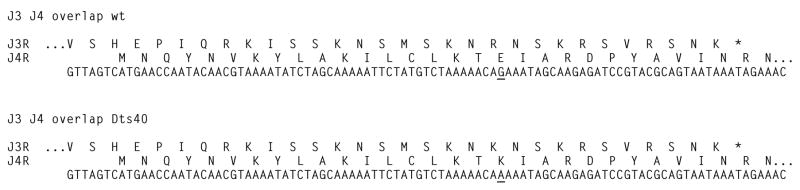Abstract
Complementation analysis of the combined Condit/Dales collection of vaccinia virus temperature sensitive mutants has been reported (Lackner et al., 2003), however not all complementation groups have previously been assigned to single genes on the viral genome. We have used marker rescue to map at least one representative of each complementation group to a unique viral gene. The final combined collection contains 124 temperature sensitive mutants affecting 38 viral genes, plus five double mutants.
Introduction
Poxviruses are unusual amongst viruses in that they carry out their replication entirely in the cell cytoplasm using a double stranded DNA molecule as a genome (reviewed in Moss, 2007). To accomplish this feat, poxviruses use a relatively large genome (~200 genes) to encode an entire complement of enzymes required for both mRNA synthesis and DNA replication, thus bypassing a strict requirement for nuclear enzymes. The most notorious of poxviruses is variola, the causative agent of smallpox. The laboratory prototype for the study of poxviruses is vaccinia virus, the poxvirus that was used as a live vaccine for eradication of smallpox. Despite the eradication of smallpox, interest in poxviruses persists because of their unusual structure, replication cycle and assembly, their utility as tools for understanding basic mechanisms of nucleic acid metabolism, the profound insights they provide into viral strategies to combat the host immune response, and the potential for deliberate release of smallpox as a bioterrorist weapon.
The study of poxvirus biology has benefitted significantly from both classical and reverse genetic analysis. Classical genetic analysis has consisted primarily of brute force isolation, mapping and characterization of temperature sensitive mutants of the virus (Condit and Niles, 1990). A burst of classical genetic analysis in the late 1970’s yielded four significant collections of temperature sensitive mutants of vaccinia virus from the Condit, Dales, Drillien and Ensinger laboratories (Condit and Motyczka, 1981; Condit et al., 1983; Dales et al., 1978; Ensinger, 1982; Drillien et al., 1982; Drillien and Spehner, 1983). In 1986, several mutants from the Ensinger collection were pooled with mutants from the Condit collection during studies on the highly conserved 16 kb Hind III DNA fragment of the vaccinia genome (Seto et al., 1987). More recently, the Condit and Dales collections were polished and pooled by performing complementation analysis among mutants in the Dales collection and between mutants in the two collections (Lackner et al., 2003). The resulting collection contained 129 temperature sensitive mutants sorted into 53 complementation groups. To maximize the utility of this combined collection, we have now conducted marker rescue analysis on at least one member of each complementation group, so that each complementation group is now identified with a unique vaccinia gene. In the course of the mapping analysis, we discovered that some of the previous complementation results represented false positives so that some mutants which were thought to comprise new complementation groups in fact mapped to genes containing previously mapped temperature sensitive mutants. In addition, five mutants proved to be double mutants. The final pooled collection contains 124 usable temperature sensitive mutants which map to 38 vaccinia genes. The analysis identifies temperature sensitive mutants in eight new genes, and adds new temperature sensitive alleles to many other genes of interest. The collection represents a valuable toolbox for study of viral functions affecting gene expression, DNA replication and virus structure and assembly.
Results and Discussion
Mapping of mutants by marker rescue
Our prior complementation analysis of temperature sensitive mutant viruses from the Condit and Dales mutant collections revealed numerous mutants whose map positions in the vaccinia genome were unknown or uncertain (Tables 1 and 2). Specifically, the analysis revealed 21 complementation groups (U1 – U21) comprising 24 viruses whose map positions were totally unknown and six complementation groups [A(8–17), A(25–29), E(2–8)a, E(2–8)b, F(11–17), G(6–8)] representing 14 viruses that had previously been only coarsely mapped to a multi-gene region of the viral genome. Furthermore, we reported map positions of three mutants comprising two complementation groups (genes A3L and J1R) as unpublished data, and numerous unmapped mutants were assigned to specific genes based solely on complementation analysis. Since publication of this prior study, data have been published from our laboratory and from other laboratories which map to a unique gene mutants in four of the six partially mapped complementation groups [A(8–17), A(25–29), E(2–8)b, G(6–8)], confirm the mapping of the two unpublished complementation groups (A3L and J1R), and confirm the map position inferred from complementation analysis of three mutants in two genes (D4R and D5) (Table 1). Our goal in this study was to map the remaining unmapped or coarsely mapped mutants (Table 2).
Table 1.
Mutants mapped since (Lackner et al., 2003)
| Mutant | Prior status | Gene | Reference |
|---|---|---|---|
| Cts8 | A3L | A3L | (Kato et al., 2004) |
| Cts26 | A3L | A3L | (Kato et al., 2004) |
| Cts40 | A(8–17) | A13L | (Unger and Traktman, 2004) |
| Cts6 | A(25–29) | A28L | (Turner et al., 2007) |
| Cts9 | A(25–29) | A28L | (Turner et al., 2007) |
| Dts27 | D4R | D4R | (Stanitsa et al., 2006) |
| Dts12 | D5R | D5R | (Boyle et al., 2007) |
| Dts56 | D5R | D5R | (Boyle et al., 2007) |
| Cts19 | E(2–8)b | E8R | (Kato et al., 2007) |
| Dts23 | E(2–8)b | E8R | (Kato et al., 2007) |
| Dts25 | E(2–8)b | E8R | (Kato et al., 2007) |
| Cts11 | G(6–8) | G7L | (Mercer and Traktman, 2005) |
| Cts45 | J1R | J1R | (Chiu et al., 2005) |
Table 2.
Mutants mapped in this paper
| Mutant | Prior status | Genea |
|---|---|---|
| Dts77 | U17 | A3L |
| Cts13 | U3 | A10L |
| Dts2 | U4 | A10L |
| Dts48 | U13 | A20R |
| Dts16 | U6 | A29L |
| Dts17 | U6 | A29L |
| Dts15 | B1R | B1R |
| Dts36 | U10 | D1R |
| Dts50 | U10 | D1R |
| Dts95 | U21 | D6R |
| Dts61 | U15 | D11L |
| Cts52 | E(2–8)a | E6R |
| Dts41 | E(2–8) a | E6R |
| Dts80 | E(2–8) a | E6R |
| Dts18 | U7 | E9L |
| Dts20 | U8 | E9L |
| Dts83 | U20 | E9L |
| Dts97 | U8 | E9L |
| Dts19 | E11L | E11L |
| Cts30 | F(11–17) | F13L |
| Cts48 | F(11–17) | F13L |
| Dts33 | U9 | G5.5R |
| Cts41 | G(6–8) | G7L |
| Dts78 | U18 | H4L |
| Dts57 | U14 | H5R |
| Dts4 | U5 | I7L |
| Dts8 | U5 | I7L |
| Dts35 | U5 | I7L |
| Dts93 | U5 | I7L |
| Cts57 | U1 | I8R |
| Cts37 | U2 | LM12, 32 dbl |
| Dts40 | U11 | J3R-J4R dbl |
| Dts47 | U12 | LM19, 21 dbl |
| Dts71 | U16 | LM23, 26 dbl |
| Dts82 | U19 | J6R, A36R dbl |
dbl = double mutant, the smallest fragments producing rescue are indicated.
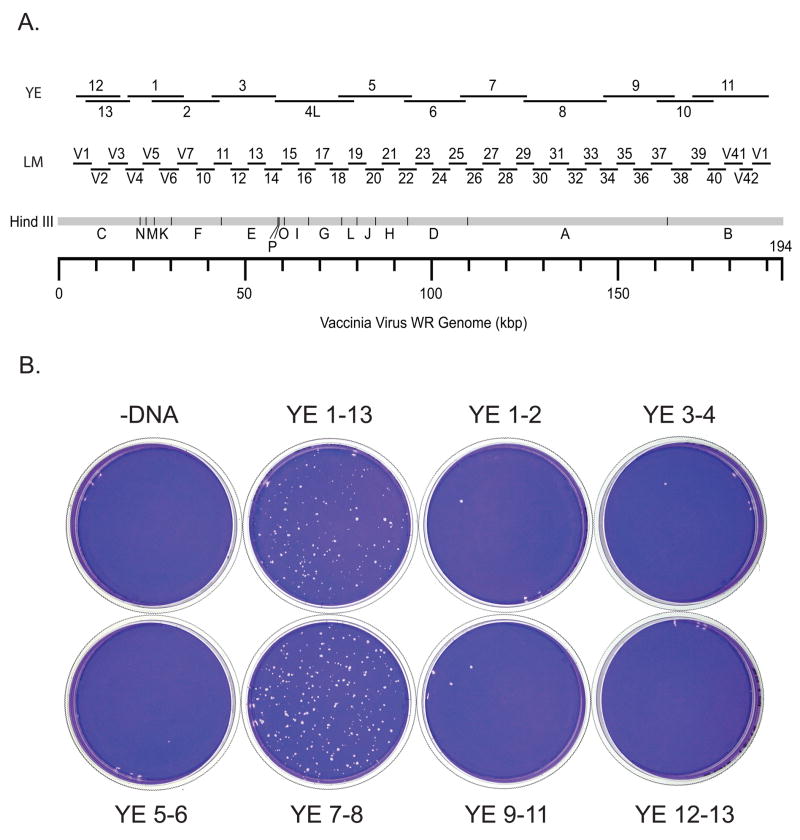
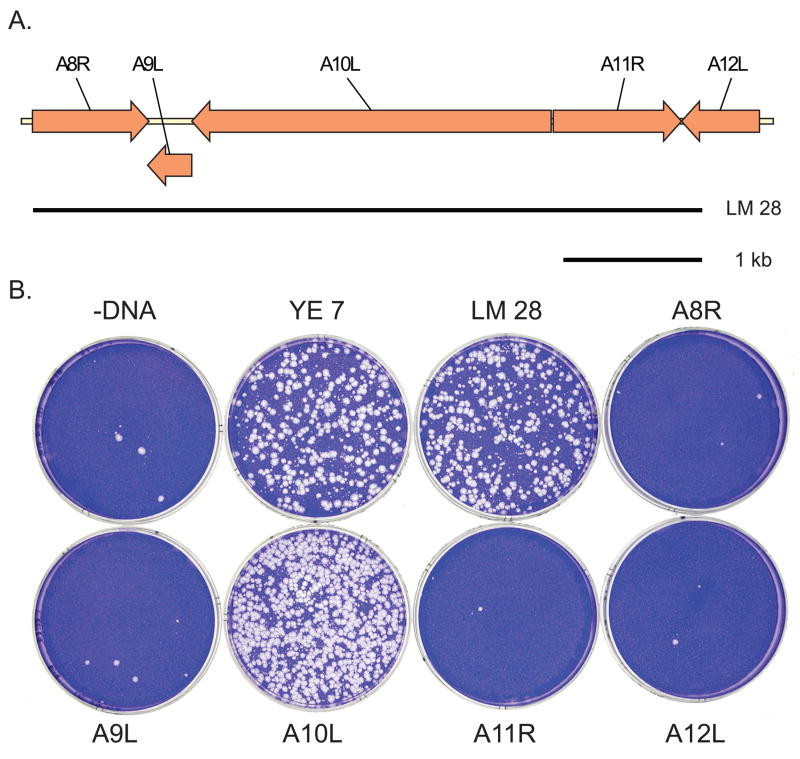
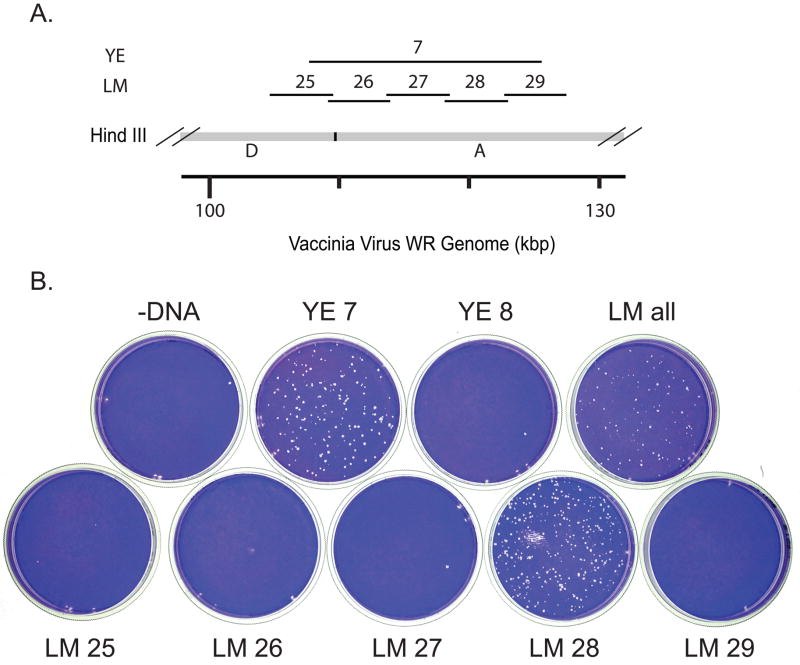
At least one representative virus of each unmapped complementation group was mapped by marker rescue. Marker rescue was done by infecting monolayers of BSC40 cells with an appropriate dilution of mutant virus, transfecting with subgenomic PCR fragments of wild type virus DNA, incubating the infected, transfected cells at the non-permissive temperature for 4 days, and staining with crystal violet to detect rescued wild type virus plaques. Mapping was done in stages using three sets of progressively smaller DNA fragments. Initial mapping used a set of 13 overlapping PCR products (YE fragments) ranging in size from 12 to 22 kb and representing virtually the entire vaccinia genome (Fig. 1A) (Yao and Evans, 2003). Refined map positions were then obtained using an appropriate selection of fragments from a set of 41 overlapping 5 kb fragments (LM fragments) representing the entire vaccinia genome (Fig . 1A) (Luttge and Moyer, 2005). Lastly mutants were mapped to unique genes using an appropriate selection of gene-sized DNA fragments. In many cases mapping data was supplemented with DNA sequence analysis of the mapped mutants (Supplemental Table 1). Figs. 1–3 show the sequential steps performed to determine the precise physical map position of Cts131, one representative unmapped virus of the collection. Fig. 1B shows a marker rescue done with pools of YE fragments, demonstrating that Cts13 maps within the region defined by fragments YE7 and YE8. Fig. 2 shows that Cts13 rescues with YE7 but not YE8. YE7 encompasses fragments LM25 through LM29 (Fig. 2A). Fig. 2B shows that Cts13 rescues exclusively with LM28. LM28 encompasses vaccinia genes A8R – A12L (Fig. 3A). Fig 3B shows that Cts13 maps to gene A10L, which encodes the major virion protein 4a.
Fig. 1. Coarse marker rescue mapping of Cts13.
A) A HindIII map of the vaccinia genome showing the positions of large sized (YE) PCR fragments and intermediate sized (LM) PCR fragments. B) Marker rescue of Cts13. Dishes were infected with Cts13, transfected with pools of PCR fragments as indicated, incubated at 39.7°C for 4 days, and stained with crystal violet.
Fig . 3. Gene specific marker rescue mapping of Cts13.
A) A map of a portion of the HindIII A fragment of the vaccinia genome. The positions of genes (arrows) and LM 28 are indicated. B) Marker rescue of Cts13. Dishes were infected with Cts13, transfected with pools of PCR fragments as indicated, incubated at 39.7°C for 4 days, and stained with crystal violet.
Fig. 2. Refined marker rescue mapping of Cts13.
A) A map of a portion of the vaccinia genome containing the HindIII D-A junction, showing the positions of YE 7, and the LM fragments contained within YE 7. B) Marker rescue of Cts13. Dishes were infected with Cts13, transfected with pools of PCR fragments as indicated, incubated at 39.7°C for 4 days, and stained with crystal violet. LM all contains LM fragments 25–34, covering YE 7 and YE 8.
The sequential marker rescue protocol was used to map 23 unmapped or partially mapped complementation groups of the combined Condit and Dales collections comprising 32 viruses, and to confirm the map positions inferred from complementation analysis for three additional mutants . For mapping mutants that had been partially mapped previously, we used an abbreviated scheme in which we determined, based on the published preliminary mapping, which long PCR products or 5-kb products could be used as positive controls in the marker rescue, and the remaining physical mapping was done as described.
The mapping results can be broken down into four categories (Table 2): 1) Five mutants representing five unmapped complementation groups proved to be double mutants (Cts37 = U2, Dts40 = U11, Dts47 = U12, Dts71 = U16, Dts82 = U19) and were therefore excluded from further analysis. The criteria for classifying these as double mutants is discussed below. 2) The map positions of three mutants inferred from prior complementation analysis was confirmed (Cts41 = G7L, Dts15 = B1R, Dts19 = E11L). 3) Surprisingly, 13 mutants comprising 9 unmapped complementation groups (U1, U15, U17, U18, U20, U21, U5, U7, U8) mapped to genes containing previously isolated temperature sensitive mutations. Thus these mutants represent false positives from the prior complementation analysis, discussed further below. 4) 14 mutants comprising 6 unmapped and 2 coarsely mapped complementation groups were mapped to genes in which classical temperature sensitive mutations have not been previously identified (genes A10L, A20R, A29L, D1R, E6R, F13L, G5.5R, and H5R). (Engineered temperature sensitive mutants have previously been isolated in A20R, D1R and H5R (Ishii and Moss, 2001; Hassett et al., 1997; DeMasi and Traktman, 2000; Punjabi et al., 2001).)
Four of the double mutants (Dts82, Cts37, Dts47, Dts71) were designated as such because each rescued with two non-overlapping DNA fragments. We interpret this to mean that each of the rescuing fragments contains a temperature sensitive allele that by itself is insufficient to confer full temperature sensitivity on virus replication, but in combination with the second allele prevents virus growth at the non-permissive temperature. The remaining double mutant, Dts40, presents a noteworthy curiosity. Fine mapping of Dts40 revealed that the mutant could be rescued with a PCR fragment containing both the J3R and the J4R genes, however the mutant could not be rescued with fragments representing the individual genes. We therefore sequenced the J3R and J4R genes from Dts40 and found a single point mutation in an 86 bp region of overlap between J3R and J4R resulting in a substitution in both J3R (R323K) and J4R (E18K) (Fig. 4). The sequence in this region of overlap is arranged such that we could not easily segregate these mutations for further analysis. In retrospect, it should theoretically have been possible to rescue the Dts40 mutation with the individual J3R or J4R gene fragments, however the mutation is located only 35 nucleotides from the C terminus of J3R and 52 nucleotides from the N terminus of J4R. Experience dictates that mutations located this close to the end of a rescuing DNA fragment rescue very poorly if at all (Turner et al., 2007). Limited additional analysis of this mutant suggests that it is not completely defective in either J3R or J4R gene activity at the non-permissive temperature. First, the Dts40 complements other mutants in J4R (Lackner et al., 2003), specifically Cts7 and Dts44, suggesting that Dts40 can supply J4R gene function (an RNA polymerase subunit, rpo22) in mixed infections under non-permissive conditions. J3R encodes a multifunctional protein that serves as a poly(A) polymerase processivity factor, a (nucleoside-2′-O-) methyltransferase and a postreplicative transcription elongation factor. Only the elongation activity is essential for virus replication, and phenotypically, J3R null mutants are dependent on the elongation enhancing drug, isatin-β-thiosemicarbazone (IBT) (Latner et al., 2002; Latner et al., 2000). However, we found that Dts40 was sensitive to IBT at both permissive and non-permissive temperatures (data not shown), suggesting that at least some J3R gene activity is expressed in Dts40 mutant infections. We hypothesize based on these observations that Dts40 is partially defective in both J3R and J4R function under non-permissive conditions, similar to other double mutants we have characterized.
Fig. 4. DNA sequence of Dts40.
The nucleotide sequence encoding the overlap between the carboxy terminus of J3R and the amino terminus of J4R is shown, with respective translations above the sequence. Wild type sequence is shown at the top and the Dts40 mutant sequence is shown at the bottom. The mutant nucleotide in the G to A transition is underlined in each sequence. The single letter amino acid code is positioned above the first nucleotide in the relevant reading frame for each peptide. Dots (...) indicate that the protein continues upstream (J3R) or downstream (J4R).
As noted above, nine of the unmapped complementation groups mapped to genes containing previously isolated temperature sensitive mutations. Thus in the prior complementation analysis these mutants complemented mutants in the same gene, that is, they were false positives. Specifically, five unmapped complementation groups, each containing a single mutant, each mapped to a different gene containing a previously mapped temperature sensitive mutation (U1 = Cts57 = I8R, U15 = Dts61 = D11L, U17 = Dts77 = A3L, U18 = Dts78 = H4L, U21 = Dts95 = D6R). One unmapped group (U5) contained four mutants that mapped to gene I7L, which contained two mutants from previous analyses. Lastly, three unmapped groups (U7, U8, U20) comprising four mutants mapped to a single gene, E9L, containing a single mutant from previous analyses. There exist two possible explanations for the false positive complementation results: 1) The complementation analysis was done using a qualitative spot test, which does not formally discriminate between complementation and recombination, thus it is possible that plaques formed during the analysis and scored as complementation actually represented wild type virus formed by intragenic recombination. Our previous experience, gained entirely with mutants in the WR strain of vaccinia, shows that intragenic recombination occurs only with a very low frequency at the non-permissive temperature, and historically, false positives have not been observed in the complementation spot test (Condit and Motyczka, 1981; Condit et al., 1983). We note that all but one of the false positive results involves mixed infections between mutants in two different virus strains, WR (Condit collection) and IHDW (Dales collection), suggesting that intragenic recombination between these strains may be more facile compared to intragenic recombination between mutants within the WR strain. 2) It is possible that the “false positives” in the complementation analysis actually represent genuine intragenic complementation. Intragenic complementation has been observed previously among mutants in gene A24R, the second largest subunit of the viral RNA polymerase (Hooda-Dhingra et al., 1990). Intragenic complementation carries interesting implications regarding the function of a gene, for example it may indicate that the gene product functions as a multimer. Discriminating among these possibilities for the mutants reported here could be accomplished ultimately using a quantitative complementation analysis, which can discriminate between complementation and intragenic recombination. Importantly, we have encountered no incidence of false negative results in the complementation analysis, that is, all non complementing mutants that have been mapped have mapped to a unique gene.
DNA sequence analysis
Seventy-two of the 129 mutants have been subjected to DNA sequence analysis (Supplementary Table 1). Of these, 54 were reported previously and 18 new sequences were determined during the course of this work. Two features of the sequence analysis are noteworthy: 1) Not all mutants contain simple missense mutations. Mutants in two genes contain frameshift (A28R) or nonsense (G3L) mutations leading to truncated protein products (Turner et al., 2007), and one mutant (Cts19 in gene E8R) changes the initiating methionine codon to a leucine codon, effectively creating a null mutation (Kato et al., 2007). Furthermore, seven mutants contain two or even three coding changes. 2) Five sibling pairs have been revealed in five different genes, A28R, A30L, D1R, E8R and I7L. This raises the possibility that the collection contains additional sibling pairs. Thus in order to reduce unnecessarily redundant effort it is advisable to obtain sequence information on mutants in a given gene prior to pursuing detailed phenotypic analysis.
DNA replication
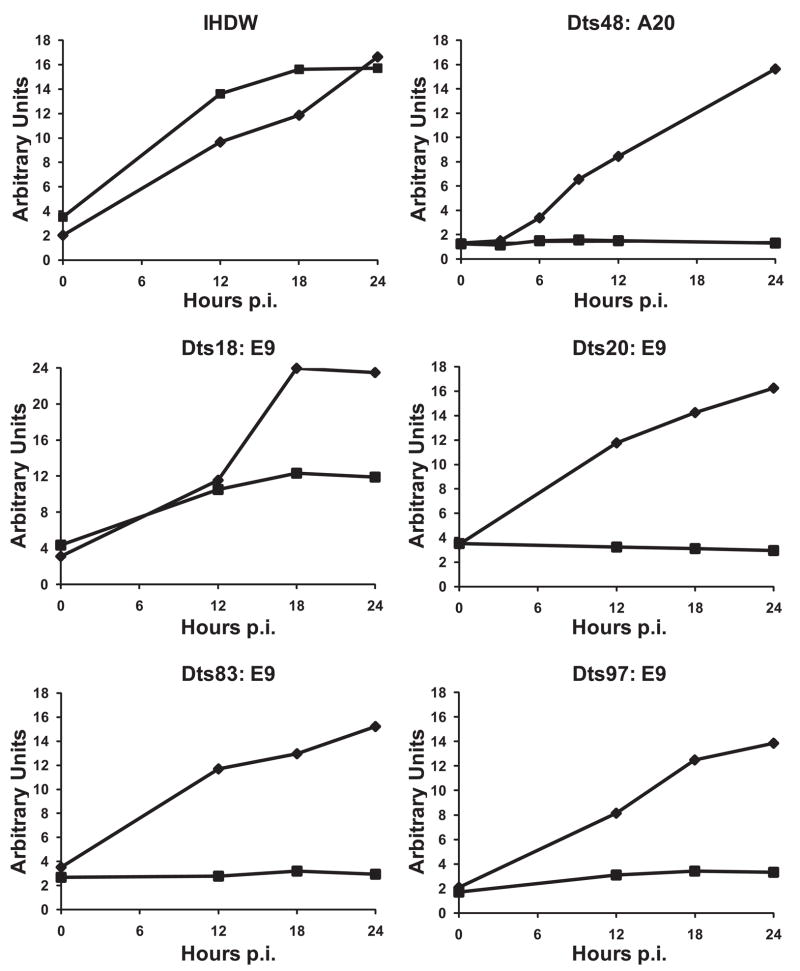
This study mapped new mutants to three genes previously shown to be required for DNA replication, A20R (DNA polymerase processivity factor), B1R (protein kinase), and E9L (DNA polymerase). We therefore assayed these mutants for DNA replication to provide additional support for the mapping assignments. The B1R protein kinase mutant Dts15 was previously reported to be DNA positive, and we confirmed this result (data not shown). Fig. 5 shows that the A20R mutant Dts48 and the E9L mutants Dts20, Dts83 and Dts97 are DNA negative as predicted. Interestingly, the E9L mutant Dts18 is DNA positive, though DNA replication is reduced by approximately one half at the non-permissive temperature relative to the permissive temperature. The DNA positive phenotype of Dts15 and Dts18 could suggest that these mutants segregate multiple functions of the affected genes. For example, Dts15 could be specifically defective in phosphorylation of targets required for virus production but not for DNA replication. Likewise, Dts18 could be specifically defective in a DNA polymerase function such as recombination that is not absolutely required for maximal DNA replication. Alternatively, these mutants may simply be phenotypically leaky. Further experiments are required to distinguish among these possibilities.
Fig. 5. Viral DNA replication in mutant infected cells.
Cells were infected with the indicated viruses at MOI = 10, incubated at 31°C (♦) or 39.7°C (■) and assayed for viral DNA by slot blot hybridization with a radiolabeled vaccinia specific DNA probe as described in materials and methods.
Summary and conclusions
The final combined collection (discounting double mutants) contains 124 temperature sensitive mutants affecting 38 viral genes (Table 3). Importantly, not all mutants assigned to a given gene have actually been mapped to that gene by marker rescue; many of the assignments are based solely on complementation analysis. While this complementation analysis has to date proven remarkably reliable, nevertheless it would be advisable to confirm any given assignment by marker rescue before investing significant effort in characterizing an unmapped allele.
Table 3.
Summary of mutants by gene
| Genea | Mutantb | Proteinc | DNAd | EMe | Function | |
|---|---|---|---|---|---|---|
| J6R | Cts51, Cts53, Cts65, Dts85 Cts27, Cts29, Cts32, Cts47, Cts62, Dts10, Dts14, | defective late | positive | viroplasm - IV | RNAP (147 kDa); rpo147 | Gene Expression |
| A24R | Dts28, Dts49, Dts52, Dts60, Dts66, Dts86, Dts90, Dts94 | defective late | positive | viroplasm - IMV | RNAP (132 kDa); rpo132 | |
| A29L | Dts16, Dts17 | not done | not done | not done | RNAP (35 kDa); rpo35 | |
| J4R | Cts7, Cts20, Dts44 | defective late | positive | viroplasm | RNAP (22 kDa); rpo22 | |
| D7R | Cts21, Ets45 | defective late | positive | viroplasm | RNAP (18 kDa); rpo18 | |
| G5.5R | Dts33 | not done | not done | not done | RNAP (7 kDa); rpo7 | |
| H4L | Cts1, Cts31, Cts55, Cts58, Dts78 | normal | positive | IMV | RAP94 | |
| D6R | Cts46, Dts95, Ets93 | normal | positive | not done | VETF subunit | |
| D11L | Cts36, Cts50, Dts61, Ets17 | defective late | positive | not done | ATPase/NPH-I | |
| D1R | Dts36, Dts50 | defective early | negative | not done | VTF and capping enzyme subunit/ VITF | |
| D12L | Dts96 | normal | positive | viroplasm | VTF and capping enzyme subunit/ VITF | |
| I8R | Cts10, Cts18, Cts38, Cts39, Cts44, Cts57, Dts67 | normal | positive | IMV | RNA helicase/NPH-II | |
| A1L | Cts63 | defective late | positive | IVN | VLTF 2 | |
| A18R | Cts4, Cts22, Cts23 | abortive late | positive | viroplasm | ATPase/DNA helicase/transcript release factor | |
| G2R | Cts56, Dts22, Dts42 | defective late | positive | viroplasm | positive transcription elongation factor | |
| E9L | Cts42, Dts18, Dts20, Dts83, Dts97 | early only | negative | negative | DNA polymerase | DNA Replication |
| A20R | Dts48 | not done | negative | not done | DNA polymerase processivity factor | |
| D5R | Cts17, Cts24, Dts12, Dts38, Dts56, Ets69 | early only | negative | not done | ATPase/DNA primase | |
| D4R | Dts27, Dts30 | early only | negative | not done | uracil DNA glycosylase | |
| B1R | Cts2, Cts3, Cts25, Dts15 | early only | negative | not done | protein kinase | |
| H5R | Dts57 | early only | negative | negative | DNA replication/transcription/morphogenesis | |
| A10L | Cts13, Cts64, Dts2 | normal | positive | crescents | p4a | Structure/Assembly |
| A3L | Cts8, Cts26, Dts77 | normal | positive | IVN | p4b | |
| E6R | Cts52, Dts41, Dts80 | normal | positive | IMV | virion core protein | |
| E8R | Cts19, Dts23, Dts25 | normal | positive | viroplasm - IMV | virion core protein | |
| E11L | Cts49, Dts19 | normal | positive | IMV | virion core protein | |
| D2L | Ets52, Ets94 | normal | positive | crescents | virion core protein | |
| D3R | Cts5, Cts35 | normal | positive | crescents | virion core protein | |
| A30L | Dts45, Dts46 | normal | not done | crescents | virion core protein | |
| G7L | Cts11, Cts41, Dts68, Dts89 | normal | positive | viroplasm | virion core protein | |
| J1R | Cts45 | normal | positive | crescents | virion membrane protein | |
| A13L | Cts40 | normal | positive | IV | membrane phosphoprotein | |
| G3L | Cts60 | normal | positive | EEV | entry/fusion complex | |
| A28L | Cts6, Cts9 | normal | positive | IMV | entry/fusion complex | |
| F10L | Cts12, Cts15, Cts28, Cts54, Cts61, Dts11 | normal | positive | viroplasm - IV | virion protein kinase | |
| D13L | Cts33, Cts43, Dts9, Dts62, Dts88, Ets101 | normal | positive | viroplasm | crescent scaffold protein | |
| I7L | Cts16, Cts34, Dts4, Dts8, Dts35, Dts93 | normal | positive | IVN | virion core proteinase | |
| F13L | Cts30, Cts48 | normal | positive | not done | EEV membrane protein | |
|
| ||||||
| LM12, 32 dbl | Cts37 | not done | not done | not done | not applicable | |
| J3R-J4R dbl | Dts40 | not done | not done | not done | not applicable | |
| LM19, 21 dbl | Dts47 | not done | not done | not done | not applicable | |
| LM23, 26 dbl | Dts71 | not done | not done | not done | not applicable | |
| J6R, A36R dbl | Dts82 | not done | not done | not done | not applicable | |
dbl = double mutant, the smallest fragments producing rescue are indicated.
genes mapped by marker rescue are underlined; all others are assigned to the indicated gene by complementation.
viral protein synthesis at the non-permissive temperature assayed by metabolic labeling and gel electrophoresis. Normal = indistinguishable from wild type; defective early = decreased synthesis of early proteins; early only = extended early protein synthesis, no late protein synthesis; defective late = decreased or delayed late protein synthesis; abortive late = late shut off of all protein synthesis. See (Condit and Motyczka, 1981; Condit et al., 1983).
viral DNA replication at the non-permissive temperature
electron microscope phenotype at the non-permissive temperature. Entries denote the most mature normal structure visualized. Abnormal structures may be present as well. Negative = no evidence of viral infection; IV = immature virions; IVN = immature virions with nucleoids; MV = mature virions. Some groups display a range of phenotypes as indicated.
The mutants in the combined collection affect genes in all of the essential aspects of viral replication, including gene expression, DNA replication and virion structure and assembly. Over time the mutants have been of value in dissecting the roles of individual genes in the virus replication cycle, and we believe that they will continue to serve this purpose indefinitely.
Material and methods
Cells and virus culture
BSC40 cells, a continuous line of African green monkey kidney cells, were grown in Dulbecco’s Modified Eagle medium (DME) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. The temperature-sensitive mutant viruses (ts) used in this study were isolated and some were characterized previously (Dales et al., 1978; Condit and Motyczka, 1981; Condit et al., 1983; Lackner et al., 2003). Wild-type virus strains WR and IHD-W and the conditions for virus culture, virus infection and plaque titration have been described in detail (Dales et al., 1978; Condit and Motyczka, 1981; Condit et al., 1983). For virological studies of the mutants, 31 and 39.7°C were used as permissive and non-permissive temperatures, respectively.
PCR based marker rescue
One-step marker rescue of mutants was carried out as a modification of the previously described protocol (Thompson and Condit, 1986). Briefly, 60-mm dishes of BSC40 cells were infected with 0.5 ml of each virus at an appropriate moi determined empirically by terminal dilution. After a 1-hour adsorption at 31°C, the inoculum was removed and replaced with 4 ml of Opti-MEM I Reduced Serum Medium (GIBCO) containing no serum. Infected monolayers were transfected with 1.5ug of pcr products corresponding to regions of vaccinia virus WR genome. Transfection was done with 200 μl of Lipofectamine reagent (Invitrogen Life Technologies)-complexed DNA which was added drop-wise to the medium. Briefly, the Lipofectamine-DNA complex was prepared by adding the DNA suspension (1.5μg of DNA in 100μl of Opti-MEM) to the lipofectamine reagent mix (14μl lipofectamine reagent and 86μl Opti-MEM). The infected-transfected cells were incubated overnight at 39.7°C and on the following day the medium was replaced with fresh media (DME) containing serum. The infected-transfected monolayers were incubated at 39.7°C for an additional 3 days. On the fourth day of infection the cells were stained with crystal violet solution and analyzed for the presence or absence of wild-type plaques. Occasionally we observed a high background of plaques in several dishes which could be attributed to contamination of a PCR product with the genomic wt viral DNA template. This background could usually be reduced or eliminated by synthesizing second generation PCR fragments using a small amount of the original PCR fragment as a template and thus effectively diluting the genomic wt DNA contamination.
PCR and primers
Mapping of mutant viruses was done by sequential marker rescues using three series of PCR products spanning all or parts of the viral genome. The first series of long PCR fragments was prepared with a set of DNA oligonucleotide primers designed by Yao and Evans (2003) and together amplified nearly all of vaccinia virus genome in a series of 13 products ranging between 12 and 22 kb. The second series of PCR fragments used to refine the mapping was generated with a set of oligonucleotide primers designed by Luttge and Moyer (2005) which amplifies the entire genome in a series of 40 5-kb products. PCR reactions were carried out using the Roche Expand long template PCR kit (Buffer system 3 for the 11–21kb series of PCR products and Buffer system 2 for the 5-kb PCR products) and these PCR amplified DNAs were purified with Amicon Microcon centrifugal filter devices (Millipore) before use in the marker rescue. The third series of PCR products was prepared using a set of primers that amplified specific individual open reading frames in a reaction containing Deep Vent DNA polymerase (New England Biolabs) and Taq polymerase. These PCR products were purified with High Pure PCR Product Purification kit (Roche) used as directed by the manufacturer.
Preparation of vaccinia virus DNA
Wild type vaccinia virus (WR) genomic DNA was prepared essentially as described previously (Condit et al., 1983; Esposito et al., 1981). Total infected cell DNA to be used for sequencing was prepared using DNeasy tissue kit (Qiagen) according to manufacturer’s instructions for isolation of DNA from animal cells in culture as previously described (Latner et al., 2000).
DNA sequence analysis
DNA sequence of specific genes from wt or mutant viruses was obtained by direct sequencing of PCR products amplified from total infected cell DNA or purified viral genomic DNA, prepared as described above. The entire open reading frame was PCR amplified using two primers that hybridize outside of the open reading frame. Sequence was obtained using the amplification primers and additional primers that hybridize within the coding sequence. Sequencing was performed by the University of Florida ICBR DNA Sequencing Core Laboratory.
Viral DNA replication analysis
Viral DNA replication was analyzed as described previously (Traktman and Boyle, 2004). Briefly, BSC40 cells were infected at moi = 10 and incubated at 31°C or 39.7°C. At various times post infection cells were harvested by scraping and centrifugation. The cells were washed once with phosphate buffered saline (PBS) and resuspended in a solution of 10x SSC (1.5 M NaCl, 150 mM sodium citrate) and 1 M ammonium acetate. Samples were subject to three cycles of freeze/thawing to disrupt the cells and were stored at −20°C. The samples were applied to a Nytran Supercharge nylon transfer membrane (Schleicher & Schuell) on a Minifold II Slot-Blotter apparatus (Schleicher & Schuell). Before removing the membrane from the slot blot apparatus, the DNA was denatured with a solution of 0.5 M NaOH/1.5 M NaCl and then neutralized with two washes of 10X SSC. The membrane was prehybridized at 42°C in a hybridization oven (Labnet International, Inc) for at least two hours in a buffer containing 6X SSC, 50% formamide, 0.5% SDS, 5X Denhardts solution (0.1% BSA, 0.1% polyvinylpyrolidone, 0.1% Ficoll), and 100μg/ml denatured salmon sperm DNA. After pre-hybridization, 2.25×106 cpm of the randomly labeled (DECAprime II kit (Ambion)) vaccinia HindIII E fragment probe was added to fresh hybridization solution and incubated with the membranes overnight at 42°C. The membranes were washed three times with 2x SSC at room temperature followed by two washes with 0.2x SSC/0.1% SDS at 55°C. The membranes were exposed to film and were then quantified with a phosphor screen (Molecular Dynamics) and analyzed by a Storm phosphorimager (Molecular Dynamics) and the ImageQuant software program (Molecular Dynamics).
Supplementary Material
Acknowledgments
We thank Jason Mercer, Jeremy Nichols and Paula Traktman for sharing unpublished data on the mapping and characterization of the Dts14 and Cts41 mutants. We thank Paula Traktman for comments on the manuscript. This work was supported by NIH grant R01 AI055560 to RCC.
Footnotes
Mutants prefaced with a “C”, “D” or “E” are from the Condit, Dales or Ensinger collections, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Boyle KA, Arps L, Traktman P. Biochemical and genetic analysis of the vaccinia virus d5 protein: Multimerization-dependent ATPase activity is required to support viral DNA replication. J Virol. 2007;81:844–859. doi: 10.1128/JVI.02217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Szajner P, Moss B, Chang W. Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly. J Virol. 2005;79:8046–8056. doi: 10.1128/JVI.79.13.8046-8056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit RC, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Condit RC, Niles EG. Orthopoxvirus genetics. Curr Top Microbiol Immunol. 1990;163:1–39. doi: 10.1007/978-3-642-75605-4_1. [DOI] [PubMed] [Google Scholar]
- Dales S, Milovanovitch V, Pogo BG, Weintraub SB, Huima T, Wilton S, McFadden G. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology. 1978;84:403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- DeMasi J, Traktman P. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J Virol. 2000;74:2393–2405. doi: 10.1128/jvi.74.5.2393-2405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillien R, Spehner D. Physical mapping of vaccinia virus temperature-sensitive mutations. Virology. 1983;131:385–393. doi: 10.1016/0042-6822(83)90506-8. [DOI] [PubMed] [Google Scholar]
- Drillien R, Spehner D, Kirn A. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology. 1982;119:372–381. doi: 10.1016/0042-6822(82)90096-4. [DOI] [PubMed] [Google Scholar]
- Ensinger MJ. Isolation and genetic characterization of temperature-sensitive mutants of vaccinia virus WR. J Virol. 1982;43:778–790. doi: 10.1128/jvi.43.3.778-790.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito J, Condit R, Obijeski J. The preparation of orthopoxvirus DNA. J Virol Methods. 1981;2:175–179. doi: 10.1016/0166-0934(81)90036-7. [DOI] [PubMed] [Google Scholar]
- Hassett DE, Lewis JI, Xing X, DeLange L, Condit RC. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology. 1997;238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- Hooda-Dhingra U, Patel DD, Pickup DJ, Condit RC. Fine structure mapping and phenotypic analysis of five temperature-sensitive mutations in the second largest subunit of vaccinia virus DNA-dependent RNA polymerase. Virology. 1990;174:60–69. doi: 10.1016/0042-6822(90)90054-u. [DOI] [PubMed] [Google Scholar]
- Ishii K, Moss B. Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants. J Virol. 2001;75:1656–1663. doi: 10.1128/JVI.75.4.1656-1663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Condit RC, Moussatche N. The vaccinia virus E8R gene product is required for formation of transcriptionally active virions. Virology. 2007;367:398–412. doi: 10.1016/j.virol.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Strahl AL, Moussatche N, Condit RC. Temperature-sensitive mutants in the vaccinia virus 4b virion structural protein assemble malformed, transcriptionally inactive intracellular mature virions. Virology. 2004;330:127–146. doi: 10.1016/j.virol.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Lackner CA, D'Costa SM, Buck C, Condit RC. Complementation analysis of the Dales collection of vaccinia virus temperature-sensitive mutants. Virology. 2003;305:240–259. doi: 10.1006/viro.2002.1745. [DOI] [PubMed] [Google Scholar]
- Latner DR, Thompson JM, Gershon PD, Storrs C, Condit RC. The positive transcription elongation factor activity of the vaccinia virus J3 protein is independent from its (nucleoside-2′-O-) methyltransferase and poly(A) polymerase stimulatory functions. Virology. 2002;301:64–80. doi: 10.1006/viro.2002.1538. [DOI] [PubMed] [Google Scholar]
- Latner DR, Xiang Y, Lewis JI, Condit J, Condit RC. The vaccinia virus bifunctional gene J3 (nucleoside-2′-O-)- methyltransferase and poly(A) polymerase stimulatory factor is implicated as a positive transcription elongation factor by two genetic approaches. Virology. 2000;269:345–355. doi: 10.1006/viro.2000.0243. [DOI] [PubMed] [Google Scholar]
- Luttge BG, Moyer RW. Suppressors of a host range mutation in the rabbitpox virus serpin SPI-1 map to proteins essential for viral DNA replication. J Virol. 2005;79:9168–9179. doi: 10.1128/JVI.79.14.9168-9179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Traktman P. J Virol. Vol. 79. 2005. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins; pp. 7146–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2905–2945. [Google Scholar]
- Punjabi A, Boyle K, DeMasi J, Grubisha O, Unger B, Khanna M, Traktman P. Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: temperature-sensitive mutants have a DNA-minus phenotype and are defective in the production of processive DNA polymerase activity. J Virol. 2001;75:12308–12318. doi: 10.1128/JVI.75.24.12308-12318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto J, Celenza LM, Condit RC, Niles EG. Genetic map of the vaccinia virus HindIII D Fragment. Virology. 1987;160:110–119. doi: 10.1016/0042-6822(87)90051-1. [DOI] [PubMed] [Google Scholar]
- Stanitsa ES, Arps L, Traktman P. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J Biol Chem. 2006;281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Condit RC. Marker rescue mapping of vaccinia virus temperature-sensitive mutants using overlapping cosmid clones representing the entire virus genome. Virology. 1986;150:10–20. doi: 10.1016/0042-6822(86)90261-8. [DOI] [PubMed] [Google Scholar]
- Traktman P, Boyle K. Methods for analysis of poxvirus DNA replication. Methods Mol Biol. 2004;269:169–186. doi: 10.1385/1-59259-789-0:169. [DOI] [PubMed] [Google Scholar]
- Turner PC, Dilling BP, Prins C, Cresawn SG, Moyer RW, Condit RC. Vaccinia virus temperature-sensitive mutants in the A28 gene produce non-infectious virions that bind to cells but are defective in entry. Virology. 2007 doi: 10.1016/j.virol.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger B, Traktman P. Vaccinia virus morphogenesis: a13 phosphoprotein is required for assembly of mature virions. J Virol. 2004;78:8885–8901. doi: 10.1128/JVI.78.16.8885-8901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao XD, Evans DH. High-frequency genetic recombination and reactivation of orthopoxviruses from DNA fragments transfected into leporipoxvirus-infected cells. J Virol. 2003;77:7281–7290. doi: 10.1128/JVI.77.13.7281-7290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.