Abstract
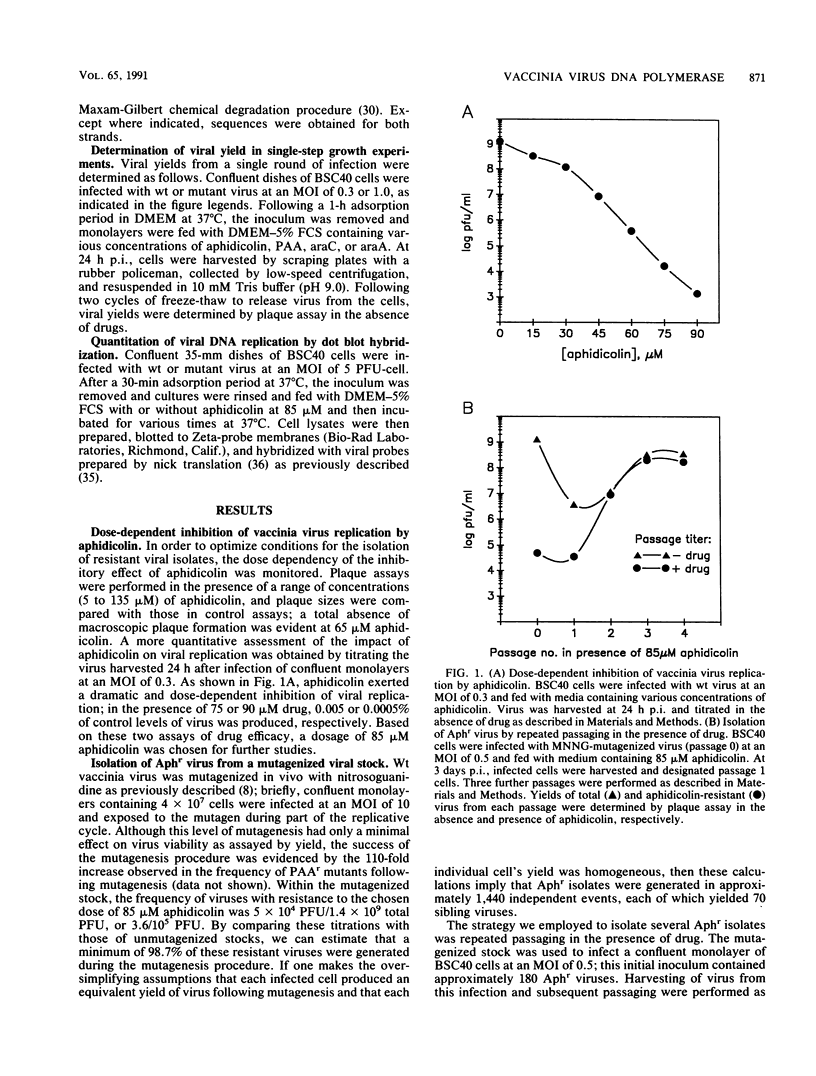
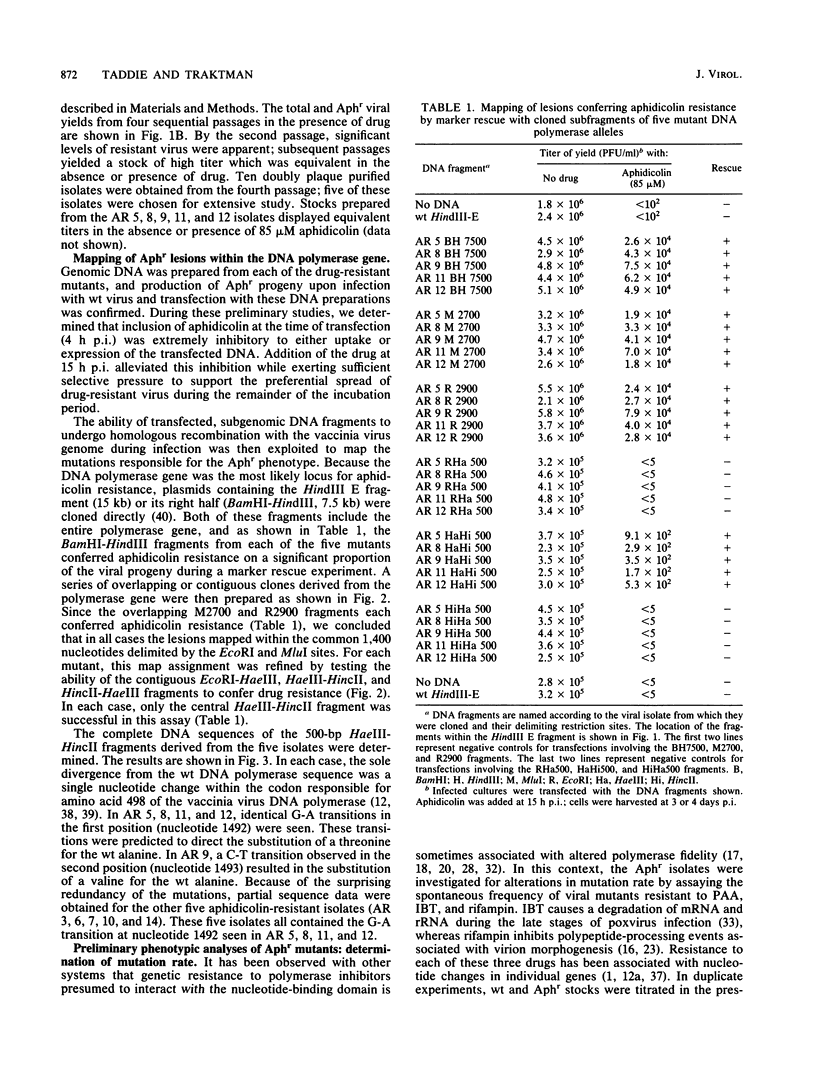
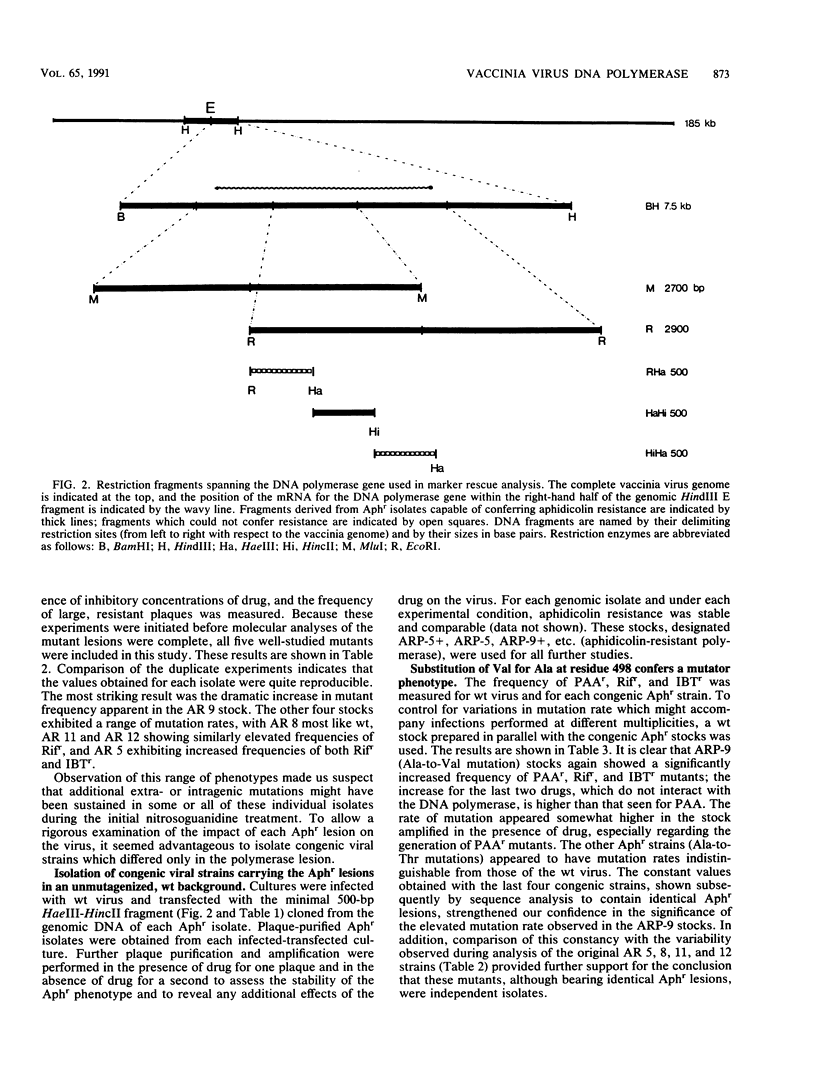
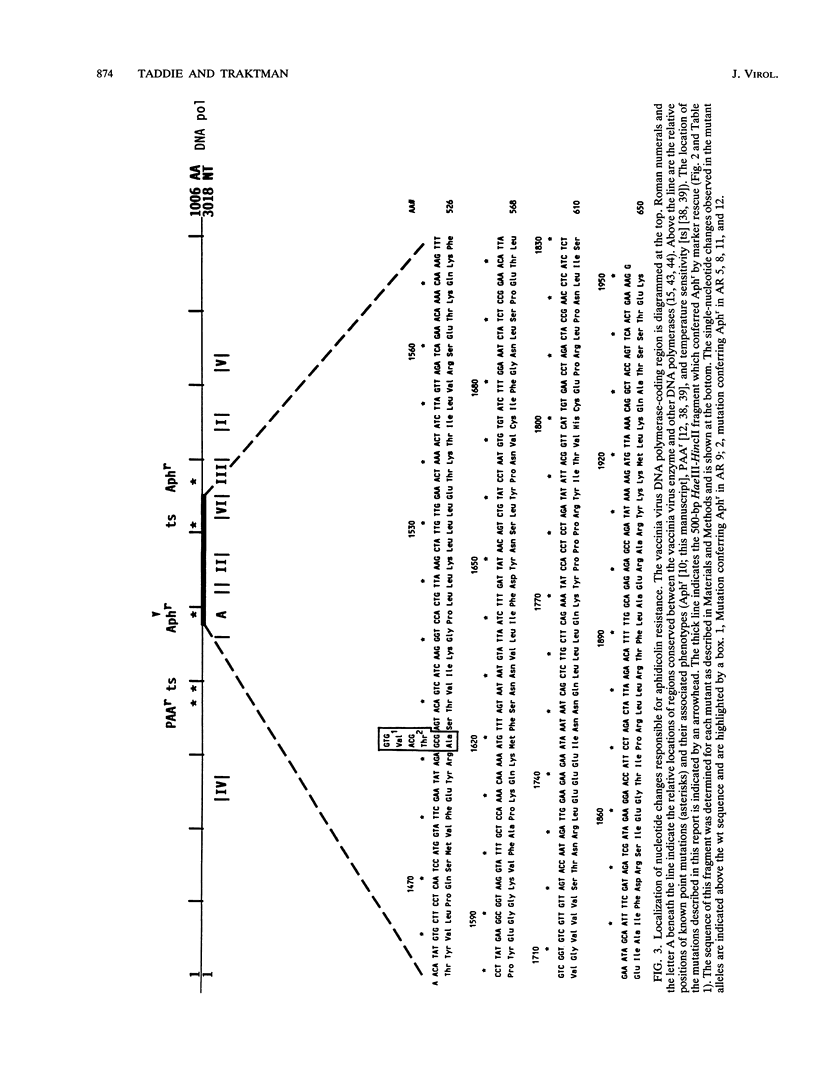
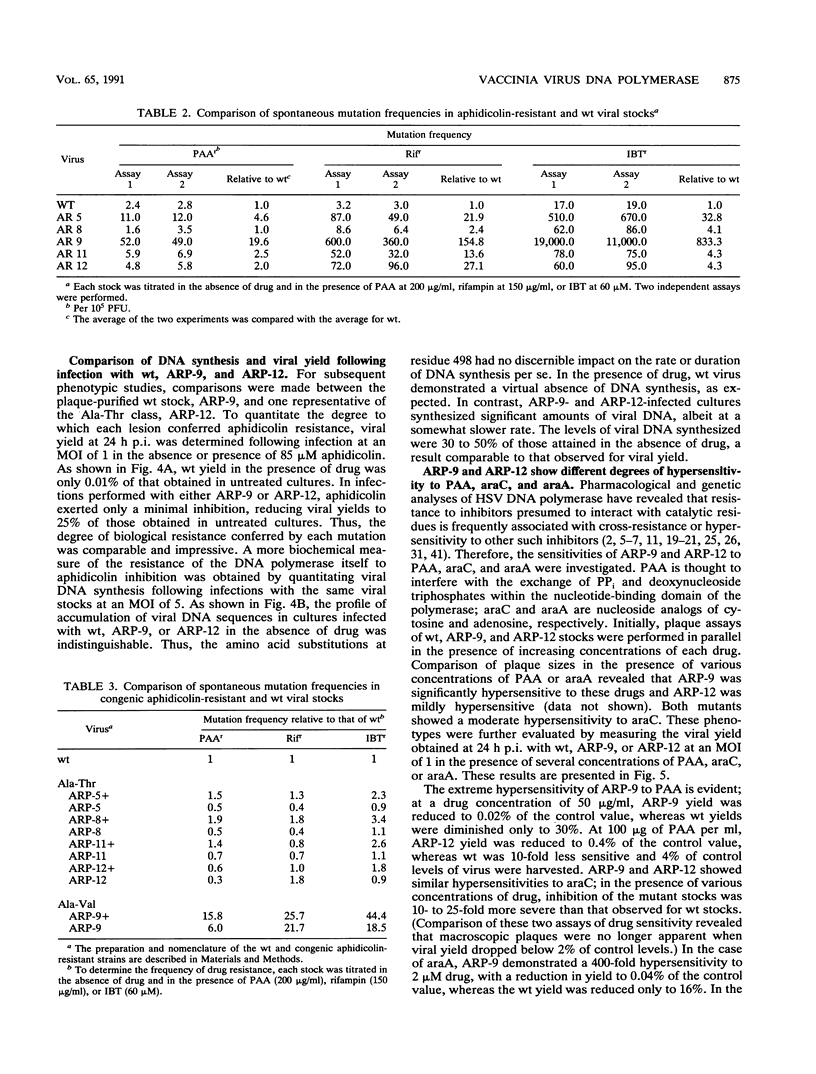
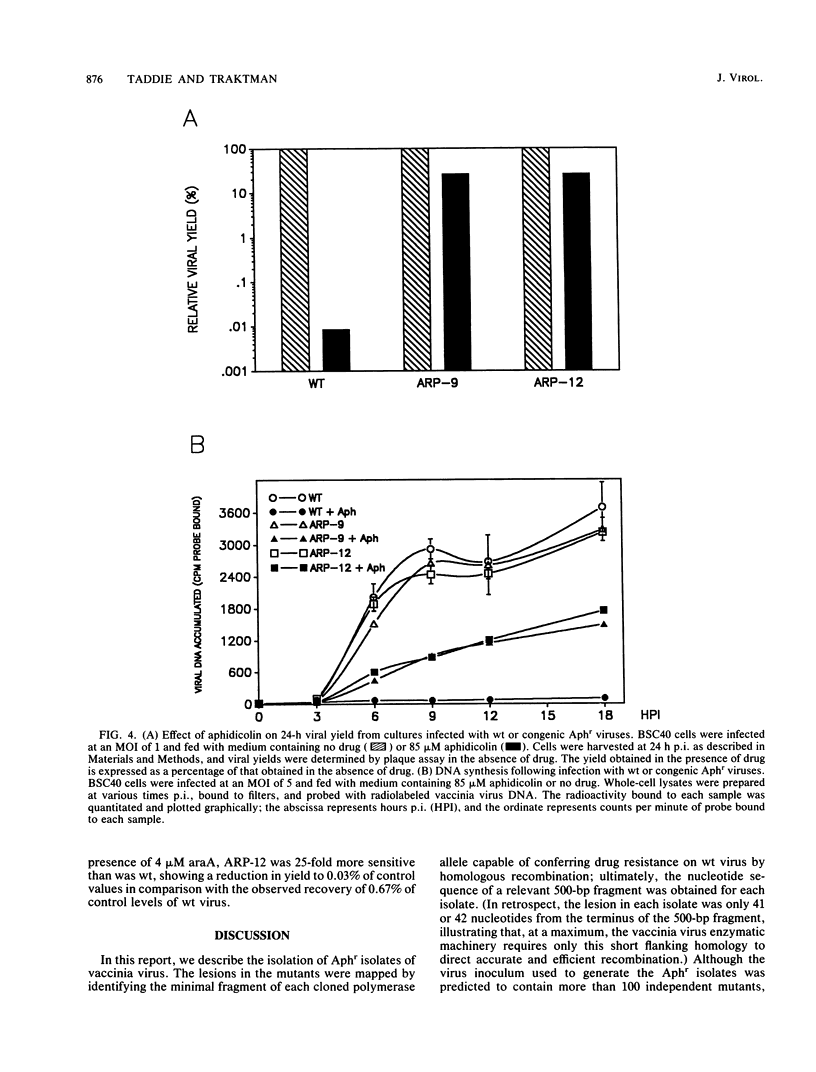
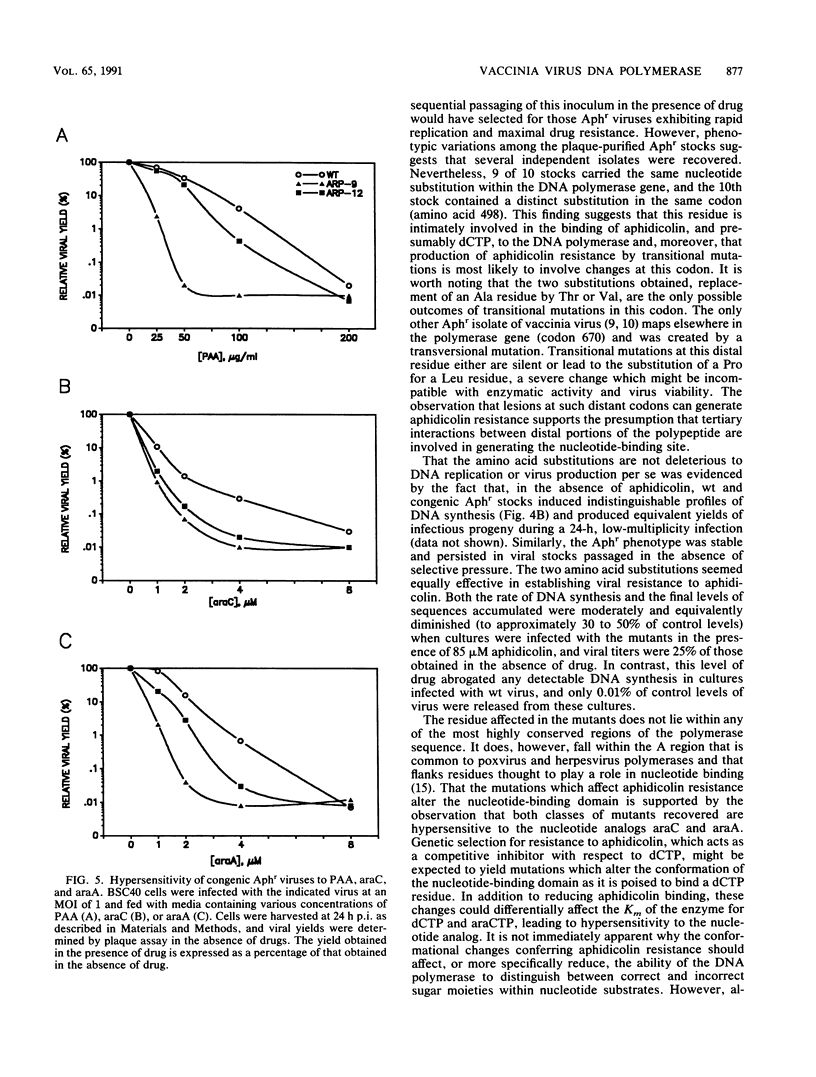
We determined that 85 microM aphidicolin was sufficient to block macroscopic plaque formation by vaccinia virus and to cause a 10(4)-fold reduction in viral yield from a wild-type infection. A chemically mutagenized viral stock was passaged sequentially in the presence of drug, and plaque-purified viral stocks resistant to aphidicolin were isolated and characterized. By use of a marker rescue protocol, the lesion in each mutant was found to map within the same 500-bp fragment within the DNA polymerase gene. All of the mutants were found to contain a single nucleotide change in the same codon. In nine of these mutants, the alanine residue at position 498 was changed to a threonine, whereas a 10th mutant sustained a valine substitution at this position. Congenic viral strains which carried the Aphr lesion in an unmutagenized wild-type background were isolated. The Thr and Val mutations were found to confer equivalent levels of drug resistance. In the presence of drug, viral yields were 25% of control levels, and the levels of viral DNA synthesized were 30 to 50% of those seen in control infections. The two mutations also conferred an equivalent hypersensitivity to the cytosine analog 1-beta-D-arabinofuranosylcytosine (araC); strains carrying the Thr mutation were moderately hypersensitive to the pyrophosphate analog phosphonoacetic acid and the adenosine analog araA, whereas the Val mutation conferred acute hypersensitivity to these inhibitors. The Val mutation also conferred a mutator phenotype, leading to a 20- to 40-fold increase in the frequency of spontaneous mutations within the viral stock.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldick C. J., Jr, Moss B. Resistance of vaccinia virus to rifampicin conferred by a single nucleotide substitution near the predicted NH2 terminus of a gene encoding an Mr 62,000 polypeptide. Virology. 1987 Jan;156(1):138–145. doi: 10.1016/0042-6822(87)90444-2. [DOI] [PubMed] [Google Scholar]
- Bastow K. F., Derse D. D., Cheng Y. C. Susceptibility of phosphonoformic acid-resistant herpes simplex virus variants to arabinosylnucleosides and aphidicolin. Antimicrob Agents Chemother. 1983 Jun;23(6):914–917. doi: 10.1128/aac.23.6.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Fleming H. E., Jr, Leslie L. K., Retondo M. J. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J Virol. 1985 Feb;53(2):477–488. doi: 10.1128/jvi.53.2.477-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Furman P. A., Aschman D. P., Schaffer P. A. Mutations in the herpes simplex virus DNA polymerase gene conferring hypersensitivity to aphidicolin. Nucleic Acids Res. 1983 Aug 11;11(15):5287–5297. doi: 10.1093/nar/11.15.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D. M., Furman P. A., Gelep P. T., Schaffer P. A. Mutations in the herpes simplex virus DNA polymerase gene can confer resistance to 9-beta-D-arabinofuranosyladenine. J Virol. 1982 Mar;41(3):909–918. doi: 10.1128/jvi.41.3.909-918.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981 Aug;113(1):224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- DeFilippes F. M. Effect of aphidicolin on vaccinia virus: isolation of an aphidicolin-resistant mutant. J Virol. 1984 Nov;52(2):474–482. doi: 10.1128/jvi.52.2.474-482.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippes F. M. Site of the base change in the vaccinia virus DNA polymerase gene which confers aphidicolin resistance. J Virol. 1989 Sep;63(9):4060–4063. doi: 10.1128/jvi.63.9.4060-4063.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Bastow K. F., Cheng Y. Characterization of the DNA polymerases induced by a group of herpes simplex virus type I variants selected for growth in the presence of phosphonoformic acid. J Biol Chem. 1982 Sep 10;257(17):10251–10260. [PubMed] [Google Scholar]
- Earl P. L., Jones E. V., Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E., Traktman P. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J Virol. 1987 Oct;61(10):3152–3162. doi: 10.1128/jvi.61.10.3152-3162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K. B., Cheng Y. C. Mutually exclusive inhibition of herpesvirus DNA polymerase by aphidicolin, phosphonoformate, and acyclic nucleoside triphosphates. Antimicrob Agents Chemother. 1985 Apr;27(4):445–448. doi: 10.1128/aac.27.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Bastow K. F., Cheng Y. C., Coen D. M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimley P. M., Rosenblum E. N., Mims S. J., Moss B. Interruption by Rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970 Oct;6(4):519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Coen D. M., Fisher B. L., Weisslitz M., Randall S., Almy R. E., Gelep P. T., Schaffer P. A. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984 Jan 15;132(1):26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Hall J. D., Furman P. A., St Clair M. H., Knopf C. W. Reduced in vivo mutagenesis by mutant herpes simplex DNA polymerase involves improved nucleotide selection. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3889–3893. doi: 10.1073/pnas.82.11.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Wang Y. S., Pierpont J., Berlin M. S., Rundlett S. E., Woodward S. Aphidicolin resistance in herpes simplex virus type I reveals features of the DNA polymerase dNTP binding site. Nucleic Acids Res. 1989 Nov 25;17(22):9231–9244. doi: 10.1093/nar/17.22.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Woodward S. Aphidicolin resistance in herpes simplex virus type 1 appears to alter substrate specificity in the DNA polymerase. J Virol. 1989 Jun;63(6):2874–2876. doi: 10.1128/jvi.63.6.2874-2876.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Purifoy D. J., Young D., Gopal R., Cammack N., O'Hare P. Single mutations at many sites within the DNA polymerase locus of herpes simplex viruses can confer hypersensitivity to aphidicolin and resistance to phosphonoacetic acid. J Gen Virol. 1984 Jan;65(Pt 1):1–17. doi: 10.1099/0022-1317-65-1-1. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Moss B. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc Natl Acad Sci U S A. 1970 Jul;66(3):677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf C. W., Weisshart K. The herpes simplex virus DNA polymerase: analysis of the functional domains. Biochim Biophys Acta. 1988 Dec 20;951(2-3):298–314. doi: 10.1016/0167-4781(88)90100-5. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Darby G. Selection and characterisation of acyclovir-resistant herpes simplex virus type 1 mutants inducing altered DNA polymerase activities. Virology. 1985 Oct 30;146(2):262–271. doi: 10.1016/0042-6822(85)90009-1. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Darby G. Susceptibility to other antiherpes drugs of pathogenic variants of herpes simplex virus selected for resistance to acyclovir. Antimicrob Agents Chemother. 1986 May;29(5):894–898. doi: 10.1128/aac.29.5.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987 Jan;6(1):169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. K., Chang C. C., Trosko J. E., Dube D. K., Martin G. M., Loeb L. A. Mammalian mutator mutant with an aphidicolin-resistant DNA polymerase alpha. Proc Natl Acad Sci U S A. 1983 Feb;80(3):797–801. doi: 10.1073/pnas.80.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Kim C. I., Kobayashi H., Kanehiro H., Hirokawa H. Aphidicolin-resistant DNA polymerase of bacteriophage phi 29 APHr71 mutant is hypersensitive to phosphonoacetic acid and butylphenyldeoxyguanosine 5'-triphosphate. Virology. 1990 Sep;178(1):337–339. doi: 10.1016/0042-6822(90)90416-o. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Suzuki S., Yamauchi M., Maeno K., Yoshida S. Characterization of an aphidicolin-resistant mutant of herpes simplex virus type 2 which induces an altered viral DNA polymerase. Virology. 1984 May;135(1):87–96. doi: 10.1016/0042-6822(84)90119-3. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Yoshida S., Tsurumi T., Yamamoto N., Maeno K. Drug-resistant mutants of herpes simplex virus type 2 with a mutator or antimutator phenotype. Microbiol Immunol. 1985;29(4):377–381. doi: 10.1111/j.1348-0421.1985.tb00837.x. [DOI] [PubMed] [Google Scholar]
- Pacha R. F., Condit R. C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985 Nov;56(2):395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrali-Noy G., Spadari S. Mechanism of inhibition of herpes simplex virus and vaccinia virus DNA polymerases by aphidicolin, a highly specific inhibitor of DNA replication in eucaryotes. J Virol. 1980 Nov;36(2):457–464. doi: 10.1128/jvi.36.2.457-464.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel R. E., Anderson M. K., Evans E., Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J Virol. 1990 Feb;64(2):574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Tartaglia J., Paoletti E. Physical mapping and DNA sequence analysis of the rifampicin resistance locus in vaccinia virus. Virology. 1985 Dec;147(2):394–404. doi: 10.1016/0042-6822(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Traktman P., Kelvin M., Pacheco S. Molecular genetic analysis of vaccinia virus DNA polymerase mutants. J Virol. 1989 Feb;63(2):841–846. doi: 10.1128/jvi.63.2.841-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P., Sridhar P., Condit R. C., Roberts B. E. Transcriptional mapping of the DNA polymerase gene of vaccinia virus. J Virol. 1984 Jan;49(1):125–131. doi: 10.1128/jvi.49.1.125-131.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P. The enzymology of poxvirus DNA replication. Curr Top Microbiol Immunol. 1990;163:93–123. doi: 10.1007/978-3-642-75605-4_4. [DOI] [PubMed] [Google Scholar]
- Tsurumi T., Maeno K., Nishiyama Y. A single-base change within the DNA polymerase locus of herpes simplex virus type 2 can confer resistance to aphidicolin. J Virol. 1987 Feb;61(2):388–394. doi: 10.1128/jvi.61.2.388-394.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


