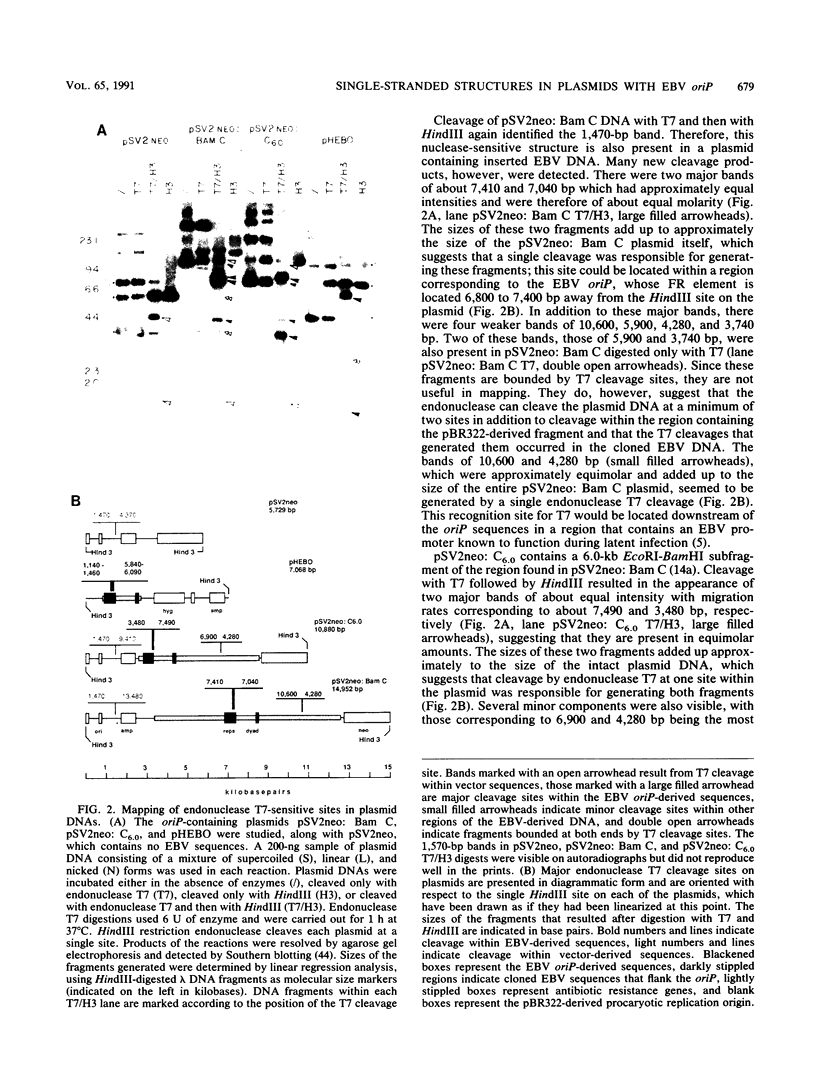
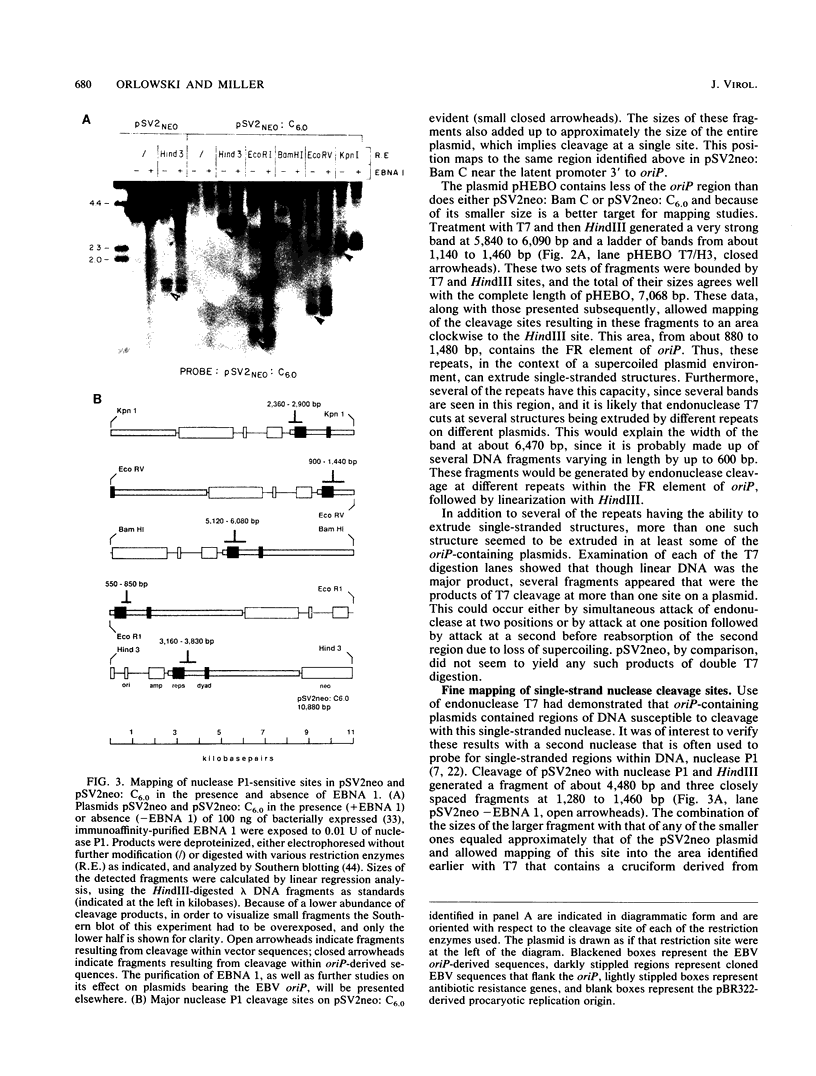
Abstract
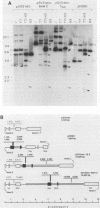
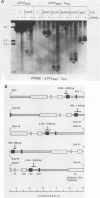
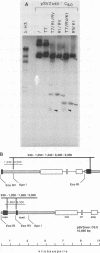
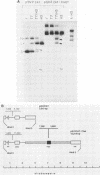
The Epstein-Barr virus (EBV) latent origin of plasmid replication (oriP) contains two essential regions, a family of repeats with 20 imperfect copies of a 30-bp sequence and a dyad symmetry element with four similar 30-bp repeats. Each of the repeats has an internal palindromic sequence and can bind EBNA 1, a protein that together with oriP constitutes the only viral element necessary for EBV maintenance and replication. Using single-strand-specific nucleases, we have probed plasmids containing oriP-derived sequences for the presence of secondary structural elements. Multiple single-stranded structures were detected within the oriP region. Of the two essential elements of oriP, the family of repeats seemed to extrude these structures at a much higher frequency than did sequences within the dyad symmetry region. Though negative supercoiling was found to stabilize the single-stranded structures, they showed significant stability even after linearization of the oriP plasmids. Two major single-stranded structures detected involved approximately 12 bp of DNA. These loci could be transiently unwound regions that form because of negative supercoiling and the high A + T content of this region of DNA, or they could be cruciform structures extruded within the palindromic sequences of oriP that may be important sites for protein-DNA interactions in the EBV oriP.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987 May;61(5):1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Theoretical analysis of competitive conformational transitions in torsionally stressed DNA. J Mol Biol. 1981 Jul 25;150(1):43–68. doi: 10.1016/0022-2836(81)90324-7. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Bodescot M., Perricaudet M., Farrell P. J. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J Virol. 1987 Nov;61(11):3424–3430. doi: 10.1128/jvi.61.11.3424-3430.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Camilloni G., Della Seta F., Grazia Ficca A., Di Mauro E. Purified Saccharomyces cerevisiae RNA polymerase II interacts homologously with two different promoters as revealed by P1 endonuclease analysis. Mol Gen Genet. 1986 Aug;204(2):249–257. doi: 10.1007/BF00425506. [DOI] [PubMed] [Google Scholar]
- Fischer D. K., Robert M. F., Shedd D., Summers W. P., Robinson J. E., Wolak J., Stefano J. E., Miller G. Identification of Epstein-Barr nuclear antigen polypeptide in mouse and monkey cells after gene transfer with a cloned 2.9-kilobase-pair subfragment of the genome. Proc Natl Acad Sci U S A. 1984 Jan;81(1):43–47. doi: 10.1073/pnas.81.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque R. J. Nucleotide sequence of the region encompassing the JC virus origin of DNA replication. J Virol. 1983 Apr;46(1):170–176. doi: 10.1128/jvi.46.1.170-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradoville L., Grogan E., Taylor N., Miller G. Differences in the extent of activation of Epstein-Barr virus replicative gene expression among four nonproducer cell lines stably transformed by oriP/BZLF1 plasmids. Virology. 1990 Oct;178(2):345–354. doi: 10.1016/0042-6822(90)90331-k. [DOI] [PubMed] [Google Scholar]
- Hampar B., Tanaka A., Nonoyama M., Derge J. G. Replication of the resident repressed Epstein-Barr virus genome during the early S phase (S-1 period) of nonproducer Raji cells. Proc Natl Acad Sci U S A. 1974 Mar;71(3):631–633. doi: 10.1073/pnas.71.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing J. C., Levine A. J. The Epstein-Barr virus nuclear antigen (BamHI K antigen) is a single-stranded DNA binding phosphoprotein. Virology. 1985 Aug;145(1):105–116. doi: 10.1016/0042-6822(85)90205-3. [DOI] [PubMed] [Google Scholar]
- Hsieh T. S., Wang J. C. Thermodynamic properties of superhelical DNAs. Biochemistry. 1975 Feb 11;14(3):527–535. doi: 10.1021/bi00674a011. [DOI] [PubMed] [Google Scholar]
- Jones C. H., Hayward S. D., Rawlins D. R. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J Virol. 1989 Jan;63(1):101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni J. M., Kornberg A. Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell. 1984 Aug;38(1):183–190. doi: 10.1016/0092-8674(84)90539-7. [DOI] [PubMed] [Google Scholar]
- Karlin S. Significant potential secondary structures in the Epstein-Barr virus genome. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6915–6919. doi: 10.1073/pnas.83.18.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D. Changes in site specificity of single-strand-specific endonucleases on supercoiled PM2 DNA with temperature and ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7071–7086. doi: 10.1093/nar/12.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Eddy M. J. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989 Dec 20;8(13):4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The kinetic properties of cruciform extrusion are determined by DNA base-sequence. Nucleic Acids Res. 1985 Mar 11;13(5):1443–1465. doi: 10.1093/nar/13.5.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Adams A., Bjursell G., Bornkamm G. W., Kaschka-Dierich C., Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976 Apr 15;102(3):511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Lupton S., Levine A. J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985 Oct;5(10):2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Nonoyama M., Pagano J. S. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat New Biol. 1972 Aug 9;238(84):169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- Orlowski R., Polvino-Bodnar M., Hearing J., Miller G. Inhibition of specific binding of EBNA 1 to DNA by murine monoclonal and certain human polyclonal antibodies. Virology. 1990 Jun;176(2):638–642. doi: 10.1016/0042-6822(90)90036-q. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Fontaine A. A native cruciform DNA structure probed in bacteria by recombinant T7 endonuclease. J Biol Chem. 1987 Aug 15;262(23):11364–11368. [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Platt J. R. POSSIBLE SEPARATION OF INTERTWINED NUCLEIC ACID CHAINS BY TRANSFER-TWIST. Proc Natl Acad Sci U S A. 1955 Mar 15;41(3):181–183. doi: 10.1073/pnas.41.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins D. R., Milman G., Hayward S. D., Hayward G. S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985 Oct;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Reisman D., Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986 Nov;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985 Aug;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sexton C. J., Pagano J. S. Analysis of the Epstein-Barr virus origin of plasmid replication (oriP) reveals an area of nucleosome sparing that spans the 3' dyad. J Virol. 1989 Dec;63(12):5505–5508. doi: 10.1128/jvi.63.12.5505-5508.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. E., Levinger L. F., Carter C. W., Jr Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979 Feb;29(2):657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C. E., Stahl M. M., Stahl F. W. Large palindromes in the lambda phage genome are preserved in a rec+ host by inhibiting lambda DNA replication. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1624–1628. doi: 10.1073/pnas.84.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sugden B., Marsh K., Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985 Feb;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989 Jun;63(6):2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. Influence of cation size and charge on the extrusion of a salt-dependent cruciform. J Mol Biol. 1987 Jan 20;193(2):397–404. doi: 10.1016/0022-2836(87)90227-0. [DOI] [PubMed] [Google Scholar]
- Summers W. P., Grogan E. A., Shedd D., Robert M., Liu C. R., Miller G. Stable expression in mouse cells of nuclear neoantigen after transfer of a 3.4-megadalton cloned fragment of Epstein-Barr virus DNA. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5688–5692. doi: 10.1073/pnas.79.18.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen D. G., Haines L. L., Hansen U. M., Martin R. G., Livingston D. M. Formation of a cruciform structure at the simian virus 40 replication origin abolishes T-antigen binding to the origin in vitro. J Virol. 1985 Oct;56(1):293–297. doi: 10.1128/jvi.56.1.293-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. 1985 Feb 28-Mar 6Nature. 313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Frappier L., Khoury M., Price G. B. Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. EMBO J. 1988 Jun;7(6):1837–1844. doi: 10.1002/j.1460-2075.1988.tb03016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]