Abstract
Retinoids, vitamin A (retinol) and its metabolic derivatives, are required for normal vertebrate development. In murine embryonic stem (ES) cells, which remain undifferentiated when cultured in the presence of LIF (leukemia inhibitory factor), little metabolism of exogenously added retinol takes place. After LIF removal, ES cells metabolize exogenously added retinol to 4-hydroxyretinol and 4-oxoretinol and concomitantly differentiate. The conversion of retinol to 4-oxoretinol is a high-capacity reaction because most of the exogenous retinol is metabolized rapidly, even when cells are exposed to physiological (≈1 μM) concentrations of retinol in the medium. No retinoic acid or 4-oxoRA synthesis from retinol was detected in ES cells cultured with or without LIF. The cytochrome P450 enzyme CYP26 (retinoic acid hydroxylase) is responsible for the metabolism of retinol to 4-oxoretinol, and CYP26 mRNA is greatly induced (>15-fold) after LIF removal. Concomitant with the expression of CYP26, differentiating ES cells grown in the absence of LIF activate the expression of the differentiation marker gene FGF-5 whereas the expression of the stem cell marker gene FGF-4 decreases. The strong correlation between the production of polar metabolites of retinol and the differentiation of ES cells upon removal of LIF suggests that one important action of LIF in these cells is to prevent retinol metabolism to biologically active, polar metabolites such as 4-oxoretinol.
Leukemia inhibitory factor (LIF) is a member of the IL-6 family of cytokines, which exhibit a variety of effects on different cell types, including embryonic stem (ES) cells, primordial germ cells, neurons, adipocytes, and osteoblasts. The actions of LIF are mediated through a specific LIF receptor (LIF-R) that heterodimerizes with the IL-6 signal-transducing receptor component gp130 (1, 2). Signal transduction through LIF involves the activation of both the Jak/STAT (signal transducer and activator of transcription; specifically STAT3) and Ras/mitogen-activated protein kinase pathways (3–7).
The physiological importance of LIF and the LIF receptor for normal development has been shown in mice with targeted disruptions in these genes (8–10). Mice lacking both alleles of the LIF gene exhibited growth retardation and female sterility. Female mice that lack both alleles of the LIF gene can produce fertile oocytes, but the blastocysts cannot implant (11). Mice that lack both alleles of the LIF-R were not able to survive past 1 day postnatally and exhibited several defects, including abnormal placentation, decreased fetal bone volume, and decreased motor neuron number (9, 10). An increase in LIF mRNA occurs in the uterine endometrial glands of mice on the fourth day of gestation, just before blastocyst implantation (12). LIF mRNA also is expressed in the differentiated trophectoderm cells of murine blastocysts, whereas LIF-R mRNA is expressed in the inner cell mass (embryonic ectoderm) but not in the trophectoderm; these data suggest that LIF affects the maintenance of the pluripotency of the early mouse embryo (13). LIF also functions to prevent the differentiation of cultured murine embryonic stem cells, which are analogous in many respects to the inner cell mass cells of the blastocyst (14).
The metabolism of retinol is complicated and varies depending on the cell type. In many cell types, including epithelial cells, retinol is metabolized primarily via esterification to retinyl esters (refs. 15 and 16; for review, see ref. 17). In other cell types, retinol is metabolized to retinoic acid and/or 14-hydroxyretroretinol and anhydroretinol (17–20). F9 teratocarcinoma cells, the MCF-7 and T47D human breast carcinoma cell lines, and NB-4 human promyelocytic leukemia cells metabolize retinol to 4-oxoretinol under certain culture conditions (21–23). We demonstrated previously that 4-hydroxyretinol and 4-oxoretinol are biologically active retinoids that can activate transcription through the retinoic acid receptors (RARs), but not through the retinoid X receptors (RXRs) (21). 4-Oxoretinol causes teratogenic effects similar to those caused by RA in Xenopus embryos (21). Additionally, 4-oxoretinol is a more potent activator of gene transcription than 4-hydroxyretinol, especially with respect to RARγ (21).
In this paper we characterize the metabolism of retinol to 4-oxoretinol in ES cells stimulated to differentiate by culture in the absence of LIF. We demonstrate that upon LIF removal, ES cells quantitatively metabolize vitamin A (retinol) into the more biologically active metabolites 4-hydroxyretinol and 4-oxoretinol.
Materials and Methods
Cell Culture.
CCE WT embryonic stem cells, grown in monolayer culture, were maintained in DMEM supplemented with 10% FCS, 2 mM glutamine, 1 mM MEM nonessential amino acids (Life Technologies, Grand Island, NY), 10 mM pyruvate, 0.0008% 2-mercaptoethanol, 1,000 units/ml penicillin, and 1,000 μg/ml streptomycin. Four days before addition of the radiolabel, cells were trypsinized and seeded at a density of approximately 5 × 104 cells/60-mm dish with or without 1,000 units/ml LIF (Life Technologies). New LIF was added to the “+LIF” cultures every 24 h.
At 96 h, the medium was removed and 2 ml of radiolabeling medium, consisting of supplemented DME plus 5% FCS and 50 nM [3H]retinol (specific activity, 35.2 μCi/nmol; 1Ci = 37 GBq) or 100 nM [3H]retinol and 900 nM unlabeled retinol (total retinol concentration of 1 μM) was added. A control of labeling medium without cells also was included. Cell and media samples (0.5 ml) were harvested at various time points as described (16). All samples were frozen at −70°C until extraction, and all retinoid manipulations were performed under subdued light. Cell number for each treatment was determined from duplicate dishes during the incubation period.
For the photodiode array experiments to examine the endogenous retinoids in the cells, CCE WT cells were plated at a density of 1 × 106 cells/150-mm dish in supplemented medium with or without 1,000 units/ml LIF. Twenty-four hours later, fresh medium was added to all plates. LIF was added to the appropriate plates every 24 h for the next 72 h. After 96 h, the medium was changed and cells were incubated with supplemented DME, containing 5% FCS and 1 μM unlabeled retinol, for 16 h.
Retinoid Extraction and HPLC Analysis.
Retinoids were extracted by the method described previously (16, 24). Retinoids were separated by using a Waters Millennium System. The samples (100 μl) were loaded onto a 5-μm analytical reverse-phase C-18 column (Vydac, Hesperia, CA) at a flow rate of 1.5 ml/min. The retinoids were separated by using a gradient from 100% 15 mM ammonium acetate in water, pH 6.5, to 100% acetonitrile over 60 min. The elution of the nonlabeled retinoid standards included in the samples was monitored at 340 nm, and a Packard A-500 radiochromatography detector was used to detect radiolabeled retinoids. The identities of the retinoids were determined by coelution of the radiolabeled samples with the known retinoid standards included in the samples and UV absorption spectra comparison of nonradiolabeled samples, analyzed via PDA, with known standards. To assess the retinol metabolism quantitatively, each peak (retinol) or group of peaks (polar metabolites) was integrated and the total retinoid concentration was calculated as described (16).
Diazomethane Treatment of Extracted Medium Samples.
A medium sample from CCE ES cells, cultured for 96 h without LIF and cultured in the presence of 50 nM [3H]retinol for 16 h as described above, was mixed with a standard of nonradioactive 4-oxoRA at a concentration of 40 μM. This sample then was divided and diazomethane (10 μl) was added to one-half of the sample (250 μl) until the sample turned yellow. The other half of the sample was left untreated. Both samples then were analyzed by HPLC.
Cloning of the Full-Length CYP26 cDNA.
Expressed sequence tag (EST) clones that partially encoded the enzyme CYP26 (25) were used to screen a cDNA library prepared from mRNA isolated from murine F9 cells cultured with RA, cAMP, and theophylline and ligated into a λgt10 vector (26). A clone containing all but the first 24 bases from the 5′ most segment of the cDNA encoding CYP26 (RA hydroxylase) was obtained from the library and subcloned into pBluescript. The full-length cDNA clone was synthesized by using PCR to attach the remaining 5′ portion of the cDNA. The upstream primer was 5′-AAAAAGGATCCGAATTCATGGGGCTCCCGGCGCTGCTGGCCAGTGCGCTCTGCACCTTCGTGCTGCCGCTGCTG (EcoRI and BamHI sites followed by nucleotides 1–57). The downstream primer was GCTCGCCGCAGCTTAGCCACTGCTCCA (inverse complement to nucleotides 491–507). Ten nanograms of plasmid containing the incomplete clone of CYP26 in pBluescript were incubated with 1 μg/μl of upstream primer, 0.5 μg/μl of downstream primer, NTPs, and 2.5 units of Amplitaq DNA polymerase in 1× reaction buffer. The approximately 500-bp reaction product was isolated and ligated into the BamHI site of the vector and the internal BlpI site in CYP26, and its DNA sequence was confirmed (27).
RNA Isolation and Northern Blot Analysis.
Total RNA was isolated from cells cultured with and without LIF as described above. At 96 h the culture medium was removed, and supplemented medium, containing 5% FBS and 50 nM or 1 μM retinol, was added. After 16 h, cells were harvested and the RNA was isolated by using RNA Stat-60 (Tel-Test, Friendswood, TX). Additional cells were incubated with supplemented medium, containing 5% FBS and 50 nM or 1 μM retinol and 2 μg/ml actinomycin D, for 4 or 8 h before cell harvest. RNA was electrophoresed through 1.2% agarose/2.2 M formamide gels, transferred to nylon filters, and cross-linked to the filters with a UV-Stratalinker (Stratagene). The cDNA probes were labeled via the random primer method (Boehringer Mannheim) with [α-32P]dCTP. The CYP26 probe used was isolated from the cDNA library screening described above. The cDNAs encoding mouse fibroblast growth factor 4 (FGF-4) and FGF-5 were obtained from Gail Martin (28), and mouse GAPDH cDNA was obtained from Ambion (Austin, TX).
Blots were prehybridized and hybridized overnight at 42°C in 50% (wt/vol) deionized formamide/0.2% BSA/0.2% polyvinyl pyrrolidone/2% Ficoll (molecular weight 400,000 g/mol)/50 mM Tris⋅HCl, pH 7.5/0.1% sodium pyrophosphate/1% SDS/10% dextran sulfate/100 μg/ml salmon sperm DNA. After hybridization, blots were washed once at 42°C with 2× SSC/0.1% SDS/0.8% sodium pyrophosphate for 15 min and at 42°C and 65°C for 20 min with 2× SSC/1% SDS. The final wash was performed at room temperature for 15 min in 0.5× SCC. All autoradiographs were quantitated by using a PhosphorImager (Molecular Dynamics).
Transient Transfection of CYP26 in COS Cells.
The full-length CYP26 cDNA then was subcloned into the EcoRI site of pSG5 for expression in COS cells. One day before transfection, 2 × 106 COS cells were plated per 100-mm dish. On the day of transfection, 20 μg of pSG5 containing CYP26 was added to 1.9 ml of PBS. pSG5 lacking an insert (20 μg) was added to another 1.9 ml as a control. DEAE-dextran solution (100 μl of a 10-mg/ml stock) was added to the tubes, COS cells were rinsed with PBS, and the DNA was added to each plate. After a 30-min incubation at 37°C, medium containing 80 μM chloroquine was added to the cells for 2.5 h. New medium containing 10% DMSO then was added for 2.5 min at room temperature (29). This medium was removed and the cells were incubated with 50 nM [3H]retinol for 1–12 h. The media and cell samples were processed for HPLC analysis as described above.
Results
Retinol Metabolism in ES Cells Cultured in the Absence of LIF.
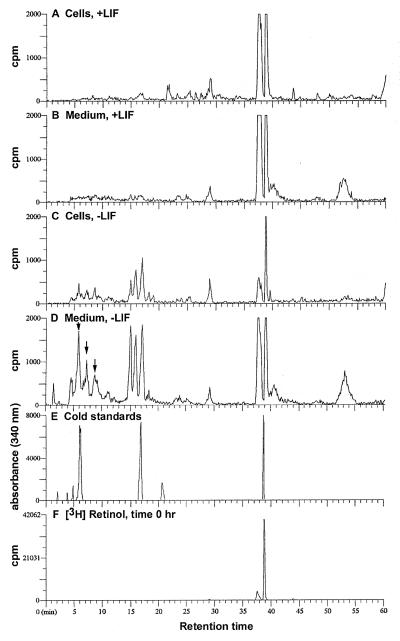
When CCE wild-type ES cells were cultured in the presence of LIF, little metabolism of [3H]retinol was observed (Fig. 1 A and B). In contrast, when ES cells were cultured without LIF for 4 days, exogenously added 50 nM [3H]all-trans retinol was almost completely converted to more polar metabolites, primarily [3H]all-trans-4-hydroxyretinol and [3H]all-trans 4-oxoretinol, which were eluted at 15.0 and 16.8 min, respectively (Fig. 1 C and D). Additional, more polar metabolites of [3H]retinol, eluting between minutes 4 and 10, also appeared primarily in the medium of cells cultured without LIF (see arrows, Fig. 1D). The identities of the radiolabeled compounds [3H]4-hydroxyretinol and [3H]4-oxoretinol were confirmed by comparing their wavelength spectra with those of authentic, nonradiolabeled standards for 4-hydroxyretinol and 4-oxoretinol and by photodiode array analysis of endogenous retinoids extracted from CCE cells cultured without LIF (ref. 21; Fig. 2). Similar patterns of [3H]retinol metabolism were observed when AB1, E14, and J1 ES cell lines were cultured with or without LIF (data not shown). Thus, the dramatic increase in [3H]retinol metabolism to more polar metabolites upon LIF removal observed in the CCE ES cells is a general property of ES cells.
Figure 1.
Metabolism of [3H]retinol by CCE ES cells. CCE wild-type ES cells were cultured in DMEM supplemented with 10% FCS (30). Cells were cultured for 4 days plus or minus 1,000 units/ml LIF (Life Technologies). LIF was added every 24 h to the +LIF samples. After 4 days, new DMEM containing 5% FCS and 50 nM [3H]retinol was added for various times (16 h is shown). [The concentration of nonradioactive retinol in this 5% FCS was ≈50 nM (X. Guo and L.J.G., unpublished data).] The retinoids were extracted (16) and separated by HPLC (21). The identities of the retinoids were determined by coelution of the radiolabeled samples with known retinoid standards included in the samples and UV absorption spectra comparison of nonradiolabeled samples, analyzed via photodiode array, with known standards. Radiolabeled retinoids extracted from ES cells (A) or from the medium (B) of ES cells cultured with LIF for 4 days and with 50 nM [3H]retinol for 16 h. Radiolabeled retinoids extracted from cells (C) or from the medium (D) of ES cells cultured for 4 days without LIF, followed by culture with 50 nM [3H]retinol for 16 h. The all-trans-4-hydroxyretinol elutes at 15.0 min, 13-cis-4-hydroxyretinol elutes at 15.9 min, and the all-trans-4-oxoretinol elutes at 16.8 min. The arrows in D indicate unidentified, more polar [3H]retinol metabolites in the medium of cells −LIF. (E) Retinoid standards are as follows: all-trans 4-oxoRA, 6.2 min; all-trans-4-oxoretinol, 16.8 min; all-trans RA, 20.6 min; all-trans retinol, 38.7 min. (F) [3H]Retinol extracted at time zero from a control plate including medium and FCS but no cells. This experiment, including multiple time points, was performed five times with similar results.
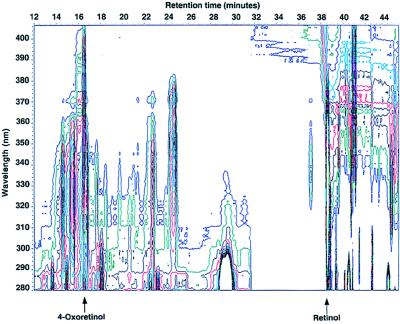
Figure 2.
Contour plot of the photo diode array analysis of retinoids present in ES cell extracts. CCE ES cells (1 × 106 cells per dish; 10 dishes) were grown for 96 h without LIF. Nonradiolabeled retinol (1 μM) was added for 16 h. The retinoids were extracted from the cell samples and analyzed via HPLC and photodiode array. The arrows at 16.6 min and 38.5 min indicate the all-trans 4-oxoretinol and all-trans retinol, respectively.
Retinol Is Not Metabolized to RA in ES Cells.
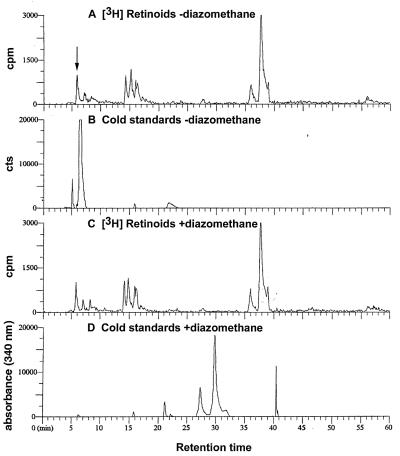
It also should be noted that no detectable [3H]RA or [3H]4-oxoRA was produced from [3H]retinol by ES cells cultured either in the presence or absence of LIF (Fig. 1). No peaks were observed in Fig. 1 A–D, where standards for RA (20.5 min) and 4-oxoretinoic acid (6.2 min, Fig. 1E) elute from the HPLC column. Diazomethane also was utilized to prove that no RA or 4-oxoRA was present. Diazomethane converts RA to methyl retinoate, a more lipophilic compound, with a retention time of 40.5 min, which is different from that of RA (22). Diazomethane also converts 4-oxoRA to methyl-4-oxoretinoate, which elutes at 30 min (Fig. 3). When the sample from CCE cells cultured without LIF was treated with diazomethane, none of the radioactive metabolites of [3H]retinol was shifted (Fig. 3 A vs. C). In contrast, nonradiolabeled 4-oxoRA was completely shifted by the diazomethane treatment (Fig. 3 B vs. D). This experiment conclusively shows that there is no detectable [3H]RA or [3H]4-oxoRA produced from [3H]retinol by the ES cells. [There is a more polar metabolite of [3H]retinol that elutes about 30 sec before 4-oxoRA (see arrow in Fig. 3A), but this clearly is not identical to 4-oxoRA because this peak was not shifted by diazomethane treatment (Fig. 3 A vs. C).]
Figure 3.
Diazomethane treatment of extracts. One sample from CCE ES cells, cultured without LIF, as described above, containing radiolabeled metabolites of retinol, was mixed with a standard of nonradioactive 4-oxoRA. This sample then was divided and diazomethane was added to one-half of the sample, and the other half was left untreated. Both samples then were analyzed by HPLC. (A) One-half of the sample of [3H]retinoids from the CCE cells without diazomethane treatment. (B) The nonradiolabeled 4-oxoRA standard (6.4 min) measured at wavelength 340 nm without diazomethane treatment. (C) One-half of the sample shown in A after treatment with diazomethane. (D) One-half of the sample shown in B after treatment with diazomethane. The standards in B and D are: 4-oxoRA, 6.4 min; all-trans RA, 21.3 min. Methyl 4-oxoretinoate, 30.0 min, and methyl retinoate, 40.5 min, are only in diazomethane-treated samples.
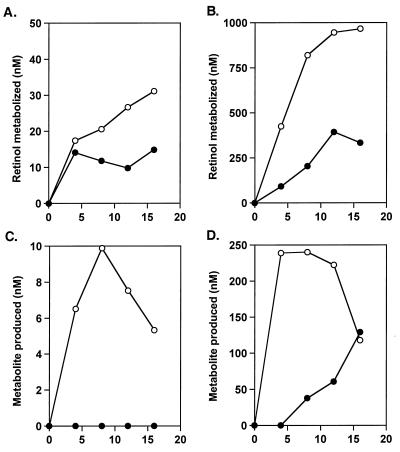
Time Course of 4-Oxortetinol Production from Retinol.
CCE cells were cultured with or without LIF, followed by culture in the presence of either 50 nM (Fig. 4 A and C) or 1 μM [3H]retinol (Fig. 4 B and D). A dose of 50 nM retinol does not, by itself, induce ES cells to differentiate; it is lower than the physiological concentration of retinol. The concentration of retinol in 100% serum is 1–2 μM (31); thus, cells also were cultured in this higher concentration of [3H]retinol to determine the capacity of the cells to metabolize retinol under physiologically relevant conditions. Even when high, physiological concentrations of [3H]retinol (31) were added exogenously; almost all of the 1 μM [3H]retinol was metabolized to more polar compounds, including [3H]4-hydroxyretinol and [3H]4-oxoretinol, within 8–12 h (Fig. 4 B and D). Thus, the metabolism of retinol to 4-hydroxyretinol and 4-oxoretinol by the ES cells cultured without LIF is a high-capacity reaction. The [3H]4-hydroxyretinol and [3H]4-oxoretinol levels peaked in these cells at 4–8 h and then declined at 12 and 16 h both as a result of further metabolism of 4-oxoretinol to aqueous soluble metabolites and the lack of remaining [3H]retinol substrate in the cells without LIF (Fig. 4 C and D); for example, note that by 8 h, approximately 800 nM, or 80% of the 1 μM [3H]retinol added exogenously to the cells cultured without LIF, had been metabolized to 4-hydroxyretinol and 4-oxoretinol (Fig. 4B).
Figure 4.
Metabolism of [3H]retinol in CCE ES cells cultured with or without LIF. The tracings are ●, +LIF; ○, −LIF. Total amounts of [3H]retinol metabolized over time in cells cultured with or without LIF in the presence of 50 nM [3H] retinol (A) or 1 μM [3H]retinol (B) for 4, 8, 12, and 16 h. Total amounts of [3H]4-hydroxyretinol and [3H]4-oxoretinol produced from [3H]retinol by the CCE cells cultured with or without LIF in 50 nM [3H]retinol (C) or 1 μM [3H]retinol (D). The concentrations of retinol and polar metabolites, y axis; time in hours, x axis. This experiment was repeated three times for the 8- and 16-h time points and twice for the 4- and 12-h time points with results within 10%. One representative experiment is shown here.
The P450 Enzyme CYP26 Converts Retinol into 4-Oxoretinol in COS cells and Is Expressed in Differentiating ES Cells.
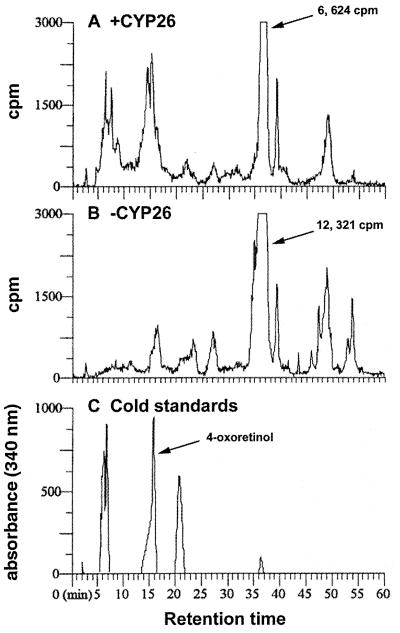
Inhibitors of P450 enzymes such as SKF525A block the conversion of retinol to 4-hydroxyretinol and 4-oxoretinol (data not shown). Although previous research has shown the ability of CYP26 to convert RA to 4-oxoRA, it was reported that this enzyme does not metabolize retinol to 4-oxoretinol (32–34). Our data indicate that the cytochrome P450 enzyme CYP26 (RA hydroxylase) (25, 32–34) catalyzes the conversion of retinol to 4-oxoretinol. COS cells transfected with a mammalian expression vector driving expression of the full-length murine CYP26 cDNA quantitatively metabolized [3H]retinol to [3H]4-oxoretinol and [3H]4-hydroxyretinol, whereas mock-transfected COS cells did not metabolize [3H]retinol to [3H]4-oxoretinol (Fig. 5 +CYP26, is a 1-h time point, and B, vector control, is a 12-h time point).
Figure 5.
Transfection of COS cells with CYP26. COS cells were transiently transfected with the full-length murine CYP26 cDNA (A) or vector pSG5 alone (B). [3H]Retinol was added to the cells for various times [1 h is shown for +CYP26 cells, 12 h is shown for −CYP26 (vector only) cells], and retinoids were extracted. The numbers to the right of the arrows indicate quantitation of the [3H]retinol remaining. The full-length CYP26 cDNA sequence was verified by DNA sequence analysis. The peak in A at 15.8 min elutes at the same position as the 4-oxoretinol standard (C). All-trans RA, 21 min; all-trans retinol, 36.5 min (C). The elution times of the retinoid standards in this experiment are slightly different from those in Fig. 1 because a different reverse-phase column was used for this experiment.
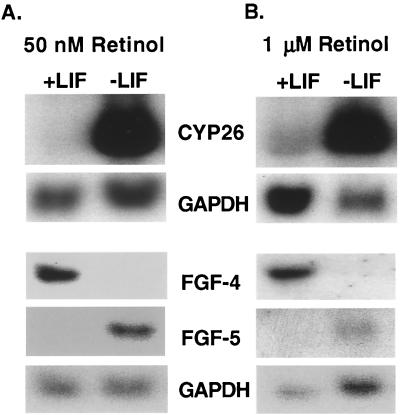
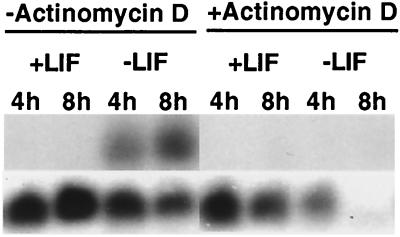
FGF-4 is a marker of undifferentiated ES cells whereas FGF-5 is expressed in differentiated ES cells (28). When CCE ES cells were induced to differentiate by the removal of LIF, the level of FGF-4 mRNA was reduced and the level of FGF-5 mRNA increased (Fig. 6). In addition, CYP26 mRNA was highly induced (>15-fold) when ES cells were cultured without LIF (Fig. 6), consistent with the massive conversion of [3H]retinol to more polar metabolites (Fig. 1). This increase in CYP26 mRNA could be the result of increased stability of the mRNA or increased gene transcription after LIF removal. To determine which was the case, ES cells were cultured without LIF, with 50 nM retinol, and with or without actinomycin D for 4 or 8 h (Fig. 7). Addition of actinomycin D, an inhibitor of RNA transcription, to ES cells that had been cultured in the absence of LIF prevented the increase in CYP26 mRNA, suggesting that the removal of LIF results in an increase in the rate of transcription of the CYP26 gene (Fig. 7, lanes 3 and 4 vs. 7 and 8). These results suggest that one action of LIF is to prevent the transcription of the P450 CYP26 gene in embryonic stem cells and, by analogy, in the pluripotent inner cell mass cells of the mammalian blastocyst.
Figure 6.
Northern blot analysis. Northern blot analysis of CYP26 (RA hydroxylase), FGF-4, FGF-5, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs. CCE cells were cultured as in Fig. 1, and 50 nM retinol (A) or 1 μM retinol (B) was added 16 h before RNA harvest. The Northern analyses were performed three times with different RNA preparations with similar results.
Figure 7.
Actinomycin D treatment of CCE. Cells were cultured as in Fig. 1, and 50 nM retinol, plus or minus 2 μg/ml actinomycin D, was added for 4 or 8 h before cell harvest and RNA preparation. RNA was hybridized with CYP26 and GAPDH. Lane 8 had a lower signal because the cells were beginning to be affected by actinomycin D toxicity at this 8-h time point (+Actinomycin D, −LIF). This experiment was performed twice with identical results.
Discussion
We have shown that when either 50 nM or 1 μM all-trans retinol is added to ES cells cultured without LIF, more than 95% of this retinol is metabolized to the polar metabolites all-trans 4-hydroxyretinol, 13-cis-4-hydroxyretinol, and all-trans 4-oxoretinol and to even more polar metabolites eluting between minutes 4–10 (Figs. 1 and 4). Some of these polar retinol metabolites are secreted into the medium (Fig. 1). Only trace amounts of retinyl esters are synthesized from retinol in the ES cells (data not shown). This quantitative conversion of retinol to 4-hydroxyretinol and 4-oxoretinol, even when 1 μM retinol is added exogenously, indicates that this is a high-capacity reaction. In contrast, in ES cells cultured without LIF, [3H]retinol is not metabolized to retinoids with acid moieties such as retinoic acid and 4-oxoretinoic acid (Figs. 1–3). These data indicate that metabolism of retinol to retinaldehyde, and the subsequent metabolism of retinaldehyde to RA, does not occur in all cell types and is not a property of embryonic stem cells.
Retinol is the form of vitamin A to which cells are generally exposed in the animal, because retinol is the form of vitamin A that is transported through the blood bound to the serum retinol-binding protein (for review, see ref. 35). The retinol concentration in 100% serum is 1–2 μM (31). Concomitant with the increase in retinol metabolism to 4-oxoretinol in the ES cells cultured without LIF, a decrease in the expression of stem cell markers, such as FGF-4 mRNA (Fig. 6) (28), and an increase in the expression of markers of differentiation, such as FGF-5 mRNA (Fig. 6), occur. Because no retinoic acid was added exogenously and ES cells do not metabolize retinol to RA (Figs. 1–3), our data suggest that rather than RA, the metabolism of retinol to 4-hydroxyretinol and 4-oxoretinol is involved in the activation of the differentiation program in these cells cultured without LIF. It is likely that the transactivation of the retinoic acid receptors by 4-oxoretinol (21, 36) is involved in regulating some aspects of the differentiation pathway of ES cells that occurs upon LIF removal.
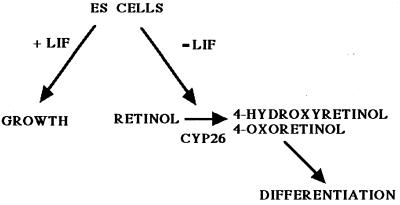
We present data that demonstrate that the enzyme that metabolizes retinol to 4-hydroxyretinol and 4-oxoretinol is CYP26 (Fig. 5). This cytochrome P450 gene CYP26 (RA hydroxylase) was cloned recently (25, 32–34), and the CYP26 enzyme was shown to metabolize RA to 4-oxoretinoic acid. A degradative function, i.e., the removal or inactivation of RA, has been suggested for this enzyme (32, 34, 37). It was reported that retinol was not a substrate for this enzyme (32, 34), but our data indicate instead that CYP26 is responsible for the production of a more bioactive retinoid, 4-oxoretinol, from retinol in some cell types (Figs. 5 and 6). Although previous studies showed that RA is preferred over retinol as a substrate for CYP26 (34), these studies were conducted by using microsomal fractions whereas in the experiments reported here, retinol metabolism in cultured cells was assessed. Even if CYP26 has a higher Km for retinol than for RA, a much higher concentration of retinol is present in serum and in many cell types (for review, see ref. 35), making the metabolism of retinol by CYP26 in specific cell types physiologically relevant. In fact, for maximal induction of the CYP26 mRNA, the cells must be cultured in the absence of LIF plus exogenous retinol (Fig. 6). More 4-oxoretinol is produced when exogenous retinol is added to the medium to supplement the amount of retinol in the 10% FBS, which is ≈50 nM (X. Guo and L.J.G., unpublished data). The retinoic acid concentration in 10% FBS is <2 nM, for comparison (A.C.C. and L.J.G., unpublished data). Our hypothesis is that the absence of LIF leads to an increase in the transcription and/or activity of CYP26. Some retinol is metabolized to 4-oxoretinol, and then the 4-oxoretinol produced from retinol further transcriptionally activates the CYP26 gene via a DNA response element in the promoter, leading to the production of greater amounts of 4-oxoretinol (Fig. 8). Additional experiments are required to prove this hypothesis.
Figure 8.
Model of interaction of LIF with ES cells.
Because 4-oxoretinaldehyde and 4-oxoretinol are the major retinoids detected in developing Xenopus embryos and RA was not detected in these embryos (36) or in murine ES cells (Fig. 1), our results suggest that during early embryonic development the regulation of the conversion of retinol to 4-oxoretinol is critical for appropriate cell differentiation. Thus, one of the important actions of LIF in the embryo may be to prevent the metabolism of retinol via CYP26 to more biologically active metabolites, such as 4-oxoretinol, in cells not ready to undergo differentiation. It is not known whether the LIF signal transduction pathway acts to repress the CYP26 gene itself or indirectly prevents the activation of the CYP26 gene by keeping the ES cells in an undifferentiated state. However, the experiments described in this report suggest both a new function for LIF in the early embryo, the control of retinol metabolism, and a molecular link between the LIF and the retinoid intracellular-signaling pathways.
Acknowledgments
We thank Taryn Resnick for editorial assistance and Dr. John Wagner for scientific advice. This research was supported by National Institutes of Health Grant R01CA77509 to L.J.G.
Abbreviations
- ES
embryonic stem
- FGF
fibroblast growth factor
- LIF
leukemia inhibitory factor
- LIF-R
leukemia inhibitory factor receptor
- RA
all-trans retinoic acid
References
- 1.Ip N Y, Nye S H, Boulton T G, Davis S, Taga T, Li Y, Birren S J, Yasukawa K, Kishimoto D, Anderson D J, et al. Cell. 1992;69:1121–1132. doi: 10.1016/0092-8674(92)90634-o. [DOI] [PubMed] [Google Scholar]
- 2.Koshimizu U, Taga T, Watanabe M, Saito M, Shirayoshi Y, Kishimoto T, Nakatsuji N. Development. 1996;122:1235–1242. doi: 10.1242/dev.122.4.1235. [DOI] [PubMed] [Google Scholar]
- 3.Ernst M, Oates A, Dunn A R. J Biol Chem. 1996;271:30136–30143. doi: 10.1074/jbc.271.47.30136. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H, Burdon T, Chambers I, Smith A. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeuf H, Hauss C, De Graeve F, Baran N, Kedinger C. J Cell Biol. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raz R, Lee C-K, Cannizaro L A, D’Eustachio P, Levy D E. Proc Natl Acad Sci USA. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escary J L, Perreau J, Dumenil D, Ezine S, Brulet P. Nature (London) 1993;363:361–364. doi: 10.1038/363361a0. [DOI] [PubMed] [Google Scholar]
- 9.Ware C B, Horowitz M C, Renshaw B R, Hunt J S, Liggitt D, Koblar S A, Gliniak B C, McKenna H J, Papayannopoulou T, Thoma B, et al. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Sendtner M, Smith A. Nature (London) 1995;378:724–727. doi: 10.1038/378724a0. [DOI] [PubMed] [Google Scholar]
- 11.Stewart C L, Kaspar P, Brunet L J, Bhatt M, Gadi I, Köntgen F, Abbondanzo S J. Nature (London) 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 12.Bhatt H, Brunet L J, Stewart C L. Proc Natl Acad Sci USA. 1991;88:11408–11412. doi: 10.1073/pnas.88.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichols J, Davidson D, Taga T, Yoshida K, Chambers I, Smith A. Mech Dev. 1996;57:123–131. doi: 10.1016/0925-4773(96)00531-x. [DOI] [PubMed] [Google Scholar]
- 14.Rathjen P D, Nichols J, Toth S, Edwards D R, Heath J K, Smith A G. Genes Dev. 1990;4:2308–2318. doi: 10.1101/gad.4.12b.2308. [DOI] [PubMed] [Google Scholar]
- 15.Bhat P V, Jetten A M. Biochim Biophys Acta. 1987;922:18–27. doi: 10.1016/0005-2760(87)90240-2. [DOI] [PubMed] [Google Scholar]
- 16.Guo X, Gudas L J. Cancer Res. 1998;58:166–176. [PubMed] [Google Scholar]
- 17.Blaner W S, Olson J A. In: The Retinoids: Biology, Chemistry, and Medicine. Sporn M B, Roberts A B, Goodman D S, editors. New York: Raven; 1994. pp. 229–256. [Google Scholar]
- 18.Kurlandsky S B, Xiao J H, Duell E A, Voorhees J J, Fisher G J. J Biol Chem. 1994;269:32821–32827. [PubMed] [Google Scholar]
- 19.Buck J, Derguini F, Levi E, Nakanishi K, Hammerling U. Science. 1991;254:1654–1656. doi: 10.1126/science.1749937. [DOI] [PubMed] [Google Scholar]
- 20.Buck J, Grün F, Kimura S, Noy N, Derguini F, Hammerling U. J Exp Med. 1993;178:675–680. doi: 10.1084/jem.178.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Achkar C C, Derguini F, Blumberg B, Langston A, Levin A A, Speck J, Evans R M, Bolado J, Jr, Nakanishi K, Buck J, Gudas L J. Proc Natl Acad Sci USA. 1996;93:4879–4884. doi: 10.1073/pnas.93.10.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen A C, Guo X, Derguini F, Gudas L J. Cancer Res. 1997;57:4642–4651. [PubMed] [Google Scholar]
- 23.Faria T F, Rivi R, Derguini F, Pandolfi P P, Gudas L J. Cancer Res. 1998;58:2007–2013. [PubMed] [Google Scholar]
- 24.McClean S W, Ruddel M E, Gross E G, DiGiovanna J J, Peck G L. Clin Chem. 1982;28:693–696. [PubMed] [Google Scholar]
- 25.Ray W J, Bain G, Yao M, Gottlieb D I. J Biol Chem. 1997;272:18702–18708. doi: 10.1074/jbc.272.30.18702. [DOI] [PubMed] [Google Scholar]
- 26.LaRosa G J, Gudas L J. Proc Natl Acad Sci USA. 1988;85:329–333. doi: 10.1073/pnas.85.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger F S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hébert J M, Basilico C, Goldfarb M, Haub O, Martin G R. Dev Biol. 1990;138:454–463. doi: 10.1016/0012-1606(90)90211-z. [DOI] [PubMed] [Google Scholar]
- 29.Chen M, Achkar C, Gudas L J. Mol Pharmacol. 1994;46:88–96. [PubMed] [Google Scholar]
- 30.Chen A C, Gudas L G. J Biol Chem. 1996;271:14971–14980. doi: 10.1074/jbc.271.25.14971. [DOI] [PubMed] [Google Scholar]
- 31.Smith F R, Goodman D S. J Clin Invest. 1971;50:2426–2436. doi: 10.1172/JCI106741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White J A, Beckett-Jones B, Guo Y-D, Dilworth F J, Bonasoro J, Jones G, Petkovich M. J Biol Chem. 1997;272:18538–18541. doi: 10.1074/jbc.272.30.18538. [DOI] [PubMed] [Google Scholar]
- 33.White J A, Guo Y-D, Baetz K, Beckett-Jones B, Bonasoro J, Hsu K E, Dilworth F J, Jones G, Petkovich M. J Biol Chem. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- 34.Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H. EMBO J. 1997;16:4163–4173. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blomhoff R, Green M H, Berg T, Norum K R. Science. 1990;250:399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- 36.Blumberg B, Bolado J, Jr, Derguini F, Craig A G, Moreno T A, Charkaravarti D, Heyman R A, Buck J, Evans R M. Proc Natl Acad Sci USA. 1996;93:4873–4878. doi: 10.1073/pnas.93.10.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iulianella A, Beckett B, Petkovich M, Lohnes D. Dev Biol. 1999;205:33–48. doi: 10.1006/dbio.1998.9110. [DOI] [PubMed] [Google Scholar]