Abstract
Introduction
Estrogens play a pivotal role in the initiation and progression of breast cancer. The genes that mediate these processes are not fully defined, but potentially include the known mammary oncogene MYC. Characterization of estrogen-target genes may help to elucidate further the mechanisms of estrogen-induced mitogenesis and endocrine resistance.
Methods
We used a transcript profiling approach to identify targets of estrogen and c-Myc in breast cancer cells. One previously uncharacterized gene, namely HBV pre-S2 trans-regulated protein 3 (HSPC111), was acutely upregulated after estrogen treatment or inducible expression of c-Myc, and was selected for further functional analysis using over-expression and knock-down strategies. HSPC111 expression was also analyzed in relation to MYC expression and outcome in primary breast carcinomas and published gene expression datasets.
Results
Pretreatment of cells with c-Myc small interfering RNA abrogated estrogen induction of HSPC111, identifying HSPC111 as a potential c-Myc target gene. This was confirmed by the demonstration of two functional E-box motifs upstream of the transcription start site. HSPC111 mRNA and protein were over-expressed in breast cancer cell lines and primary breast carcinomas, and this was positively correlated with MYC mRNA levels. HSPC111 is present in a large, RNA-dependent nucleolar complex, suggesting a possible role in ribosomal biosynthesis. Neither over-expression or small interfering RNA knock-down of HSPC111 affected cell proliferation rates or sensitivity to estrogen/antiestrogen treatment. However, high expression of HSPC111 mRNA was associated with adverse patient outcome in published gene expression datasets.
Conclusion
These data identify HSPC111 as an estrogen and c-Myc target gene that is over-expressed in breast cancer and is associated with an adverse patient outcome.
Introduction
Breast cancer is the major contributor to cancer incidence and mortality in women in the Western world. Although the genetic and environmental factors that lead to the initiation of breast cancer remain unclear, it is known that exposure to estrogens plays a crucial role in the development and progression of this disease [1]. It has been proposed that the causative link between estrogen and breast cancer is due to its potent mitogenic and antiapoptotic effects [2]. However, it is not fully understood how these effects are mediated at the molecular level. Such insight may provide clues to the mechanisms of estrogen-induced mitogenesis and cell survival, or resistance to endocrine therapies, or identify potential novel therapeutic targets for breast cancer, particularly in the settings of endocrine insensitivity and resistance. Thus, the identification and characterization of estrogen target genes is a major research priority.
The majority of breast cancers (about 75%) are estrogen receptor (ER)-positive, and estrogen is a potent mitogen for human breast cancer cells in vitro. The proliferation of ER-positive MCF-7 breast cancer cells in culture is inhibited by antiestrogens, and this effect is reversed by estrogen. Estrogen and antiestrogens regulate cell cycle entry and rates of progression during early G1 phase [3-5], and this is effected by modulation of G1 cyclin gene expression and activation of cyclin-dependent kinases 2 and 4 [6,7]. In addition, there is now evidence of converging activation of downstream estrogen signaling through crosstalk with growth factor-activated tyrosine kinase receptors [8]. Thus, there are compelling data suggesting that estrogen can mediate its growth effects by influencing the expression and function of genes critical to cell proliferation, by both 'genomic' and 'nongenomic' (cytoplasmic signaling) mechanisms [9].
One of the earliest transcriptional responses to estrogen is increased MYC expression [10]. Myc is a nuclear transcription factor that exhibits high-affinity and site-specific DNA-binding activity when complexed with its cellular partner Max, and it is rate-limiting for cell cycle progression through G1 phase [11], mediated in part through its effects on activation of cyclin-dependent kinases [12,13]. Inhibition of c-Myc expression abrogates estrogen-stimulated breast cancer cell proliferation [14], and blocks cell cycle progression leading to a G1 arrest [15]. Estrogen-regulated induction of MYC may play a critical role in the initiation of breast tumorigenesis, because MYC was the first mammary oncogene to be demonstrated by transgenesis [16]. These data strongly implicate c-Myc as an important mediator of the mitogenic function of estrogen, with a potential role in the initiation and progression of breast cancer. This concept is supported by studies demonstrating that Myc over-expression confers resistance to antiestrogens in vitro [17,18], and that inducible expression of c-Myc can replace estrogen in reinitiating cell cycle progression in antiestrogen-arrested breast cancer cells [12].
Because c-Myc can mimic the effects of estrogen on cell cycle progression in MCF-7 cells [12], we examined the transcriptional response to estrogen and to inducible c-Myc to identify novel targets of both estrogen and c-Myc in breast cancer cells (Musgrove EA, Sergio CM, Butt AJ, Sutherland RL; unpublished data). Here, we report an initial characterization of one such gene, namely HBV pre-S2 trans-regulated protein 3 (HSPC111). These studies reveal that HSPC111 is a direct transcriptional target of Myc, which is localized in the nucleolus and is over-expressed in several common cancers. Furthermore, elevated expression of HSPC111 is associated with reduced survival in breast cancer patients.
Materials and methods
Breast cancer cell lines and tissue samples
The human breast cancer cell line, MCF-7, was routinely maintained in RPMI-1640 medium supplemented with 10% fetal calf serum, 10 μg/ml insulin and 2.92 mg/ml glutamine under standard conditions. The human breast cancer mRNA samples utilized in this study have previously been described [19].
Quantitative real-time PCR
Total RNA was isolated using the RNAeasy kit (Qiagen Pty Ltd, Victoria, Australia) from cells pretreated with ICI 182780 (7α-[9-(4,4,5,5,5-pentafluoropentylsulfinyl) nonyl] estra-1,3,5,(10)-triene-3,17β-diol), which was a kind gift from Dr Alan Wakeling (Astra-Zeneca Pharmaceuticals, Alderly Park, Cheshire, UK), and then treated with 17β-estradiol, zinc, or vehicle. Quantitative real-time PCR was performed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using Taq-Man® probes for HSPC111 (Applied Biosytems). Data analyses were performed using the ΔCt method with RPLP0 (Applied Biosystems) as internal loading control. Fold changes in gene expression were calculated relative to the 0 hours time point. For correlation experiments, total RNA from a panel of breast cancer cell lines was isolated and quantitative real-time PCR was performed using Taq-Man® probes for MYC and HSPC111. Correlation was performed using standard linear regression analysis.
Immunoblot analysis
Cell lysates were collected as described previously [6]. Antibodies used were HSPC111 (see below) or V5 (Invitrogen Life Technologies Inc., Carlsbad, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Ambion, Austin, TX) or actin (Sigma, St Louis, MO, USA) was used as loading control.
Constructs
The sequence between -799 and +43 base pairs (bp) of the HSPC111 promoter was amplified by nested PCR from MCF-7-derived genomic DNA. The resulting 842 bp fragment was cloned into pGL3-Basic reporter construct (Promega, Madison, WI, USA).
Luciferase reporter assays
MCF-7 cells were transfected using Lipofectamine 2000 (Invitrogen) with luciferase reporter construct, renilla luciferase reporter construct, pRLSV40 (Promega), and either the c-Myc expression plasmid pCDNA3.1-cMyc or pcDNA3.1. Transfected cells were stimulated with increasing concentrations of zinc (up to 80 μmol/l) for 6 hours before harvesting. Luciferase activity was assayed 24 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega) and normalized to renilla luciferase activity. All values are relative to the activity of the pGL3-Basic reporter.
Electrophoretic mobility shift assays (EMSAs)
The sequences of the oligonucleotides used to investigate the three putative c-Myc binding sites in the HSCP111 promoter were as follows: HSPCsite1(TOP): 5'-CTAGGAGGCCCATGTGTCGCTG-3' ; HSPCsite1(BOT): 5'-CTAGCAGCGACACATGGGCCTC-3' ; HSPCsite2(TOP): 5'-CTAGGGCTCACACCTGTAATCC-3' ; HSPCsite2(BOT): 5'-CTAGGGATTACAGGTGTGAGCC-3' ; HSPCsite3(TOP): 5'-CTAGGCGGATCACCTGAGGTCA-3' ; HSPCsite3(BOT): 5'-CTAGTGACCTCAGGTGATCCGC-3' ; CAD(TOP): 5'-CTAGGTTAGCCACGTGGACCGA-3' ; and CAD(BOT): 5'-CTAGTCGGTCCACGTGGCTAAC-3'. The annealed oligonucleotides were radiolabeled with [α-32P]dCTP using Klenow fragment. Electrophoretic mobility shift assays were performed using nuclear extracts from MCF-7 cells. Equal amounts of nuclear extracts were incubated with the radiolabeled oligonucleotides following standard protocols, resolved on a 5% acrylamide gel and visualized by autoradiography. Competition assays were performed using 100-fold excess of competitor unradiolabeled oligonucleotides. The following oligonucleotides were used as nonspecific competitor oligonucleotides: 5'-CTAGTCTACTCCACTGCTGTCTATC-3' and 5'-CTAGGATAGACAGCAGTGGAGTAGA-3'.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed on chromatin from MCF-7/MycWT cells using a ChIP Assay Kit (Upstate Biotechnology, Millipore Corp. Billerica, MA, USA), following the manufacturer's instructions. Complexes were immunoprecipitated with c-Myc antibodies (9E10, C-33; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or a nonspecific PICK-1 antibody (Santa Cruz Biotechnology). The oligonucleotides used to detect the putative c-Myc binding sites in the HSPC111 promoter were as follows: HSPC111-ChIP P1: 5'-GAGTTTATTAAGCAGGGGAGTGGAG-3' ; HSPC111-CHIP P2: 5'-CCGCAGAAATGATTCCAAAACC-3', for site 1; HSPC111-CHIP P3: 5'-GTTGGTCAGGCTGGTCTTGAAC-3' ; HSPC111-ChIP P4: 5'-CGGACTTTGGAGTGGTGCTTAG-3', for site 3. For the analysis using quantitative real-time PCR, the following oligonucleotides were used: HSPC111-QPCR P1: 5'-TCCGCAGAAATGATTCCAAAA-3' ; and HSPC111-QPCR: P2 5'-AAGGGTCACTTCCTCCCCAG-3'.
Stable transfection
Full-length HSPC111 cDNA was generated by reverse transcription PCR from MCF-7 cells, and cloned into pDONR221 (Invitrogen). Constructs were recombined with the Gateway destination vector pcDNA3.1/nV5-pDEST (amino-terminal V5 fusion; Invitrogen), and then transfected into MCF-7 using Fugene-6 transfection reagent (Roche Applied Science, Indianapolis, IN, USA). Clones (MCF/HSPC) were selected and expanded in the presence of Geneticin (800 μg/ml; Invitrogen). Cells transfected with a pcDNA3.1/nV5-pDEST-LacZ vector were used as control (MCF/LacZ). MCF-7 cells inducibly expressing c-Myc wild-type (MCF/MycWT) or empty vector control cells were generated as previously described [12].
HSPC111-specific antibody production
Amino-terminal HIS-tagged HSPC111 was expressed in Escherichia coli BL21 (DE3) pLysS. Cultures were lyzed and proteins elutions were pooled for polyclonal antibody production in rabbits. Initial bleeds were purified using a protein A column and optimized for immunoblot analysis. Antibody specificity was confirmed in MCF/HSPC-NV5 cells by comparison with V5-tagged protein detected by immunocytochemisty and immunoblotting. The antibody detected both endogenous and V5-tagged HSPC-111 protein.
Cell proliferation and S phase analysis
HSPC111-expressing cells and LacZ controls were plated at 1 × 105 (day 0) and subsequently harvested and counted up to day 5. Exponentially growing MCF-7 cells expressing HSPC111 or LacZ controls were treated with 1 μmol/l 4-hydroxytamoxifen (Sigma), 10 nmol/l ICI 182780 or vehicle (ethanol) for 48 hours. Cells were harvested and S phase was analyzed by propidium iodide staining and flow cytometry.
Small interfering RNA
Small interfering (si)RNAs (siMyc17: D-003282-17-0050; siHSPC2: D-016096-02-0050; siHSPC4: D-016096-04-0050; siCONTROL RISC-Free siRNA: D-001220-01-20; and siRNA nontargeting control 2: D-001210-02-20) were purchased from Dharmacon (Lafayette, CO, USA) and transfected using Lipofectamine 2000 (Invitrogen). For estrogen 'rescue' experiments, cells were pretreated with ICI 182780 (10 nmol/l) at 24 hours after transfection and 48 hours later were treated with vehicle (ethanol) or 17β estradiol (100 nmol/l).
Immunofluorescence
Parental MCF-7 cells or those expressing V5-tagged HSPC111 were stained with anti-HSPC111, anti-V5 (Invitrogen), anti-nucleophosmin or anti-fibrillarin (Santa Cruz Biotechnology) antibodies and DAPI (4,6-diamidino-2-phenylindole), and were visualized using confocal microscopy.
Sucrose density gradient fractionation
Nuclear extracts from exponentially growing MCF-7 cells were separated by sucrose density gradient fractionation as described previously [20]. The gradients were analyzed through a UV monitor for continuous measurement of the absorbance at 254 nm and fractions collected. For immunoblot analysis, proteins from each fraction were precipitated with cold trichloroacetic acid at a final concentration of 10%.
Survival analyses
Datasets from two breast cancer cohorts using two different methodologies to analyze global gene expression were accessed. The first (referred to as the Uppsala cohort) is a group of 236 breast cancer patients [21] whose tumor RNA was analyzed using Affymetrix Genechip® (Affymetrix Inc., Santa Clara, CA, USA) HGU133A and B microarrays (NCBI GEO accession GSE3494; files were GSE3494-GPL96_series_matrix.txt.gz [HG U133A] and GSE3494-GPL97_series_matrix.txt.gz [HGU133B]). The second (from The Nederlands Kanker Instituut and designated the NKI cohort [22]) contained 295 cases that were assessed using Rosetta NKI spotted oligonucleotide arrays [23]. Datasets from both published series had complete data for clinicopathological variables and ER, progesterone receptor and HER2/neu status, as well as disease-specific survival. Univariate and multivariate analyses were performed as previously described [24] to assess the association of HSPC111 and MYC expression with survival using Statview 5.0 Software (Abacus Systems, Berkeley, CA, USA). P < 0.05 was considered statistically significant. The outcome variables were assessed as time to event, which was defined as the difference between the time of diagnosis and the time of death from breast cancer. Kaplan-Meier analysis was used for univariate analysis and to plot survival curves. Cox proportional hazards models were used to estimate hazard ratio (and its 95% confidence interval [CI]) associated with each risk factor and covariate and were also used for multivariate analyses.
Results
Identification of HSPC111 as an estrogen-regulated c-Myc target
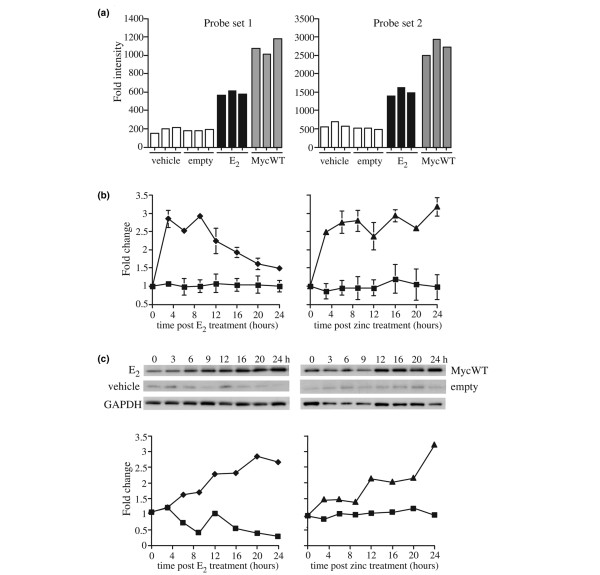
We have established an in vitro model to identify novel, estrogen-regulated targets of c-Myc in breast cancer cells [12]. RNA was collected from MCF-7 cells 6 hours after treatment with estrogen or after induction of c-Myc, and differential gene expression was determined using Affymetrix GeneChip Arrays (HG-U133 Plus V2.0) and Bayesian linear modeling methods in the limma package [25]. Candidate genes were selected from the list of probes that were significantly upregulated by both estrogen and c-Myc. The previously uncharacterized gene HSPC111 was among the most highly induced mRNAs identified in this analysis. Figure 1a shows the intensity of two probe sets for HSPC111 that were significantly upregulated by both estrogen and c-Myc.
Figure 1.
Identification of HSPC111 as an estrogen-regulated target of c-Myc. Cells were pretreated with ICI 182780 for 48 hours. Parental MCF-7 cells were then treated with either 17β-estradiol (diamonds) or vehicle (squares), and MCF-7/MycWT (triangles) and empty vector (squares) cells were treated with zinc. (a) HSPC111 mRNA expression in two probe sets from HG-U133 Plus V2.0 microarray platforms, 6 hours after treatment with estradiol (E2) or vehicle, or after expression of c-Myc (MycWT) or zinc treatment of empty vector cells (empty). (b) RNA was isolated at various time points as indicated and analyzed in triplicate by reverse transcription PCR with HSPC111-specific primers. Expression of HSPC111 is presented normalized to RPLP0. (c) immunoblot analysis of endogenous HSPC111 expression in whole cell lysates at time points up to 24 hours. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. Representative blots and densitometric analyses from three independent experiments are shown.
We confirmed the upregulation of HSPC111 by estrogen and c-Myc over a time course of treatment with 17β-estradiol or induction of c-Myc expression. MCF-7 cells were treated with 17β-estradiol or zinc (Myc or empty vector), and HSPC111 mRNA and protein expression was determined by quantitative real-time PCR and immunoblot, respectively. HSPC111 mRNA was rapidly induced (within 3 hours) after estrogen treatment or induced c-Myc expression compared with controls (Figure 1b), and reached a maximal 2.5-fold to 3-fold increase. Immunoblot analysis (Figure 1c) showed a similar increase in expression of endogenous HSPC111 protein after treatment with estrogen or induced c-Myc expression.
Estrogen's effects on HSPC111 expression are dependent upon Myc
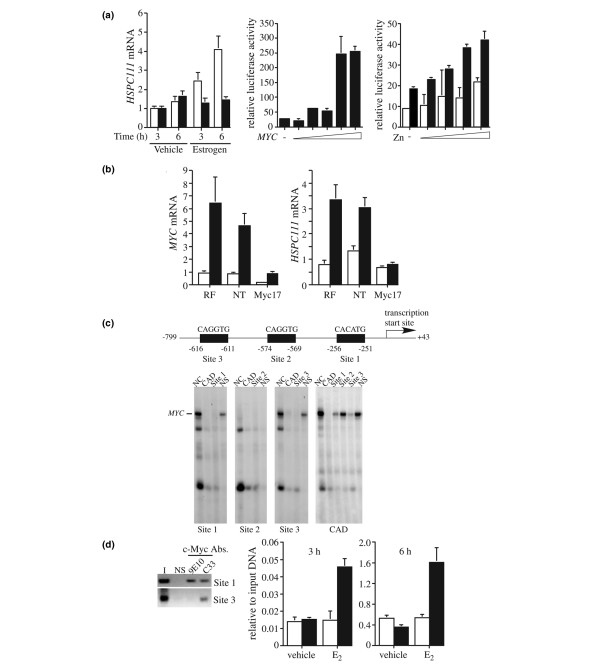
Initial experiments indicated that estrogen induction of HSPC111 mRNA was dependent on ongoing protein synthesis, because it did not occur in the presence of cycloheximide (Figure 2a). We then considered whether the effects of estrogen on HSPC111 expression are mediated via c-Myc. First, we determined whether c-Myc stimulated transcription from the HSCP111 promoter. A luciferase reporter construct containing 800 bp upsteam of the HSPC111 transcriptional start site was transfected into MCF-7 cells with increasing amounts of a c-Myc expression vector (Figure 2a). Co-expression of c-Myc resulted in a greater than sevenfold increase in luciferase activity. Increased luciferase expression was also observed in MCF-7/Myc cells upon zinc treatment compared with empty vector controls, particularly at higher concentrations of zinc (Figure 2a).
Figure 2.
Estrogen regulation of HSPC111 expression is dependent upon direct transcriptional activation by Myc. (a) Left panel: MCF-7 cells were arrested with ICI 182780 for 48 hours and then treated with cycloheximide (black bars) or control (white bars) before addition of estrogen or vehicle for 3 and 6 hours; levels of HSPC111 mRNA were determined by quantitative real-time PCR. Middle panel: MCF-7 cells were transfected with an HSPC111-luciferase reporter construct in the presence of increasing amounts of the c-Myc expression construct pCDNA3.1-cMyc. Right panel: MCF-7/MycWT (black bars) and empty vector controls (white bars) were transfected with the HSPC111-luciferase reporter construct and stimulated with increasing concentrations of zinc. Values are expressed as means ± standatrd deviation of triplicate samples from three independent experiments. (b) Left panel: MCF-7 cells were transfected with Myc-specific small interfering (si)RNA (siMyc17), RISC-free (RF), or nontargeting (NT) siRNA controls, or mock transfected with no siRNA. Transfected cells were arrested with ICI 182780 for 48 hours. Levels of MYC and HSPC111 mRNA were determined by quantitative real-time PCR after 24 hours of treatment with estradiol (black bars) or vehicle control (white bars). (c) Schematic showing the structure of the HSPC111 proximal promoter with the location of putative Myc-binding sites (E-boxes). Electrophoretic mobility shift assays demonstrate specific binding of c-Myc to E-boxes within the HSPC111 promoter. Radiolabeled oligonucleotides, as indicated above each gel, were incubated with nuclear extract from MCF-7 cells. Lane NC indicates no competitor oligonucleotides were added. The nonlabeled competitor oligonucleotides are indicated below each lane. Lane NS indictates nonspecific competitor oligonucleotide. (d) Chromatin was obtained from MCF-7/MycWT cells after 6 hours of treatment with zinc, and immunoprecipitated with c-Myc-specific or nonspecific (NS) antibodies as indicated. Left panel: Chromatin immunoprecipitation (ChIP) assay demonstrating the binding of c-Myc to the endogenous HSPC111 promoter. Lane I contains input chromatin that was not immunoprecipitated. Specific regions were then amplified by PCR using primers specific for site 1 or site 3, as indicated. Right panel: ChIP assay demonstrating the recruitment of c-Myc to the endogenous HSPC111 promoter in response to treatment with estradiol (E2) at 3 and 6 hours. Chromatin was immunoprecipitated with either a c-Myc-specific (C33; black bars) or a nonspecific (white bars) antibody and analyzed by quantitative real-time PCR using primers specific for site 1.
To determine whether estrogen's upregulation of HSPC111 is mediated via Myc, we examined the effects of estrogen on HSPC111 expression in the presence of Myc siRNA. MCF-7 cells were transfected with Myc-specific (siMyc17), RISC-free (RF), or nontargeting (NT) siRNA, and then arrested with ICI 182780 for 48 hours and stimulated with estradiol for 24 hours, when levels of MYC and HSPC111 mRNA expression were determined. The stimulation of MYC mRNA and protein was attenuated in the presence of siMyc17 (Figure 2b and data not shown). However, although HSPC111 expression was elevated in controls, there was no significant estrogen-mediated stimulation of HSPC111 mRNA in the siMyc17-treated cells (Figure 2b), indicating that estrogen stimulation of HSPC111 expression is dependent upon Myc expression increasing above the level achieved in the presence of siMyc.
HSPC111 is a direct transcriptional target of c-Myc
The rapid upregulation of HSPC111 mRNA by c-Myc suggested that it may be a direct transcriptional target of Myc. Analysis of the human HSPC111 genomic sequence revealed three sequences similar to the E-box consensus (CACATG) upstream of the transcriptional start site (Figure 2c). To determine whether c-Myc was able to bind to any of these putative E-boxes, a series of electrophoretic mobility shift assays were performed using double-stranded, radiolabeled oligonucleotides that encompass each of these sites or a known Myc consensus site from the carbamoyl phosphate synthetase-aspartate transcarbamylase-dihydroorotase (CAD) promoter. A band was identified in extracts incubated with radiolabeled oligonucleotides corresponding to sites 1, 3 and CAD, but not site 2 (Figure 2c). A competition assay with radiolabeled CAD oligonucleotide confirmed this as a Myc-specific band. Site 1 and 3 oligonucleotides were able to compete for c-Myc binding to a greater extent than either site 2 or a nonspecific oligonucleotide.
To establish whether the endogenous HSPC111 promoter is bound by c-Myc, ChIP assays were performed. Myc expression was induced in MCF-7/MycWT cells by zinc 6 hours before the ChIP assay being performed. Complexes were immunoprecipitated with c-Myc-specific antibodies raised to different epitopes or a nonspecific antibody, and then amplified by PCR using primers specific for site 1 or site 3. Both sites were successfully amplified from the chromatin that was immunoprecipitated with c-Myc antibodies but not with the nonspecific antibody (Figure 2d), suggesting that sites 1 and 3 in the HSPC111 promoter are bona fide c-Myc binding sites. We further determined whether stimulation with estrogen results in the recruitment of c-Myc to the endogenous HSPC111 promoter. Uninduced MCF-7/MycWT cells were treated with either 17β-estradiol or vehicle for 3 and 6 hours before performance of the ChIP assay with site 1 specific primers. Stimulation of the cells with estrogen resulted in a 4.5-fold enhancement in the binding of c-Myc to the HSPC111 promoter (Figure 2d).
HSPC111 localizes to the nucleolus
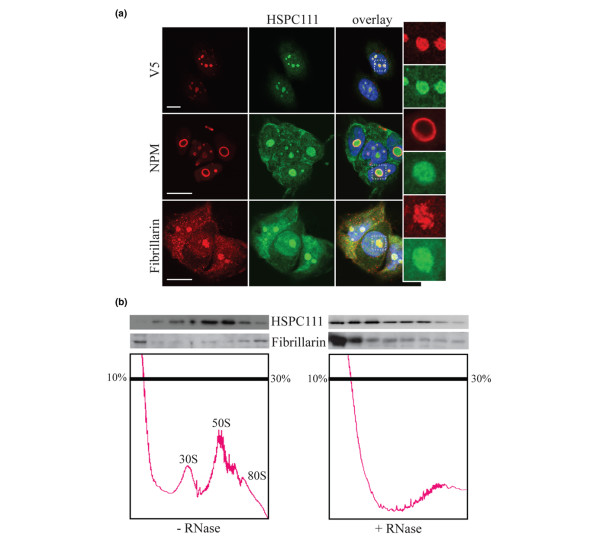
Next, we performed a series of experiments aimed at delineating a possible function for HSPC111. HSPC111 has been identified as a nucleolar protein of undefined function [26,27]. We examined this more closely in parental MCF-7 cells and those expressing V5-tagged HSPC111 by indirect immunofluorescence using antibodies against endogenous HSPC111 protein and the V5 tag. Figure 3a demonstrates prominent immunoreactivity of both endogenous and tagged HSPC111 in the nucleolus. Markers of different functional components of the nucleolus were used to define further its subnucleolar localization and hence provide insight into its possible function. The granular region of the nucleolus contains maturing ribosomes and can be identified by immunostaining with nucleophosmin (NPM)/B23, which is an abundant nucleolar phosphoprotein involved in mediating pre-rRNA processing [28]. Figure 3a shows no colocalization of HSPC111 and NPM/B23 within the nucleolus, indicating that HSPC111 is not present in the granular region and that HSPC111 and NPM/B23 probably reside within distinct protein complexes. Antibodies against fibrillarin were then used to label the dense fibrillar components, the site of newly synthesized preribosomal RNA [29]. Dual staining for fibrillarin and HSPC111 expression again demonstrated that each protein resided in distinct compartments of the nucleolus (Figure 3a), which suggests that HSPC111 is not localized in the dense fibrillar component.
Figure 3.
HSPC111 resides in high molecular weight protein complexes in the nucleolus. (a) Detection of endogenous and tagged HSPC111 by indirect immunofluorescence. Upper panels: Immunostaining of MCF-7/HSPC-NV5 cells with purified antibodies against endogenous protein (HSPC111; green) and the V5 tag (V5; red). Middle panels: Parental MCF-7 cells were stained with anti-HSPC111 (green) and anti-nucleophosmin (NPM; red) antibodies. Lower panels: Parental MCF-7 cells stained with anti-HSPC111 (green) and anti-fibrillarin (red) antibodies. DNA was counterstained with DAPI (4,6-diamidino-2-phenylindole; blue). Images are representative of at least two independent experiments. Bar = 10 μm. (b) Nuclear extracts of MCF-7 cells treated with or without RNase A were fractionated on sucrose density gradients. The trace from continuous monitoring of absorbance at 254 nm is shown. Fractions were precipitated and immunoblotted for HSPC111 and fibrillarin.
HSPC111 is a component of a large ribonucleoprotein particle
The nucleolus contains many large, multiprotein complexes with a variety of roles, principally in ribosomal biosynthesis. We next addressed whether HSPC111 might be contained within a ribonucleoprotein complex. Nuclear extracts from MCF-7 cells were subjected to sucrose density gradient fractionation to separate various nucleoprotein complexes, and then immunoblotted for HSPC111 expression. A significant proportion of HSPC111 was found in fractions of the gradient that also contained the 30S and 50S pre-preribosomal particles [29]. In contrast, fibrillarin was present in both the low molecular weight fractions at the top of the gradient and at the bottom of the gradient, near the 80S pre-preribosomal particle (Figure 3b). Following treatment with RNaseA to disrupt ribonucleoprotein particles, the sedimentation of both HSPC111 and fibrillarin predominantly shifted toward the top of the gradient (Figure 3b). The loss of pre-ribosomal peaks confirmed the disruption of ribonucleoprotein particles. Together these data indicate that, like fibrillarin, HSPC111 resides in a large multiprotein complex that requires RNA for its integrity.
Effects of modulation of HSPC111 expression on cell cycle progression
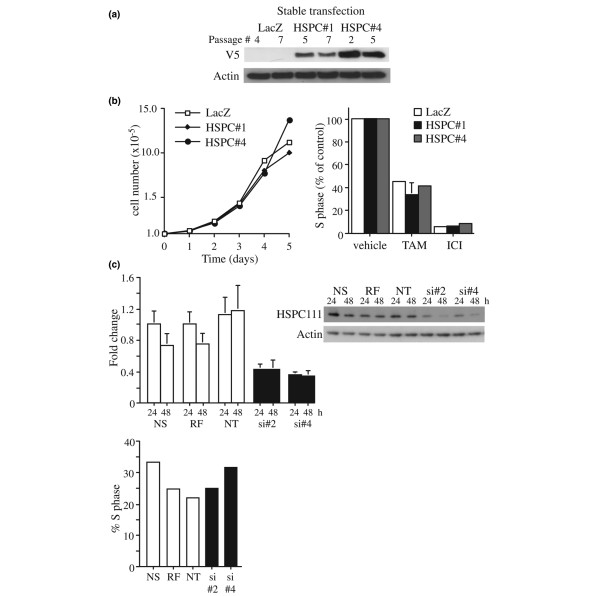
Given the proliferative role of Myc in our model and the well characterized link between RNA biosynthesis and cell proliferation [30], we asked whether HSPC111 could recapitulate this aspect of the Myc phenotype. This was examined using two clones of MCF-7 cells constitutively expressing HSPC111 (clones 1 and 4) and LacZ vector alone controls (Figure 4a). Cell proliferation assays exhibited no significant differences in cell number between HSPC111-expressing cells and controls up to 5 days after seeding (Figure 4b). We also examined the effect of HSPC111 expression on antiestrogen-induced cell cycle arrest. HSPC111-expressing cells were treated with tamoxifen or ICI 182780 for 48 hours then cell cycle progression was examined by flow cytometric analysis of propidium iodide stained cells. Treatment of LacZ control cells with antiestrogens resulted in an accumulation of cells in G1 phase and a decrease in the percentage of cells in S phase. Constitutive expression of HSPC111 did not significantly alter this response pattern (Figure 4b).
Figure 4.
Effects of modulation of HSPC111 expression on cell proliferation. (a) Immunoblot analysis of whole cell lysates from MCF-7 clones stably expressing HSPC111 (HSPC#1 and HSPC#4) or LacZ controls at various passages after transfection. Blots were analyzed for expression of tagged HSPC111 protein using V5 antibody or actin as a loading control. (b) Left panel: Growth curves of HSPC111 over-expressing clones and LacZ controls. Right panel: Stable transfectants were treated with tamoxifen (TAM), ICI 182780 (ICI), or vehicle for 48 hours, and then S phase was determined by flow cytometric analysis of propidium iodide-stained cells. (c) Upper panel: Endogenous HSPC111 mRNA and protein expression in MCF-7 cells 24 and 48 hours after transfection with HSPC111-specific small interfering (si)RNA (siHSPC2 and siHSPC4) determined by quantitative real-time PCR and immunoblot analysis with HSPC111 antibody, respectively. NS indicates mock transfection with no siRNA, RF indicates RISC-free control siRNA, and NT indicates nontargeting control siRNA. Lower panel: S phase was determined 48 hours after transfection by flow cytometric analysis of propidium iodide-stained cells.
The effect of decreased HSPC111 expression on the proliferation of MCF-7 cells was examined using HSPC111-specific siRNAs. Transfection of MCF-7 cells with two HSPC111-specific siRNAs (siHSPC2 and siHSPC4) resulted in a significant decrease in HSPC111 mRNA and protein expression at 24 and 48 hours compared with controls (Figure 4c). However, inhibition of endogenous HSPC111 expression had no significant effect on cell cycle progression in proliferating cells (Figure 4c). Thus, we concluded that HSPC111 did not have a significant role in mediating Myc-induced proliferation.
HSPC111 and c-Myc are over-expressed and correlated in human breast cancer cell lines and tissues
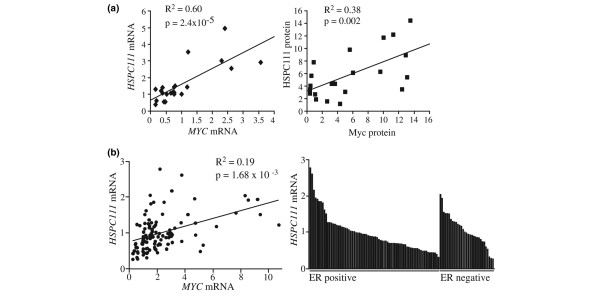
To further investigate HSPC111 as a Myc target gene in human breast cancer, we examined the expression of HSPC111 and Myc in a panel of breast cancer cell lines and primary breast cancers. A significant positive correlation between HSPC111 and Myc expression was observed at both the mRNA and protein level in 16 breast cancer cell lines (Figure 5a). We extended this study to examine the relationship between HSPC111 and MYC mRNA expression in a cohort of 105 primary breast carcinomas [19]. Comparative analysis of HSPC111 and MYC mRNA expression revealed a weak positive correlation in this breast cancer cohort (r2 = 0.19, indicating that MYC contributed only about 20% of the variance in HSPC111 levels; Figure 5b). Furthermore, HSPC111 mRNA expression was distributed similarly across both ER-positive and ER-negative breast cancers (mean relative expression 0.94 ± 0.06 and 1.03 ± 0.08 respectively; Figure 5b).
Figure 5.
Correlation between HSPC111 and c-Myc expression in human breast cancer cell lines and tumor samples. (a) Expression of HSPC111 and Myc mRNA and protein was determined by quantitative real-time PCR and immunoblot, respectively, in a panel of 16 breast cancer cell lines with either estrogen receptor (ER)-negative or ER-positive status. (b) Correlation between HSPC111 and MYC mRNA expression in primary breast cancers, and distribution of HSPC111 mRNA expression levels amongst ER-negative and ER-positive cancers.
We then utilized the cancer-profiling database Oncomine™ [31,32] and our own published studies [33,34] to further explore the relative expression of HSPC111 in normal versus cancer tissues from a range of published gene expression arrays. This confirmed that HSPC111 mRNA was significantly over-expressed in two other breast cancer cohorts [35,36] and in other steroid-regulated cancers, such as prostate [33,37] and ovarian carcinoma [34,38]. In addition, studies in other cancer types such as testis [39], liver [40], colon [41], and pancreas [42] revealed over-expression of HSPC111. Interestingly, some but not all of these studies showed a concurrent over-expression of MYC [37,39], suggesting that HSPC111 is regulated both by Myc-dependent and -independent pathways.
HSPC111 expression is associated with poor survival in breast cancer patients
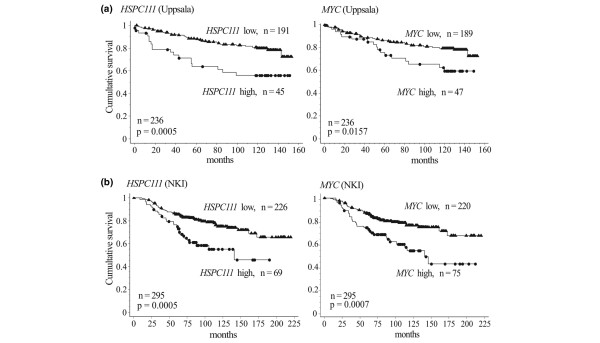
Datasets from two well characterized breast cancer cohorts were examined to assess whether the documented HSPC111 over-expression in breast cancer was associated with disease outcome. From the Uppsala cohort [21], two probesets that assessed expression of HSPC111 on the Affymetrix HGU133 microarrays satisfied quality control criteria for analysis. For both probesets, high expression of HSPC111 mRNA was associated with poor survival when modeled as a continuous variable using Cox proportional hazards analysis (probeset 203023_at: hazard ratio = 2.30, 95% CI 1.23 to 4.30; P = 0.0091; probeset 214011_s_at: hazard ratio = 3.13, 95% CI 1.62 to 6.06; P = 0.0007). None of the four probesets assessing MYC expression were associated with outcome when modeled as continuous variables (P > 0.13).
To dichotomize data for both cohorts, a cutpoint was selected to identify approximately the upper quartile of HSPC111 expressors in both studies. As shown in Figure 6a, Kaplan-Meier analysis demonstrated that high expression of HSPC111 was associated with poor survival for both probesets (203023_at: 45/236 [19.1%] with high expression; P = 0.0005; 214011_s_at: 66/236 [28%]; P = 0.0137). Similarly, approximately the highest quartile was selected for MYC, reflecting the expected proportion of cancers that would probably have amplification of the gene. The only probeset that demonstrated an association between high MYC expression and a poor survival was 224340_at (47/236 [19.9%], P = 0.0157). Multivariate modeling identified that HSPC111 expression was an independent prognostic factor with the final resolved model shown in Table 1. HSPC111 was independent of the influence of high MYC expression on survival (shown as a bivariate model in Table 1).
Figure 6.
High HSPC111 expression is associated with poor survival in breast cancer cohorts. Kaplan-Meier survival curves of the relationship between HSPC111 and MYC mRNA expression and survival in two independent publicly available breast cancer cohorts: (a) Uppsala cohort and (b) the Nederlands Kanker Instituut (NKI) cohort.
Table 1.
Multivariate analysis of clinicopathological parameters and HSPC111 expression in two breast cancer cohorts
| Variable | Hazard ratio (95% confidence interval) | P |
| Uppsala cohort | ||
| Tumor size > 20 mm | 3.20 (1.65 to 6.21) | 0.0006 |
| Lymph node positive | 3.13 (1.79 to 5.46) | < 0.0001 |
| HER2 high expression | 1.82 (1.02 to 3.27) | 0.0444 |
| HSPC111 high expression | 2.29 (1.30 to 4.03) | 0.0043 |
| HSPC111 high expression | 2.51 (1.43 to 4.41) | 0.0014 |
| MYC high expression | 1.89 (1.06 to 3.36) | 0.0305 |
| NKI cohort | ||
| Tumour size > 20 mm | 1.90 (1.10 to 3.28) | 0.0223 |
| Tumour grade > 2 | 2.04 (1.22 to 3.44) | 0.0070 |
| ER positive | 0.67 (0.38 to 1.18) | 0.1636 |
| PR positive | 0.56 (0.32 to 1.00) | 0.0490 |
| HER2 high expression | 1.46 (0.87 to 2.43) | 0.1493 |
| HSPC111 high expression | 1.29 (0.79 to 2.12) | 0.3074 |
| HSPC111 high expression | 1.89 (1.17 to 3.08) | 0.0097 |
| MYC high expression | 1.83 (1.14 to 2.93) | 0.0118 |
NKI, Nederlands Kanker Instituut.
In the dataset from The Nederlands Kanker Instituut cohort (NKI [22]), one probe assessed expression of HSPC111. High expression of HSPC111 mRNA was associated with poor survival when modeled as a continuous variable using Cox proportional hazard analysis (hazard ratio = 16.6, 95% CI 3.25 to 85.26; P = 0.0007). High MYC expression was also associated with poor survival (hazard ratio = 2.63, 95% CI 1.16 to 5.92; P = 0.02). Adopting a similar strategy for dichotomization of gene expression variables in this cohort demonstrated that, as with the Uppsala cohort, high HSPC111 expression (which occurred in 69/295 [23.4%] cases) was associated with poor survival (P = 0.0005), and high MYC expression (in 75/295 [25.4%] cases) was also associated with poor survival (P = 0.0007). Multivariate modeling demonstrated that in this cohort, HSPC111 expression was not an independent prognostic factor with the final resolved model shown in Table 1. However, as with the Uppsala cohort, HSPC111 was independent of the influence of high MYC expression on survival, which is shown as a bivariate model in Table 1.
Discussion
Although it is now well established that the mitogenic effects of estrogen play a pivotal role in the initiation and progression of breast cancer, how these effects are mediated at the molecular level remains to be fully elucidated. The transcription factor c-Myc is a prominent player in the response of breast cancer cells to estrogen, mimicking the effects of estrogen on cell cycle progression [12] and conferring resistance to antiestrogens in vitro [14,15,43]. Thus, identification and characterization of key downstream effectors of estrogen and Myc action will not only provide a greater insight into estrogen effects on mitogenesis and survival, but could also lead to an enhanced understanding of the mechanisms governing endocrine resistance [8].
In a search for estrogen-target genes that are regulated secondarily to estrogen's induction of c-Myc, we identified a novel gene of unknown function, namely HSPC111, which was among the most highly regulated estrogen and Myc target genes in our model [12] (Musgrove EA, Sergio CM, Butt AJ, Sutherland RL; unpublished data). HSPC111 is rapidly (within 3 hours) upregulated in response to treatment with estrogen (about threefold) and induction of Myc (about fourfold). However, the response to estrogen is abrogated in the presence of Myc siRNA, providing strong evidence that estrogen stimulates HSPC111 expression via its well documented upregulation of Myc. This conclusion is further supported by our demonstration of functional E-boxes in the HSPC111 promoter, and Myc-responsive promoter activity, identifying HSPC111 as a direct transcriptional target of Myc. Although gene expression profiling has recently identified HSPC111 as a target of estrogen [44] and Myc [45], this is the first report demonstrating that estrogen's effects on HSPC111 are dependent upon a direct transcriptional activation by Myc.
Although HSPC111 is a previously uncharacterized protein, it is known to reside in the nucleolus [46]. In an attempt to elucidate a cellular role for HSPC111, we further investigated its subcellular localization. The nucleolus is the center of ribosomal biosynthesis and assembly [29]. HSPC111 did not colocalize with either NPM/B23 or fibrillarin, both of which are known to play a role in ribosomal biosynthesis. However, sucrose density fractionation demonstrated that HSPC111 is part of a RNA-dependent complex sedimenting in the 40 to 80S region, which also contains preribosomal ribonucleoprotein particles [29]. In addition to driving cell division, Myc plays a crucial role in controlling cell growth and protein synthesis [47]. Thus, the acute transcriptional regulation of HSPC111 by Myc may represent part of a coordinated stimulation of ribosome biogenesis [47], occurring concurrently with its stimulation of cell proliferation. However, whether HSPC111 has a role in the ribosomal biosynthesis pathway is not clear from these studies and requires further investigation.
Recent studies have emphasized an important link between nucleolar function, in particular ribosomal biogenesis, and cell cycle control, and several genes coordinately regulate both processes. For example, disruption of the nucleolar PeBoW complex, consisting of Pes1, Bop1 and WDR12, blocks both rRNA processing and cell cycle progression [48,49]. Given the proliferative role of Myc in our model and our data suggesting HSPC111 interacts with RNA in the nucleolus, we questioned whether HSPC111 might play a role in Myc's effects on cell cycle progression. However, modulation of HSPC111 expression had no effect on cell proliferation end-points. We detected no effect of constitutive HSPC111 expression on proliferation, and although it is possible that the level of over-expression achieved was not sufficient for a detectable increase in proliferation rate, HSPC111 expression was not required for cell cycle progression, and neither was its downregulation required for antiestrogen inhibition of proliferation. These data are supported by Schlosser and coworkers [45], who identified HSPC111 as a Myc target gene in the human B-cell line P493-6 under conditions in which Myc induces cell growth but not cell proliferation [50]. Furthermore, they suggested that, even if HSPC111 does play a role in rRNA synthesis, there is either an element of functional redundancy in its role or it is not rate-limiting for cell cycle progression. Indeed, although adequate cell growth is essential for proliferation, it is not sufficient, requiring additional proliferative signals for cell cycle progression to proceed [51]. These data emphasize the complexity of the Myc phenotype, even within the relative restrictions of our model system, and support the concept that the coordinated regulation of multiple effector genes is required to recapitulate Myc functions [52].
To address further a potential role of HSPC111 in cancer, we initially identified a strong positive correlation between MYC mRNA and both HSPC111 mRNA and protein in breast cancer cell lines, raising the possibility that HSPC111 expression might be a useful surrogate marker of Myc over-expression in breast cancer. However, the relationship at the mRNA level was less robust in primary breast cancer (r2 = 0.19 versus 0.60 for primary cancer versus cell lines). An extension of this analysis to published datasets from a number of other cancers identified that elevated expression of HSPC111 was a feature of several cancers including those of the breast, prostate, ovary, testis, liver, colon, and pancreas [31,41], but this was not always associated with MYC over-expression. Thus, HSPC111 over-expression appears to be a common feature of many cancers but its relationship to aberrant Myc function, which is only contributed in part by MYC mRNA levels, remains to be elucidated. More importantly, HSPC111 over-expression was a strong predictor of an adverse outcome in two cohorts of breast cancer patients on univariate analysis and remained significant in a multivariate model in the Uppsala cohort. These effects were independent of MYC mRNA over-expression, which is in support of our conclusions from other cancers. Whether HSPC111 over-expression is functionally associated with disease progression remains an open question. The data presented here failed to support a role in cell proliferation or endocrine sensitivity, but other aspects of the biology of tumor progression require further investigation. It is well established that aberrant cell growth (increased/dysregulated ribosome biogenesis and protein synthesis) are common features of cancer. Because our preliminary data point to nucleolar localization of HSPC111 in association with ribonucleoproteins, it may be either functionally associated with these processes or a marker of aberrant cell growth regulation in general. In any event, further investigation of the normal physiological role of HSPC111 in nucleolar function and the functional consequences of overexpression on cellular growth control are warranted.
Conclusion
In summary, we have identified HSPC111 as an estrogen-responsive, Myc target gene in breast cancer cells. Although the precise function of HSPC111 remains unclear from our studies, its over-expression is common in a number of cancer cell types, and its association with poor outcome in breast cancer cohorts warrants its further analysis as an effector of estrogen and Myc action in both normal and neoplastic growth.
Abbreviations
bp = base pairs; CAD = carbamoyl phosphate synthetase-aspartate transcarbamylase-dihydroorotase; ChIP = chromatin immunoprecipitation; CI = confidence interval; ER = estrogen receptor; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; NKI = Nederlands Kanker Instituut; NPM = nucleophosmin; PCR = polymerase chain reaction; si = small interfering.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AJB, RLS, and EAM conceived the study, participated in its design, coordination and interpretation, and drafted the manuscript. AJB additionally generated cell lines over-expressing HSPC111. CMS performed analyses of HSPC111 expression, sucrose density gradient fractionation, and analyses of cells following HSPC111 over-expression or knock-down. CKI performed analyses of HSPC111 transcriptional regulation, participated in sucrose density fractionation, measured gene expression in breast cancers, and helped to draft the manuscript. LRA performed immunolocalization experiments, participated in HSPC111 knock-down experiments, and measured gene expression in breast cancer cell lines. CMM analyzed HSPC111 expression in breast cancers. AJR generated and characterized the HSPC111 antibody. MN and TP participated in the design and interpretation of sucrose density fractionation. AVB examined HSPC111 association with disease outcome and helped to draft the manuscript. All authors read and approved the final version.
Acknowledgments
Acknowledgements
The authors thank Associate Prof Sue Henshall and Dr Pip O'Brien for assistance with analysis of data from prostate and ovarian cancers, Dr Will Hughes for assistance with confocal microscopy, and Chehani Alles and Mark Pinese for assistance with microarray data analysis. This research was supported by the National Health and Medical Research Council (NHMRC) of Australia, the RT Hall Trust, the Australian Cancer Research Foundation, and the Association for International Cancer Research. AJB, AVB and EAM are Research Fellows of the Cancer Institute NSW, and CMM and LRA are Cancer Institute NSW Research Scholars. CMM is the recipient of an NHMRC Postgraduate Scholarship. RLS is a Senior Principal Research Fellow of the NHMRC.
Contributor Information
Alison J Butt, Email: a.butt@garvan.org.au.
C Marcelo Sergio, Email: m.sergio@garvan.org.au.
Claire K Inman, Email: c.inman@garvan.org.au.
Luke R Anderson, Email: l.anderson@garvan.org.au.
Catriona M McNeil, Email: c.mcneil@garvan.org.au.
Amanda J Russell, Email: a.russell@garvan.org.au.
Marco Nousch, Email: m.nousch@victorchang.edu.au.
Thomas Preiss, Email: t.preiss@victorchang.edu.au.
Andrew V Biankin, Email: a.biankin@garvan.org.au.
Robert L Sutherland, Email: r.sutherland@garvan.org.au.
Elizabeth A Musgrove, Email: e.musgrove@garvan.org.au.
References
- Colditz GA. Relationship between estrogen levels, use of hormone replacement therapy, and breast cancer. J Natl Cancer Inst. 1998;90:814–823. doi: 10.1093/jnci/90.11.814. [DOI] [PubMed] [Google Scholar]
- Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. N Engl J Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- Sutherland RL, Hall RE, Taylor IW. Cell proliferation kinetics of MCF-7 human mammary carcinoma cells in culture and effects of tamoxifen on exponentially growing and plateau-phase cells. Cancer Res. 1983;43:3998–4006. [PubMed] [Google Scholar]
- Taylor IW, Hodson PJ, Green MD, Sutherland RL. Effects of tamoxifen on cell cycle progression of synchronous MCF-7 human mammary carcinoma cells. Cancer Res. 1983;43:4007–4010. [PubMed] [Google Scholar]
- Osborne CK, Boldt DH, Clark GM, Trent JM. Effects of tamoxifen on human breast cancer cell cycle kinetics: accumulation of cells in early G1 phase. Cancer Res. 1983;43:3583–3585. [PubMed] [Google Scholar]
- Prall OWJ, Sarcevic B, Musgrove EA, Watts CK, Sutherland RL. Estrogen-induced activation of cdk4 and cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-cdk2. J Biol Chem. 1997;272:10882–10894. doi: 10.1074/jbc.272.16.10882. [DOI] [PubMed] [Google Scholar]
- Foster JS, Henley DC, Ahamed S, Wimalasena J. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol Metab. 2001;12:320–327. doi: 10.1016/s1043-2760(01)00436-2. [DOI] [PubMed] [Google Scholar]
- Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: the potential roles of c-Myc, cyclin D1 and cyclin E. Endocr-Rel Cancer. 2005;12:S47–S59. doi: 10.1677/erc.1.00993. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Aitkenhead M, Hughes CCW, Levin ER. Integration of the non-genomic and genomic actions of estrogen. J Biol Chem. 2002;277:50768–50775. doi: 10.1074/jbc.M210106200. [DOI] [PubMed] [Google Scholar]
- Dubik D, Shiu RP. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J Biol Chem. 1988;263:12705–12708. [PubMed] [Google Scholar]
- Hanson KD, Shichiri M, Follansbee MR, Sedivy JM. Effects of c-myc expression on cell cycle progression. Mol Cell Biol. 1994;14:5748–5755. doi: 10.1128/mcb.14.9.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall OWJ, Rogan EM, Musgrove EA, Watts CK, Sutherland RL. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph B, Saffrich R, Zwicker J, Henglein B, Muller R, Ansorge W, Eilers M. Activation of cyclin-dependent kinases by Myc mediates induction of cyclin A, but not apoptosis. EMBO J. 1996;15:3065–3076. [PMC free article] [PubMed] [Google Scholar]
- Watson PH, Pon RT, Shiu RP. Inhibition of c-myc expression by phosphorothioate antisense oligonucleotide identifies a critical role for c-myc in the growth of human breast cancer. Cancer Res. 1991;51:3996–4000. [PubMed] [Google Scholar]
- Heikkila R, Schwab G, Wickstrom E, Loke SL, Pluznik DH, Watt R, Neckers LM. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature. 1987;328:445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Venditti M, Iwasiow B, Orr FW, Shiu RP. C-myc gene expression alone is sufficient to confer resistance to antiestrogen in human breast cancer cells. Int J Cancer. 2002;99:35–42. doi: 10.1002/ijc.10269. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Conrad SE. C-Myc suppresses p21WAF1/CIP1 expression during estrogen signaling and antiestrogen resistance in human breast cancer cells. J Biol Chem. 2005;280:17617–17625. doi: 10.1074/jbc.M502278200. [DOI] [PubMed] [Google Scholar]
- Hui R, Cornish AL, McClelland RA, Robertson JF, Blamey RW, Musgrove EA, Nicholson RI, Sutherland RL. Cyclin D1 and estrogen receptor messenger RNA levels are positively correlated in primary breast cancer. Clin Cancer Res. 1996;2:923–928. [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Robustness, Scalability, and Integration of a Wound-Reponse Gene Expression Signature in Predicting Breast Cancer Survival http://microarray-pubs.stanford.edu/wound_NKI/explore.html [DOI] [PMC free article] [PubMed]
- Biankin AV, Morey AL, Lee C-S, Kench JG, Biankin SA, Head DR, Hugh TB, Sutherland RL, Henshall SM. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol. 2002;20:4531–4542. doi: 10.1200/JCO.2002.12.063. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editor. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2005. pp. 397–420. [Google Scholar]
- Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez J-C, Greco A, Hochstrasser D, Diaz J-J. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- Lam YW, Trinkle-Mulcahy L, Lamond AI. The nucleolus. J Cell Sci. 2005;118:1335–1337. doi: 10.1242/jcs.01736. [DOI] [PubMed] [Google Scholar]
- Leary DJ, Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–150. doi: 10.1016/s0014-5793(01)03143-x. [DOI] [PubMed] [Google Scholar]
- Oncomine Research™ http://www.oncomine.org
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henshall SM, Afar DEH, Hiller J, Horvath LG, Quinn DI, Rasiah KK, Gish K, Willhite D, Kench JG, Gardiner-Garden M, Stricker PD, Scher HI, Grygiel JJ, Agus DB, Mack DH, Sutherland RL. Survival analysis of genome-wide gene expression profiles of prostate cancers identifies new prognostic targets of disease relapse. Cancer Res. 2003;63:4196–4203. [PubMed] [Google Scholar]
- Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, Scurry J, Scolyer RA, Davies MJ, Heinzelmann M, Kalish LH, Bali A, Kench JG, Edwards LS, Vanden Bergh PM, Hacker NF, Sutherland RL, O'Brien PM. Overexpression of the cell adhesion molecules DDR1, claudin 3 and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10:4427–4436. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- Radvanyi L, Singh-Sandhu D, Gallichan S, Lovitt C, Pedyczak A, Mallo G, Gish K, Kwok K, Hanna W, Zubovits J, Armes J, Venter D, Hakimi J, Shortreed J, Donovan M, Parrington M, Dunn P, Oomen R, Tartaglia J, Berinstein NL. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci USA. 2005;102:11005–11010. doi: 10.1073/pnas.0500904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, Smith D, Hartmann L, Fishman D, Berchuck A, Schmandt R, Whitaker R, Gershenson DM, Mills GB, Bast RC., Jr Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- Korkola JE, Houldsworth J, Chadalavada RSV, Olshen AB, Dobrzynski D, Reuter VE, Bosl GJ, Chaganti RSK. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, Van De Rijn M, Botstein D, Brown PO. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graudens E, Boulanger V, Mollard C, Mariage-Samson R, Barlet X, Gremy G, Couillault C, Lajemi M, Piatier-Tonneau D, Zaborski P, Eveno E, Auffray C, Imbeaud S. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Yoshida K, Yamashita Y, Ota J, Takada S, Kisanuki H, Koinuma K, Choi YL, Kaneda R, Iwao T, Tamada K, Sugano K, Mano H. Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci. 2005;96:387–393. doi: 10.1111/j.1349-7006.2005.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger CA, Andres AC, Groner B, van der Valk M, Lemeur M, Gerlinger P. Targeted c-myc gene expression in mammary glands of transgenic mice induces mammary tumors with constitutive milk protein gene transcription. EMBO J. 1988;7:169–175. doi: 10.1002/j.1460-2075.1988.tb02797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Ström A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, Vergara LA, Tang S, Chong A, Bajic VB, Miller LD, Gustafsson JA, Liu ET. Discovery of estrogen receptor α target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. NOPdb: Nucleolar Proteome Database. Nucleic Acids Res. 2006;34:D218–220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Trumpp A. The Myc trilogy: lord of RNA polymerases. Nat Cell Biol. 2005;7:215–217. doi: 10.1038/ncb0305-215. [DOI] [PubMed] [Google Scholar]
- Strezoska Z, Pestov DG, Lau LF. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J Biol Chem. 2002;277:29617–29625. doi: 10.1074/jbc.M204381200. [DOI] [PubMed] [Google Scholar]
- Hölzel M, Rohrmoser M, Schlee M, Grimm T, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Hiddemann W, Bornkamm GW, Eick D. Mammalian WDR12 is novel member of the Pes-1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Rothermund K, Rogulski K, Fernandes E, Whiting A, Sedivy JM, Pu L, Prochownik EV. C-Myc-independent restoration of multiple phenotypes by two c-Myc target genes with overlapping functions. Cancer Res. 2005;65:2097–2107. doi: 10.1158/0008-5472.CAN-04-2928. [DOI] [PubMed] [Google Scholar]