiled-coil domain plays a structural role in CRAC channel activation, probably by bridging important protein-protein interactions between the ER and the PM. The unusually long coiled-coil domain of STIM1 likely provides the structural features needed for connection between the two membranes to transfer the signal from the ER to the PM that activates CRAC channel. A likely candidate for such interactions is PM-STIM1 (6, 11, 13, 14). However, direct interactions with Orai1 are also possible. Furthermore, our results suggest that the Glu106 residue of Orai1 coordinates both Ca2+ and monovalent ions binding to the selectivity filter of the CRAC channel pore and is important for the channel selectivity. The nonselective Orai1E106D-CRAC channel revealed preserved inward rectification in DVF, suggesting intrinsic voltage gating in CRAC channels. Another mutation of Orai1, V102I, close to the selectivity site, modified the CRAC channel voltage dependence. The Orai1V102I-CRAC channel whole cell current was potentiated by negative voltages in a time-dependent manner. Overall the results demonstrate intrinsic voltage gating of CRAC channel with a voltage gating sensor at the selectivity filter. The CRAC channel voltage gating switch is coordinated by the Val102 residue in the first transmembrane domain of the protein. This last result reveals a mechanism of voltage gating in channels that lack the classical voltage sensor and is fundamentally important for understanding the phenomenon of voltage gating.
Acknowledgments
We thank Dr. Tobias Meyer for the gift of the YFP-STIM1 and YFP-STIM1D76A constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant AI058173 to the University of Maryland School of Medicine. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CRAC, Ca2+ release-activated Ca2+ channel; PM, plasma membrane; ER, endoplasmic reticulum; siRNA, small interfering RNA; TM, transmembrane domain; DVF, divalent cation-free solution; YFP, yellow fluorescent protein; BAPTA, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate; NMDG, N-methyl-d-glucamine.
M. Spassova, unpublished observations.
References
- 1.Parekh, A. B., and Penner, R. (1997) Physiol. Rev. 77 901–930 [DOI] [PubMed] [Google Scholar]
- 2.Prakriya, M., and Lewis, R. S. (2003) Cell Calcium 33 311–321 [DOI] [PubMed] [Google Scholar]
- 3.Parekh, A. B., and Putney, J. W., Jr. (2005) Physiol. Rev. 85 757–810 [DOI] [PubMed] [Google Scholar]
- 4.Roos, J., DiGregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., Safrina, O., Kozak, J. A., Wagner, S. L., Cahalan, M. D., Velicelebi, G., and Stauderman, K. A. (2005) J. Cell Biol. 169 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liou, J., Kim, M. L., Heo, W. D., Jones, J. T., Myers, J. W., Ferrell, J. E., and Meyer, T. (2005) Curr. Biol. 15 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spassova, M. A., Soboloff, J., He, L. P., Xu, W., Dziadek, M. A., and Gill, D. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 4040–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feske, S., Gwack, Y., Prakriya, M., Srikanth, S., Puppel, S. H., Tanasa, B., Hogan, P. G., Lewis, R. S., Daly, M., and Rao, A. (2006) Nature 441 179–185 [DOI] [PubMed] [Google Scholar]
- 8.Peinelt, C., Vig, M., Koomoa, D. L., Beck, A., Nadler, M. J., Koblan-Huberson, M., Lis, A., Fleig, A., Penner, R., and Kinet, J. P. (2006) Nat. Cell Biol. 8 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soboloff, J., Spassova, M. A., Tang, X. D., Hewavitharana, T., Xu, W., and Gill, D. L. (2006) J. Biol. Chem. 281 20661–20665 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, S. L., Yeromin, A. V., Zhang, X. H., Yu, Y., Safrina, O., Penna, A., Roos, J., Stauderman, K. A., and Cahalan, M. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9357–9362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, S. L., Yu, Y., Roos, J., Kozak, J. A., Deerinck, T. J., Ellisman, M. H., Stauderman, K. A., and Cahalan, M. D. (2005) Nature 437 902–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vig, M., Peinelt, C., Beck, A., Koomoa, D. L., Rabah, D., Koblan-Huberson, M., Kraft, S., Turner, H., Fleig, A., Penner, R., and Kinet, J. P. (2006) Science 312 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauser, C. T., and Tsien, R. Y. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3693–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mignen, O., Thompson, J. L., and Shuttleworth, T. J. (2007) J. Physiol. 579 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lis, A., Peinelt, C., Beck, A., Parvez, S., Monteilh-Zoller, M., Fleig, A., and Penner, R. (2007) Curr. Biol. 17 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liou, J., Fivaz, M., Inoue, T., and Meyer, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9301–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu, M. M., Buchanan, J., Luik, R. M., and Lewis, R. S. (2006) J. Cell Biol. 174 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams, R. T., Senior, P. V., Van Stekelenburg, L., Layton, J. E., Smith, P. J., and Dziadek, M. A. (2002) Biochim. Biophys. Acta 1596 131–137 [DOI] [PubMed] [Google Scholar]
- 19.Stathopulos, P. B., Li, G. Y., Plevin, M. J., Ames, J. B., and Ikura, M. (2006) J. Biol. Chem. 281 35855–35862 [DOI] [PubMed] [Google Scholar]
- 20.Gwack, Y., Srikanth, S., Feske, S., Cruz-Guilloty, F., Oh-Hora, M., Neems, D. S., Hogan, P. G., and Rao, A. (2007) J. Biol. Chem. 282 16232–16243 [DOI] [PubMed] [Google Scholar]
- 21.Varnai, P., Toth, B., Toth, D. J., Hunyady, L., and Balla, T. (2007) J. Biol. Chem. 282 29678–29690 [DOI] [PubMed] [Google Scholar]
- 22.Luik, R. M., Wu, M. M., Buchanan, J., and Lewis, R. S. (2006) J. Cell Biol. 174 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talley, E. M., Sirois, J. E., Lei, Q., and Bayliss, D. A. (2003) Neuroscientist 9 46–56 [DOI] [PubMed] [Google Scholar]
- 24.Yeromin, A. V., Zhang, S. L., Jiang, W., Yu, Y., Safrina, O., and Cahalan, M. D. (2006) Nature 443 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prakriya, M., Feske, S., Gwack, Y., Srikanth, S., Rao, A., and Hogan, P. G. (2006) Nature 443 230–233 [DOI] [PubMed] [Google Scholar]
- 26.Vig, M., Beck, A., Billingsley, J. M., Lis, A., Parvez, S., Peinelt, C., Koomoa, D. L., Soboloff, J., Gill, D. L., Fleig, A., Kinet, J. P., and Penner, R. (2006) Curr. Biol. 16 2073–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo, D., and Lu, Z. (2000) J. Gen. Physiol. 116 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spassova, M., and Lu, Z. (1998) J. Gen. Physiol. 112 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, Z., and MacKinnon, R. (1994) Nature 371 243–246 [DOI] [PubMed] [Google Scholar]
- 30.Bakowski, D., and Parekh, A. B. (2000) J. Physiol. 529 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermosura, M. C., Monteilh-Zoller, M. K., Scharenberg, A. M., Penner, R., and Fleig, A. (2002) J. Physiol. 539 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, R. T., Manji, S. S., Parker, N. J., Hancock, M. S., Van Stekelenburg, L., Eid, J. P., Senior, P. V., Kazenwadel, J. S., Shandala, T., Saint, R., Smith, P. J., and Dziadek, M. A. (2001) Biochem. J. 357 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel, K. A., Bartoli, K. M., Fandino, R. A., Ngatchou, A. N., Woch, G., Carey, J., and Tanaka, J. C. (2005) Investig. Ophthalmol. Vis. Sci. 46 2282–2290 [DOI] [PubMed] [Google Scholar]
- 34.Prakriya, M., and Lewis, R. S. (2002) J. Gen. Physiol. 119 487–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neher, E. (1992) Methods Enzymol. 207 123–131 [DOI] [PubMed] [Google Scholar]
- 36.Yeromin, A. V., Roos, J., Stauderman, K. A., and Cahalan, M. D. (2004) J. Gen. Physiol. 123 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zweifach, A., and Lewis, R. S. (1995) J. Gen. Physiol. 105 209–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakowski, D., and Parekh, A. B. (2002) Pfluegers Arch. Eur. J. Physiol. 443 892–902 [DOI] [PubMed] [Google Scholar]
- 39.Cordero-Morales, J. F., Cuello, L. G., and Perozo, E. (2006) Nat. Struct. Mol. Biol. 13 319–322 [DOI] [PubMed] [Google Scholar]
- 40.Cordero-Morales, J. F., Cuello, L. G., Zhao, Y., Jogini, V., Cortes, D. M., Roux, B., and Perozo, E. (2006) Nat. Struct. Mol. Biol. 13 311–318 [DOI] [PubMed] [Google Scholar]
- 41.Mignen, O., Thompson, J. L., and Shuttleworth, T. J. (2008) J. Physiol. (Lond.) 586 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]