Abstract
Background
The rapid expansion in the availability of genome and DNA sequence information has opened up new possibilities for the development of methods for detecting free-living protozoa in environmental samples. The protozoan Blepharisma japonicum was used to investigate a rapid and simple detection system based on polymerase chain reaction amplification (PCR) from organisms immobilised on FTA paper.
Results
Using primers designed from the α-tubulin genes of Blepharisma, specific and sensitive detection to the equivalent of a single Blepharisma cell could be achieved. Similar detection levels were found using water samples, containing Blepharisma, which were dried onto Whatman FTA paper.
Conclusion
This system has potential as a sensitive convenient detection system for Blepharisma and could be applied to other protozoan organisms.
Background
The detection and identification of protozoan species in environmental samples, such as water or soil, is important in many ecological, environmental, medical or population studies [1-3]. Although many of these protozoa are easily recognised by morphological criteria, microscopy procedures for detection of low concentrations of protozoa can be cumbersome, time consuming and sometimes unreliable. Furthermore, these organisms often exist in other lifecycle stages such as cysts or oocysts and identification can be very problematic.
Very recently, considerable advances have been made in generating DNA sequence data in many protozoan species [4,5] which has produced a legacy of species and genus specific DNA sequence information. Using the Polymerase Chain Reaction (PCR), which can specifically amplify large quantities of DNA from small starting quantities, sensitive identification procedures based on protozoan DNA sequence information can be developed. These type of identification approaches have been developed for several protozoan organisms, for example Cryptosporidium, Cyclospora [6], Giardia [7], Toxoplasma [8] amongst others, although not for Blepharisma japonicum. Sensitivity of detection by these methods is high, with some reported examples of detection down to a single organism [9]. Additionally, convenient methods of collecting protozoa for DNA amplification are also required. FTA paper, developed by Whatman, is a matrix that binds and lyses cells. This results in amplifiable DNA being immobilised on the paper and sections of the paper can be punched out directly for use. This method has been used in some protozoa for example Cryptosporidium [10] and offers a good method for the collection of Blepharisma.
Blepharisma japonicum is easily identified by a bright purple photopigment and is therefore a useful organism for developing and evaluating a molecular detection system. Furthermore, the development of PCR based systems offers the potential for automation and eliminates the need for laborious microscopy by skilled individuals. In this study, we have set out to investigate the sensitivity and specificity of a PCR system for the detection of this free living protozoan in water samples and to evaluate the use of Whatman FTA paper to improve the convenience of this system.
Results
Primer design
The sensitivity of the PCR detection system is dependent on the number of target sequences present in the PCR reaction. This can be enhanced by the selection as a target of a repetitive DNA sequence such as the tubulin gene family. Using the GENBANK nucleotide databases http://www.ncbi.nlm.nih.gov/PubMed/, we obtained sequences for the α-tubulin gene family from Blepharisma japonicum (Accession No. AF003250), the related genus Spirostomum (Accession No. AF003251) [11] and sequences from Paramecium tetraurelia (Accession Nos. X99490 and X99489) and Paramecium caudatum (Accession No. AB035413). Sequences were aligned using the program CLUSTAL W [12] and the alignments were scanned for regions which were unique to Blepharisma. Putative primer sites were identified manually and then checked for hairpin structures using the program PRIMER3 [13]. The sequences corresponding to primers BLEPTUBF and BLEPTUBR were checked for cross matching with other DNA sequences in the databases using the program BLAST [14]. A predicted PCR fragment size of 399 base pairs was deduced from the Blepharisma α-tubulin gene sequence.
Specificity of detection
PCR amplification of DNA extracted from Blepharisma japonicum using primers BLEPTUBF and BLEPTUBR produced a band which co-migrated with the 400 bp marker and was therefore consistent with the predicted fragment size of 399 base pairs (Figure 1). This product was consistently produced when other independent cultures of Blepharisma were also tested. To investigate the specificity of this amplification reaction, we extracted DNA from a further 18 species of protozoa (Table 1) which represented 3 protozoan phyla and also included a genus, Spirostomum, which is believed to be closely related to Blepharisma, [15]. In all cases no amplification of the 399 bp fragment was detected under the conditions used (Figure 1).
Figure 1.
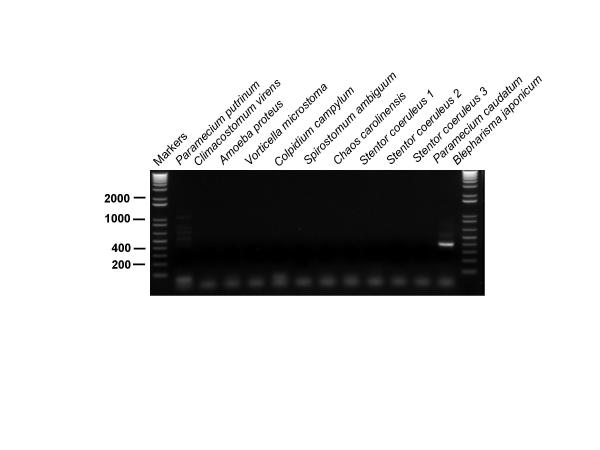
Specifity of detection of Blepharisma japonicum. Specificity of detection of Blepharisma japonicum using gel electrophoresis. Lanes denoted M contain molecular size markers – figures refer to sizes in base pairs. Protozoan species: Paramecium putrinum (lane 1), Climacostomum virens (lane 2), Amoeba proteus (lane 3), Vorticella microstoma (lane 4), Colpidium campylum (lane5), Spirostomum ambiguum (lane 6), Chaos carolinensis (lane 7), Stentor coeruleus (lanes 8 – 10), Paramecium caudatum (lane 11), Blepharisma japonicum (lane 12). (Three different isolates of Stentor – different size morphs – were incorporated).
Table 1.
Protozoan species used in this investigation. The classification indicates the diversity and relationships of species included. Protozoan classification is currently under review: this is taken from [19] and incorporates the latest information on Blepharisma [15].
| Protozoan Species | ||||
| Phylum | Class | Order | Family | Species |
| Ciliophora | Spirotrichea | Heterotrichida | Spirostomidae | Blepharisma japonicum |
| Spirostomum ambiguum | ||||
| Climacostomidae | Climacostomum virens | |||
| Stentoridae | Stentor coeruleus | |||
| Stichotrichida | Oxytrichidae | Stylonychia mytilus | ||
| Colpodea | Colpodida | Colpodidae | Colpoda steinii | |
| Nassophorea | Peniculada | Parameciidae | Paramecium caudatum | |
| Paramecium putrinum | ||||
| Paramecium bursaria | ||||
| Euplotida | Euplotidae | Euplotes patella | ||
| Oligohymenophorea | Hymenostomatida | Turaniellidae | Colpidium campylum | |
| Sessilida | Opisthonectida | Telotrochidium henneguyi | ||
| Vorticellidae | Vorticella microstoma | |||
| Litostomatea | Haptorida | Didiniidae | Didinium nasutum | |
| Lacrymariidae | Lacrymaria olor | |||
| Pharyngophorida | Tracheliidae | Dileptus anser | ||
| Rhizopoda | Lobosea | Amoebida | Amoebidae | Chaos carolinensis |
| Amoeba proteus | ||||
| Euglenozoa | Euglenea | Euglenida | Euglenidae | Euglena gracilis |
Sensitivity of detection
To investigate the sensitivity of this process for the detection of Blepharisma, we carried out serial dilutions of DNA extracted from a known number of Blepharisma which were micromanipulated into an extraction tube and counted manually prior to extraction. The results are depicted in Figure 2 which shows that an amplification product can be clearly seen in a 1:3125 dilution. This was equivalent to 0.015 of a Blepharisma cell. Detection at this level was reproducible over a number of experiments. Further dilution to 1:10000 (equivalent to 0.005 cells) occasionally produced a faint band in some experiments.
Figure 2.
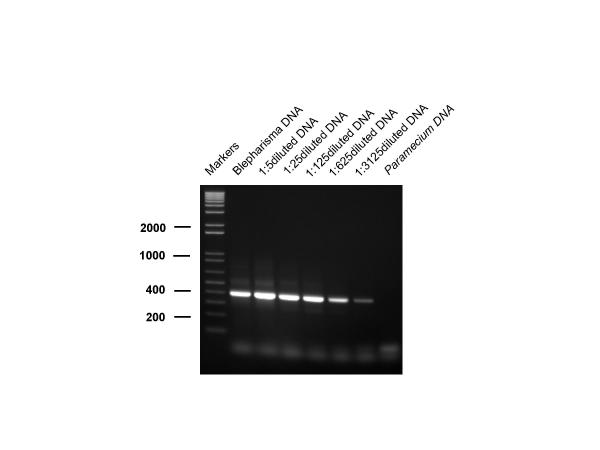
Sensitivity of detection of Blepharisma japonicum. Sensitivity of detection of Blepharisma japonicum by PCR amplification. Lanes denoted M contain molecular size markers – figures refer to sizes in base pairs. Dilutions of target DNA were amplified as follows: undiluted (lane 1), 1/5 (lane 2), 1/25 (lane 3), 1/125 (lane 4), 1/625 (lane 5), 1/3125 (lane 6). Lane 7 contains undiluted Paramecium caudatum DNA as a negative control.
Immobilisation onto Whatman FTA paper
To investigate whether Whatman FTA paper could be used as a useful medium for testing samples for Blepharisma, we micropipetted 5 μl of a Blepharisma culture containing known numbers of cells onto FTA paper and allowed to dry overnight according to the instructions. The number of Blepharisma cells was calculated for each area of FTA paper. The results showed that we could clearly detect an amplification product when the area of FTA paper was equivalent to 1.25 Blepharisma cells but not from an equivalent of 0.625 Blepharisma cells. (Figure 3). Repeated experiments demonstrated that, on average, this technique could be used to detect levels of Blepharisma down to the equivalent of a single cell. Larger samples of the FTA paper consistently inhibited the PCR reaction (eg. Figure 3, lanes 3 and 4). To investigate the robustness of the FTA detection system, we investigated the sensitivity of the system using a larger water sample volume which also contained an excess of other organisms. A 1 ml culture mixture was made which contained Paramecium caudatum, Euglena gracilis, Stentor coeruleus and a known number of Blepharisma japonicum (99 cells/ml). The tube was centrifuged for 10 minutes at 10000 rpm in a microcentrifuge to concentrate the organisms and the pellet was resuspended in 50 μl of water. 5 μl samples were spotted onto the FTA paper. Figure 4 depicts the results of an amplification using known proportions of the FTA paper. Detection was achieved from an area of FTA paper equivalent to 0.62 Blepharisma cells. This showed that, even in the presence of other organisms, this system could detect levels of Blepharisma down to just less than a single cell equivalent. A parallel sample in which the Blepharisma was replaced with an equivalent volume of water showed no amplification (Figure 4 lane 6). Several experiments, identical to those described in Figure 4, were conducted in which water taken from natural sources (River Irwell, Salford; local ponds and a canal near Salford) was used to make up the 1 ml mixed cultures of Blepharisma, Stentor and Paramecium. In all cases the results showed that Blepharisma could be specifically detected and that no inhibition was achieved using real environmental samples.
Figure 3.
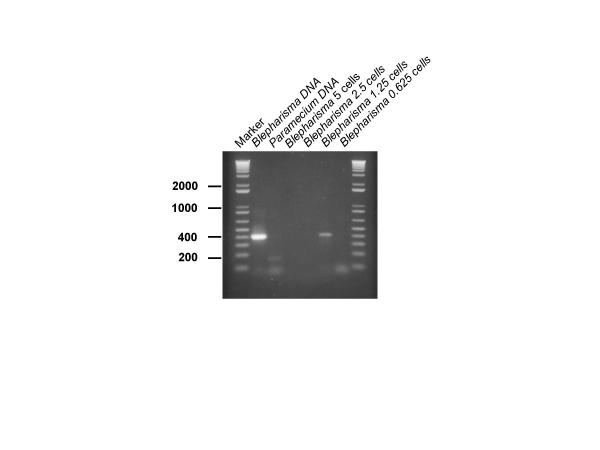
Sensitivity of detection of Blepharisma japonicum on Whatman FTA paper Sensitivity of detection of Blepharisma japonicum which has been immoblised on Whatman FTA paper. Lanes denoted M contain molecular size markers – figures refer to sizes in base pairs. Amplified products from a range of areas of FTA paper were loaded on the gel. These areas contained the following equivalent number of Blepharisma cells: 5 cells (lane 3), 2.5 cells (lane 4), 1.25 cells (lane 5), 0.625 cells (lane 6). Lane 1 contained pure Blepharisma DNA, immobilised on FTA paper, as a positive control for the PCR reaction while Lane 2 contained Paramecium caudatum DNA, on FTA paper, as a negative control. Larger areas of FTA paper (eg lanes 3 an 4) consistently inhibited the PCR reaction.
Figure 4.
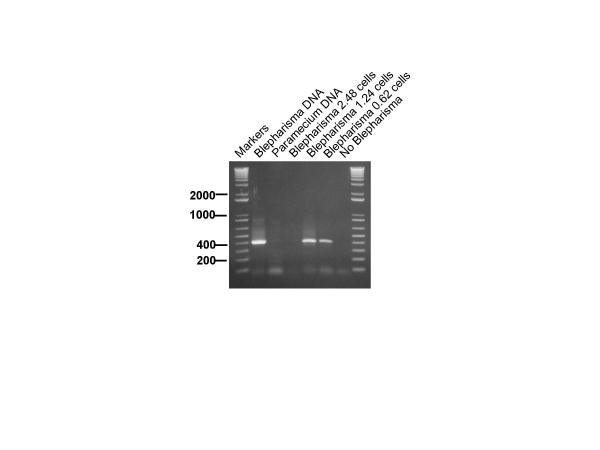
Sensitivity of detection in a complex mixture. Sensitivity of detection of Blepharisma from a complex mixture of protozoa immobilised on FTA paper. Lanes denoted M contain molecular size markers – figures refer to sizes in base pairs. Amplified products from a range of areas of FTA paper were loaded on the gel. These areas contained the following equivalent number of Blepharisma cells: 2.48 cells (lane 3), 1.24 cells (lane 4), 0.62 cells (lane 5). The amplification products run in lane 6 contained the same area of FTA paper as in lane 4 except that they were taken from the mixed culture of protozoa which did not contain Blepharisma. Lane 1 contained pure Blepharisma DNA, immobilised on FTA paper, as a positive control for the PCR reaction while Lane 2 contained Paramecium caudatum DNA, on FTA paper, as a negative control. Larger areas of FTA paper (eg lane 3) consistently inhibited the PCR reaction.
Discussion
In this paper, we have described a method for the detection of Blepharisma japonicum in water and, potentially, other environmental samples. The method is sensitive enough to detect a single organism even when immobilised on FTA paper. Furthermore, the presence of other organisms seems not to decrease the sensitivity of the assay. The sensitivity, as measured by dilution of pure Blepharisma DNA, is some 50 – 100 higher than when using FTA paper. The reasons which could explain the detection of less than a single cell are twofold. Firstly, the α-tubulin target sequence is a highly repeated gene family in many organisms. The copy number in the Blepharisma genome is not known. However, Euglena has only two copies, [16] which may indicate that this may not be the complete explanation. Secondly, ciliates possess a micro- and macronucleus, the latter containing many amplified copies of housekeeping genes such as tubulin. This natural phenomenon of in vivo DNA amplification in ciliates may have useful implications for the design of sensitive molecular detection tools.
Although specifically designed for the detection of Blepharisma, the principles of this study can be applied to the design of molecular methods for many protozoa of environmental importance. The ever increasing amount of primary DNA sequence will extend this possibility to many species. Furthermore the adoption of the Whatman FTA paper method for immobilising samples will considerably simplify collection and storage. Successful PCR amplification can be achieved from samples immobilised on Whatman FTA paper following storage at room temperature for long time periods [17].
Conclusions
Primers BLEPTUBF and BLEPTUBR can be used in a PCR detection system to specifically distinguish Blepharisma japonicum from at least 18 species of closely or distantly related protozoa. Using pure Blepharisma DNA as a substrate, the detection system can detect down to 0.015 cell equivalents. The PCR detection system can detect Blepharisma down to a single cell when using water samples immobilised on FTA paper. There is no loss of sensitivity when the PCR detection system is used to detect Blepharisma in water samples containing a complex mixture of organisms.
Methods
Protozoan species
A range of protozoan species (see Table 1) were supplied by SCIENTO (61 Bury Road, Whitefield, Manchester, UK, M45 6TB). Cultures of Blepharisma japonicum was supplied as pure cultures and checked for the presence of other species by microscopy. Other cultures were supplied as pure cultures except for a few cases where those organisms depended on smaller protozoa as a food source. All cultures were checked for the absence of Blepharisma by microscopy. Cultures were stored at 4°C for up to one week unless specifically instructed otherwise (RM, Sciento).
Extraction of DNA
Protozoan cultures were allowed to settle overnight. A sample of this settled culture (500 μl) was taken and, if desired, counts were made of the number of organisms present. (Fast moving species were slowed by addition of an equal volume of methyl cellulose). Three hundred microlitres of the settled culture was added to 700 μl of lysis buffer (0.01 M Tris pH 7.8; 0.5% SDS; 0.005 M EDTA pH8.0) and 10 μl Proteinase K (20 mg ml-1) and incubated at 56°C overnight. Five hundred microlitres of the lysate was added to an equal volume of phenol-chloroform (50:50 v/v), gently rotated for 10 minutes and centrifuged for 10 minutes at 10000 g in a microcentrifuge. The aqueous (upper) layer was recovered without disturbing the interface. This phenol-chloroform extraction was repeated twice more and the final aqueous phase added to 90 μl 3 M sodium acetate pH 5.2 and 900 μl ice cold 100% ethanol. Following an incubation overnight at -20°C, the sample was centrifuged at 10000 g for 30 minutes at 4°C in a microcentrifuge. The resulting pellet was washed in 500 μl 70% ethanol, centrifuged as described previously, resuspended in 50 μl TE Buffer (10 mM TRIS; 1 mM EDTA pH 8.0) and left overnight at 4°C to ensure complete resuspension.
PCR amplification
Blepharisma specific PCR amplification was achieved using primers BLEPTUBF (5' CTT GAA CAG ACT TAT CGC C 3') and BLEPTUBR (5' GGT TGG TAG TTG ATT CCA CAT 3'). Reactions were carried out in a volume of 25 μl containing 1 μl DNA (or 1 μl water and FTA paper), 2.5 μl 10x PCR buffer (100 mM TRIS-HCL pH 9.0; 15 mM MgCl2; 0.5 M KCl; 1% Triton X100; 0.1%w/v Bovine Serum Albumen), 0.25 μl dNTPs (100 mM), 0.5 μl BLEPTUBF (2 μM), 0.5 μl BLEPTUBR (2 μM), 0.25 μl (1.25 units) SuperTaq polymerase (HT Biotechnology, UK) and 20 μl sterile water. Amplifications were carried out in a Stratagene (UK) Robocycler using the following cycling conditions: 94°C for 5 minutes for a single cycle to ensure complete denaturation, 94°C for 1 minute, 51°C for 1 minute, 72°C for 1 minute (repeated for 35 cycles) and 72°C for 10 minutes to ensure complete extension. Amplification products were separated by gel electrophoresis using 1.2% agarose gels, stained in Ethidium Bromide and visualised using an Alphainnotech 1220 imaging system as described elsewhere [8,18].
Immobilisation on Whatman FTA paper
Five microlitres, taken from a culture which had been counted, were spotted onto Whatman FTA paper, circled with pencil to outline the sample, dried overnight and stored, dry, at room temperature until used. To prepare samples for PCR reactions, a defined area of paper was cut, with a 2 mm diameter hole punch (Whatman Bioscience), from the sample areas and treated as follows. The segment of FTA paper was placed in the PCR tube and 200 μl of FTA Purification Reagent (Whatman Bioscence) was added and incubated for 5 minutes at room temperature. The purification reagent was then removed without removing the FTA paper. Two further identical washes in purification reagent were carried out. After the final treatment, the FTA reagent was removed and the FTA paper was washed twice in TE (10 mM TRIS, 1 mM EDTA, pH 8.0) for five minutes per wash. Following the final TE wash, the TE was removed and the FTA paper allowed to dry for at least 1 hour. For PCR, the PCR reagents were added directly to the FTA paper and the amplifications carried out as described above.
Authors' contributions
GH conceived, designed and co-ordinated the study, participated the sequence alignments and primer design and drafted the manuscript. JH carried out the molecular genetic studies and participated in the sequence alignment and primer design. RM collected, prepared and curated all protozoa species cultures. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank the University of Salford and the Leverhulme Trust for funding this research and to Maria Giovetty, Jonathan Butler, Helen Murray and Will Brown for their contributions to this work.
Contributor Information
Geoff Hide, Email: g.hide@salford.ac.uk.
Jacqueline M Hughes, Email: j.m.hughes@salford.ac.uk.
Robert McNuff, Email: info@sciento.co.uk.
References
- Lunapabello VM, Aladrolubel MA, Durandebazua C. Biomonitoring of wastewaters in treatment plants using ciliates. J Industrial Microbiol. 1996;17:62–68. [Google Scholar]
- Ricci N. Protozoa as tools in the pollution assessment. Marine Pollution Bull. 1991;22:265–268. doi: 10.1016/0025-326X(91)90802-Y. [DOI] [Google Scholar]
- Monis PT, Saint CP. Development of a nested PCR assay for the detection of Cryptosporidium parvum in finished water. Water Research. 2001;35:1641–1648. doi: 10.1016/S0043-1354(00)00426-7. [DOI] [PubMed] [Google Scholar]
- Nanney DL, Park C, Preparata R, Simon EM. Comparison of sequence differences in a variable 23S rRNA domain among sets of cryptic species of ciliated protozoa. J Eukaryo Microbiol. 1998;45:91–100. doi: 10.1111/j.1550-7408.1998.tb05075.x. [DOI] [PubMed] [Google Scholar]
- Bernhard D, Schlegel M. Evolution of histone H4 and H3 genes in different ciliate lineages. J Molec Evol. 1998;46:344–354. doi: 10.1007/pl00006311. [DOI] [PubMed] [Google Scholar]
- Quintero-Betancourt W, Peele ER, Rose JB. Cryptosporidium parvum and Cyclospora cayetanensis: a review of laboratory methods for detection of these waterborne parasites. J Microbiol Methods. 2002;49:209–24. doi: 10.1016/S0167-7012(02)00007-6. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Debnath A, Sil A, De S, Chattopadhyay DJ, Das P. PCR detection of Giardia lamblia in stool: targeting intergenic spacer region of multicopy rRNA gene. Molec Cell Probes. 2000;14:181–9. doi: 10.1006/mcpr.2000.0302. [DOI] [PubMed] [Google Scholar]
- Duncanson P, Terry RS, Smith JE, Hide G. High levels of congenital transmission of Toxoplasma gondii in a commercial sheep flock. Int J Parasitol. 2001;31:1699–1703. doi: 10.1016/S0020-7519(01)00282-X. [DOI] [PubMed] [Google Scholar]
- MacLeod A, Turner CMR, Tait A. Detection of single copy gene sequences from single trypanosomes. Molec Biochem Parasitol. 1997;84:267–270. doi: 10.1016/S0166-6851(96)02810-1. [DOI] [PubMed] [Google Scholar]
- Orlandi PA, Lampel KA. Extraction-free filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J Clin Microbiol. 2000;38:2271–7. doi: 10.1128/jcm.38.6.2271-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourancheau AB, Villalobo E, Tsao N, Torres A, Pearlman RE. Protein coding gene trees in ciliates: comparison with rRNA-based phylogenies. Molec Phylogenetics Evol. 1998;10:299–309. doi: 10.1006/mpev.1998.0534. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Method Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Molec Biol. 1990;215:403–10. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Aescht E, Foissner W. Divisional morphogenesis in Blepharisma americanum, B. undulans and B. hyalinium Ciliophora: Heterotrichida. Acta Protozool. 1998;37:71–92. [Google Scholar]
- Canaday J, Tessier LH, Imbault P, Paulus F. Analysis of Euglena gracilis alpha- beta- and gamma-tubulin genes: introns and pre-mRNA maturation. Mol Genet Genomics. 2001;265:153–160. doi: 10.1007/s004380000403. [DOI] [PubMed] [Google Scholar]
- Rogers C, Burgoyne L. Bacterial typing: storing and processing of stabilised reference bacteria for polymerase chain reaction without preparing DNA – an example of an automatable procedure. Anal Biochem. 1997;247:223–227. doi: 10.1006/abio.1997.2031. [DOI] [PubMed] [Google Scholar]
- Terry RS, Smith JE, Duncanson P, Hide G. MGE-PCR: a novel approach to the analysis of Toxoplasma gondii strain differentiation using mobile genetic elements. Int J Parasitol. 2001;31:155–161. doi: 10.1016/S0020-7519(00)00158-2. [DOI] [PubMed] [Google Scholar]
- Corliss JO. Classification of protozoa and protists: the current status. In: Coombs GH, Vickerman K, Sleigh MA, Warren A, editor. In Evolutionary Relationships Among Protozoa. London: Kluwer Academic Publishers; 1998. pp. 409–447. [Google Scholar]


