Abstract
Background
In the last years several high-throughput cDNA sequencing projects have been funded worldwide with the aim of identifying and characterizing the structure of complete novel human transcripts. However some of these cDNAs are error prone due to frameshifts and stop codon errors caused by low sequence quality, or to cloning of truncated inserts, among other reasons. Therefore, accurate CDS prediction from these sequences first require the identification of potentially problematic cDNAs in order to speed up the posterior annotation process.
Results
cDNA2Genome is an application for the automatic high-throughput mapping and characterization of cDNAs. It utilizes current annotation data and the most up to date databases, especially in the case of ESTs and mRNAs in conjunction with a vast number of approaches to gene prediction in order to perform a comprehensive assessment of the cDNA exon-intron structure. The final result of cDNA2Genome is an XML file containing all relevant information obtained in the process. This XML output can easily be used for further analysis such us program pipelines, or the integration of results into databases. The web interface to cDNA2Genome also presents this data in HTML, where the annotation is additionally shown in a graphical form. cDNA2Genome has been implemented under the W3H task framework which allows the combination of bioinformatics tools in tailor-made analysis task flows as well as the sequential or parallel computation of many sequences for large-scale analysis.
Conclusions
cDNA2Genome represents a new versatile and easily extensible approach to the automated mapping and annotation of human cDNAs. The underlying approach allows sequential or parallel computation of sequences for high-throughput analysis of cDNAs.
Background
Since the completion of numerous whole-genome sequencing projects involving eukaryotic organisms such as C. elegans, D. melanogaster or A. thaliana, culminating more recently in the sequencing of several vertebrate genomes including mouse, rat, zebrafish and, of course, human [1,2] – the primary focus of research efforts has shifted to the systematic identification and characterization of structure, function and regulation of all genes and proteins encoded within these genomes [3,4].
The rate at which further eukaryotic genomes are currently being sequenced in projects spanning the globe reflects the effectiveness of both high-throughput sequencing and shotgun assembly algorithms, but is clearly outpacing the identification of genes and deciphering of gene structures. As the number of genes identified in one sequenced genome after another turn out to be lower than expected [1], it seems clear that knowledge of the genome sequence alone is not sufficient for determining the patterns of coding and non-coding regions genomes are comprised of and certainly does not resolve the role individual genes play in complex biological systems. In this context the detection of all coding regions in a genome and their transcript expression variation gains importance as a way to systematically identify and characterize gene structure, function and regulation on these genomes [4] that will serve as the basis for refined gene models and improved coding sequence annotation.
The use of full complementary DNA (cDNA) sequences, containing the complete and uninterrupted protein coding region of genes, has proven to be very effective for this purpose [5]. Thus, several high-throughtput cDNA sequencing projects have been funded worldwide with the aim of identifying and characterizing complete sequences of novel human transcripts at the cDNA level and providing a unique perspective of a genome's coding potential.
The large amount of cDNA data produced by these projects requires the development of automated tools capable of filling the gap between data collection and its annotation as well as interpretation. A required step for the large scale of coding sequences (CDS) prediction and annotation in a genome is the processing and selection of full length cDNAs from all the high-througtput-cDNAs cloned. Most of these high-throughput-cDNAs are high quality sequences; however, some of them have sequence problems, such as frameshifts and stop codon errors caused by low sequence quality, and other cDNA clones are produced from incompletely processed transcripts or have truncated inserts caused by cloning errors. This step is a time consuming task where the manual curator maps and characterises single cDNAs in order to validate them.
In collaboration with the group of Stefan Wiemann, member of the German cDNA Sequencing Consortium at the German Cancer Research Center (DKFZ), we have designed an application for automatic high-throughput mapping and characterization of cDNAs. cDNA2Genome first determines the location of the input cDNA in the human genome, avoiding ambiguous mapping, followed by an exhaustive gene structure analysis. Additionally, cDNA2Genome extracts the most recent annotation information (e.g. CDS, proteins) available in frequently updated public databases and merges it with precomputed data from the NCBI pipeline [6]. The results from individual analysis programs are then also merged and processed into a compound report.
cDNA2Genome has been implemented under the W3H task system [7]. This framework allows the combination of heterogeneous bioinformatics applications to create complex analysis task flows for high-throughput pipelining and the immediate integration of cDNA2Genome into the W2H web interface [8].
Implementation
Implementation under the W3H-Task-System
cDNA2Genome has been implemented under the W3H task system [7] which was designed to interact with the web interface W2H [8] – a free, popular web interface for sequence analysis tools.
The W3H framework reduces the amount of necessary programming skills for a task author significantlly and contains a concept of re-usability for the written code. It allows the integration of heterogeneous applications to create tailor-made analysis task flows. By specifying dependency rules between the used applications, tasks of high complexity can be designed.
However, the most important aspect of the tasks designed using this framework is that they seamlessly integrate into the web interface W2H. Due to the meta-data concept used in W2H and in the W3H task framework, knowledge of web programming is not required for bringing custom tasks onto the web.
The W3H task framework and cDNA2Genome itself are built using Object Oriented Perl programming. At the DKFZ the W3H task framework is currently used within the HUSAR environment (Heidelberg Unix Sequence Analysis Resources).
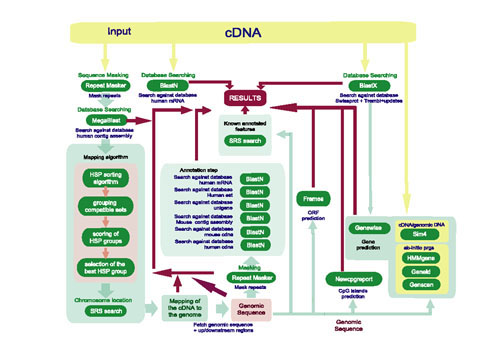
For the implementation of cDNA2Genome under the task framework it was necessary to describe the applications dependencies, the data flow (Fig. 1) and the merging of the individual outputs into a common output report. The system stores both, the results of the different applications together with newly computed results. The final output of the task is an XML file which contains all relevant information obtained. For the web user this XML output can be transformed by means of W2H's post-processing mechanism into an HTML page using XSLT (Extensible Stylesheet Language Transformations) http://www.w3.org/TR/xslt.
Figure 1.
Data flow and dependencies of applications in cDNA2Genome. programs used by cDNA2genome during the annotation process.
Raw sequence pre-processing
Depending on the analysis to be performed the cDNA sequence and the genomic sequence are masked for known repeats and low complexity sequences using RepeatMasker http://repeatmasker.genome.washington.edu/.
Databases
For querying pre-computed data available in public databases containing information about the location of human genomic contigs on specific chromosomes, and also about known annotated features in the human genome we have implemented several regularly updated databases under SRS (Sequence Retrieval System) [9]http://genius.embnet.dkfz-heidelberg.de/menu/srs/. The public databases used in cDNA2Genome are specified in Table 1.
Table 1.
Databases used in cDNA2genome
| Database type | Database |
| Genomic | Human genomic sequence contig assembly database ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens |
| Mouse genomic sequence contig assembly database ftp://ftp.ncbi.nlm.nih.gov/genomes/M_musculus | |
| Proteins | Swissprot + Trembl + updates ftp://ftp.expasy.org/databases/sp_tr_nrdb/ftp://ftp.ebi.ac.uk/pub/databases/swissprot |
| mRNA | Human mRNA database ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/RNA |
| Sequences representing a unique gene | Unigene ftp://ftp.ncbi.nih.gov/repository/UniGene/ |
| Curated sequences and descriptive information about genetic loci | LocusLink http://www.ncbi.nlm.nih.gov/LocusLink |
| Expressed Sequence Tags (ESTs) | ESTs EMBL+ ESTs Genbank +updates ftp://ftp.ebi.ac.uk/pub/databases/emblftp://ftp.ncbi.nih.gov/genbank/ |
Analysis Tools
The analysis programs used by cDNA2Genome can be divided into three main categories: database homology searches, gene finders, and sequence feature predictors (i.e., start/stop codons, open reading frames (ORFs)).
Homology searches
The homology searches are performed using the gapped BLAST (Basic Local Alignment Search Tool) [10] algorithm. For locating cDNAs in the human genome we use MegaBLAST [11]. This program is optimized for aligning sequences that differ slightly as a result of sequencing errors and handles longer DNA sequences much more efficiently than the traditional BLAST algorithm when a sufficiently large word size is used.
Gene prediction
Ab-initio prediction programs
These programs use the statistical information contained within the genomic sequence to predict gene structures. We currently use GenScan [12], HMMgene [13], and Geneid [14]. GenScan determines the gene structure under a probabilistic model specific for a given organism. HMMgene is based on Hidden Markov Models and can predict several whole or partial genes in one sequence. Geneid predicts genes in genomic eukaryotic DNA sequences scoring them using so called Position Weight Matrices.
Gene prediction based on genomic DNA-cDNA alignments
We use SIM4 [15], a local similarity program for aligning cDNA to genomic DNA. SIM4 efficiently aligns a transcribed and spliced DNA sequence (mRNA, EST) with a genomic sequence containing the corresponding gene, allowing for introns in the genomic sequence and a relatively small number of sequencing errors. This method differs from ab-initio methods in the knowledge that SIM4 contains about the gene structure, such as a model for the detection of consensus splice signals.
Gene prediction by similarity-based approaches
These are based on an alignment of a protein or cDNA sequence to the genomic sequence using the BLAST algorithm. Both the input cDNA and the genomic region it was mapped to are queried against the protein databases indicated in Table 1.
Gene prediction by combining ab-inito prediction with homology
The use of prediction programs in conjunction with other data sources can provide valuable annotation data. For that reason we use the program Genewise [16], whose particular strength is the comparison of DNA sequences at the level of their protein translation. This comparison allows the simultaneous prediction of gene structure with homology based alignment. The algorithm does not attempt to predict an entire gene. It tries to predict regions which are justified only by the protein homology. In cDNA2Genome the genomic sequence is compared to the protein most probably coded by the input cDNA.
Sequence feature predictors
The open reading frames of the cDNA sequence are predicted for the three translation frames. The minimum length of the ORF can be defined by the user (default 50 aa). Additionally, CpG rich areas are reported. By default we define CpG islands as regions longer that 200 bases when, over an average of 10 windows, the calculated GC composition is over 50% and the calculated Observed/Expected ratio is over 0.6.
Performance
cDNA2genome runs on a SUN Enterprise with six processors. The performance depends on the length of both the input cDNA and its corresponding genomic region, on the abundance of repeated elements, the parameters selected and on the overall machine load. A cDNA with a length of 585 bp (ENST00000269816 from ENSEMBL gene) mapped to a region of 3945 bp in the human chromosome 9 takes 3 min 05 seconds to run. In the case of a cDNA that codes for the Homo sapiens 5-hydroxytryptamine (serotonin) receptor 2C (HTR2C) which has a length of 4400 bp it takes 30 minutes under a normal-high machine load.
Results and Discussion
cDNA2genome is an automated task for the high-throughput mapping and annotation of cDNAs. The data flow and application dependencies can be viewed in Figure 1. The mapping of the input cDNAs to specific chromosomes is done by a MegaBLAST analysis of the cDNA sequence against the complete human genomic sequence. The output produced by the Basic Local Alignment Search Algorithm (BLAST) [10] often contains unspecific hits and/or suggests ambiguous mapping of the sequences being compared.
We have developed a method for filtering out noise and ambiguities producing the best combination of HSPs (high scoring segment pairs). The method is based on a greedy approach, in which all HSPs for each significant MegaBLAST hit are classified in groups of compatible HSPs. In a first step all HSPs located in the same strand than the input cDNA and with a percentage of identity greater than 95% are selected and ordered by position in the input cDNA. The HSPs are assigned to the same compatible group when they have a consecutive order in the cDNA and an overlap of less than 3 bp (this parameter can be modified). Additionally, they must be in successive order in the genomic sequence the cDNA has been mapped to. Overlapping HSPs are not allowed in the genomic sequence. Once the groups of compatible HSPs have been created following these constrains for each MegaBLAST hit, they are scored depending on their cDNA coverage. The group of compatible HSPs with the largest percentage of cDNA coverage is then selected for the calculation of the cDNA physical mapping in the target genomic DNA.
This filtering method allows a quick and accurate mapping of the cDNA to the given genome. Should a cDNA be mapped to more than one position in the genome, both mappings are reported, but further analysis are currently performed using only the very best hit.
In addition to the genomic sequence where the cDNA has been localized, both the immediate upstream and downstream regions are analyzed in order to identify possible promotors and CpG islands. Such regions are resistant to methylation and tend to be associated with genes which are expressed frequently. Finding a CpG island upstream of predicted exons or genes is a significant additional evidence for the validity of the prediction. The length of these regions can be specified by the user. The default values are 2500 bp upstream and 1000 bp downstream.
cDNA2Genome then uses SIM4 to determine the cDNAs exon/intron structure by performing an efficient and accurate alignment between the cDNA and the corresponding genomic sequence, under the assumption that the differences between both are limited to introns in the genomic sequence, and sequencing errors in either sequence.
Most cDNAs are currently produced by high-throughput sequencing projects. Many of these sequences are high-quality full-length cDNAs containing the complete and non-interrupted protein coding region (CDS). However, some of these cDNAs contain frameshifts and stop codons due to sequencing and cloning errors and others are derived from incompletely processed or alternatively spliced transcripts. In order to help annotators to assess the quality of their cDNAs (e.g. full-length, truncated) cDNA2Genome predicts the gene structure in the genomic region the cDNA is mapped to. For this reason, three ab-initio gene prediction programs Genscan, HMMGene and Geneid are used in the pipeline, each with different strengths and predictions due to their underlying algorithms but nevertheless all with high level of accuracy and thereby providing a exon structure prediction that is independent of homology between the cDNA and the corresponding genomic sequence
cDNA2Genome also compares the genomic DNA sequence the cDNA is mapped to, to the most probable protein coded by the input cDNA. This comparison allows the prediction of gene structure with homology based alignments. The most probable protein sequence coded by the cDNA is searched against the protein databases. The selected protein is then compared against the genomic DNA using Genewise. The potential protein sequences are identified by a search for open reading frames in each of the three forward frames of the genomic sequence.
At the same time the genomic sequence containing the mapped cDNA (Fig. 1) is screened for low complexity regions and interspersed repeats using the program RepeatMasker in conjunction with the RepBase Update database from the Genetic Information Research Institute (GIRI). The masked genomic sequence is then used as input for homology searches against the human mRNA, human EST, and Unigene databases, thereby extracting even more information that might play a role in the annotation process. Searches against the latest updates of these databases are performed to analyze the abundance of transcripts, to obtain information on a possible tissue specificity of expression and to identify putative alternative splice forms. These results are filtered using different thresholds depending on the database (Fig. 1) in order to filter out insignificant hits.
We also created a database containing all available information about known mRNAs, annotated ESTs, Single Nucleotide Polymorphism (SNP), Sequence Tagged Sequences (STS), exons, introns and genes already annotated in Locuslink. This database has been implemented using the SRS database system and it is now used to collect all known annotated features in the selected genomic area, once the cDNA is mapped to the genome.
Last but not least, the human genomic sequence the cDNA has been mapped to is compared against the mouse genomic database. Only HSPs with an identity percentage higher than 75 are included in the output, since this value is sufficiently stringent for localizing conserved exons between mouse and man.
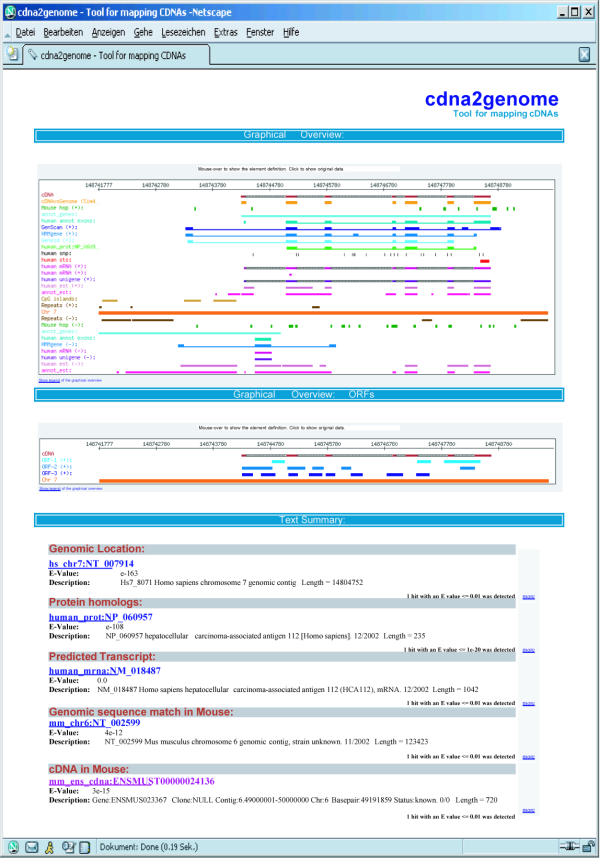
The final result of cDNA2Genome is an Extensible Markup Language (XML) file which contains all relevant information obtained by the task. This XML output can be used in successive analysis pipelines, or integrated in user databases. At the same time, the web user can easily inspect the XML output through a web browser. The result of cDNA2Genome in the web is summarized in two graphical outputs and one text output (Fig. 2). The text output shows a summary including sequence identifier, description, and scores of the most important hits found for each of the homology searches performed. The first graphical output places the cDNA at its corresponding position in a human chromosome and displays in a comprehensible way the results from each of the homology and database searches and from each of the prediction programs. The second graphical output shows open reading frames (ORFs) in each of the three forward frames with different offsets and colours for each frame. The graphical display provides an interactive graphical view of the annotations (Fig. 2) and is hyperlinked.
Figure 2.
Screenshot of the web output of cDNA2genome
The horizontal axis in the graph represents the genomic sequence where the cDNA is mapped to. Forward-strand annotations are displayed above and reverse-strand annotations below the axis. Each type of annotation is displayed in its own row, using its own colour following a specified code. The most important information concerning each feature, such as type, beginning and end can be visualized when moving the mouse over the desired object. The user has immediate access to all complete application outputs (via hyperlinks) by clicking on the corresponding part of the picture. In the case of features such as SNPs, STS and annotated genes it is possible to get the corresponding database information by clicking on their graphical representation. At the bottom of each graph there is a link to the corresponding explanatory legend.
The development of cDNA2genome under the W3H-task system allows this tool to be easily extensible. As new improved algorithms and methodologies are developed, they can be incorporated into the analysis process without redesigning of the whole task. It is also possible to incorporate specific sets of databases and arbitrary configuration parameters at the same time that new species genomes become available. This framework also allows the immediate implementation of a task in the UNIX command line environment. This command line version can be used in batch processes to allow the sequential or parallel computation of many sequences for large-scale data analysis. Web access to cDNA2genome is enabled through the W2H environment where processes can be checked at any time or results can be retrieved. Sequences and results are always stored in a private user space that can not be viewed publicly.
There are a number of bioinformatics approaches that overlap with the cDNA2Genome functionality to some extent. At the moment there are three main sites providing annotation of the human genome. These are the Ensembl site at the European Bioinformatics Institute (EBI)/Sanger Center [17], the NCBI Analysis Pipeline, and the Golden Path browser provided by the University of California Santa Cruz (UCSC) [18]. Each of them uses a different combination of resources to precompute their annotations. cDNA2Genome combines the precomputed annotation data provided by NCBI with data generated on the fly from frequently updated databases in order to fill the gap between data releases. In this way the user obtains the most recent information about the query sequence.
To make the result of the cDNA analysis more transparent we decided to use the visual concept from Golden Path (UCSC) due to the fact, that it is very intuitive and allows a quick visual assessment of the region analysed. We have the opinion that it is more useful for a scientist to see all the predictions lined up (plus the BLAST hits, ESTs, and other supporting features) and then judge whether the prediction of exons or other interesting features are likely to be true or not.
Currently we are extending the tool to other organisms as their genomes become available. In the next release, it will be possible to analyse mouse cDNAs as well. At the same time we are implementing a new gene function annotation method based on Gene Ontology (GO) [19] using a novel algorithm developed in our group (Vinayagam et al., in preparation). To provide a broader overview to the user, we will also integrate the precomputed data from the Ensembl project. Both pipelines, Ensembl and NCBI use different approaches to data analysis that result in different sets of annotated genes. For the web users we are implementing more user interaction allowing a better online direction of cDNA2Genome's analysis. For example, it will be possible to run all the processes for more than one significant mapping of the same cDNA.
cDNA2Genome is being developed in close collaboration with the group of Stefan Wiemann within the German Human Genome Project (DHGP). At the moment cDNA2Genome is used within this project for the high-throughput annotation and exon structure confirmation of full cDNAs (results to be published elsewhere).
Conclusion
cDNA2Genome represents a new versatile and easily extensible approach for the automated mapping and annotation of human cDNAs using several frequently updated databases. The underlying approach allows the sequential or parallel processing of sequences for high-throughput analysis of cDNAs. Additionally, it enhances existing annotation data through the use of the most up-to-date databases currently available. The graphical display enables users to assess the significance of the predictions just with a look into the graph. The use of standardised data formats like XML alleviates the use of cDNA2genome's results in further analysis pipelines or for their integration into databases.
Availability
CDNA2genome is available for academic users at http://genius.embnet.dkfz-heidelberg.de/menu/biounit/open-husar/. Contact: genome@dkfz.de
Author's contribution
C.V. and KHG conceived and designed the project. C.V. coded the program and wrote the manuscript. KHG implemented the databases edited and revised the manuscript and S.S oversaw this study. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank Ingo Schupp, Vladimir Kuryshev, and Stephan Wiemann from the Molecular Genome Analysis division (MGA) at the DKFZ for their valuable suggestions, Peter Ernst for many useful discussions about the design of the program and during the preparation of this manuscript, and Christopher Previti for proof-reading.
Contributor Information
Coral del Val, Email: c.delval@dkfz.de.
Karl-Heinz Glatting, Email: glatting@dkfz.de.
Sandor Suhai, Email: s.suhai@dkfz.de.
References
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Stein L. Genome annotation: from sequence to biology. Nat Rev Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- Sakaki Y, Hattori M, Toyoda A, Watanabe H, Yada T, Taylor T, Park HS, Totoki Y, Fujiyama A. [Determination of DNA sequence of the whole chromosome 21] Tanpakushitsu Kakusan Koso. 2000;45:2520–2527. [PubMed] [Google Scholar]
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Bocher M, Blocker H, Bauersachs S, Blum H, Lauber J, Dusterhoft A, Beyer A, Kohrer K, Strack N, Mewes HW, Ottenwalder B, Obermaier B, Tampe J, Heubner D, Wambutt R, Korn B, Klein M, Poustka A. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–435. doi: 10.1101/gr.GR1547R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuth JP. The NCBI. Publicly available tools and resources on the Web. Methods Mol Biol. 2000;132:301–312. doi: 10.1385/1-59259-192-2:301. [DOI] [PubMed] [Google Scholar]
- Ernst P, Glatting KH, Suhai S. A task framework for the web interface W2H. Bioinformatics. 2003;19:278–282. doi: 10.1093/bioinformatics/19.2.278. [DOI] [PubMed] [Google Scholar]
- Senger M, Flores T, Glatting K, Ernst P, Hotz-Wagenblatt A, Suhai S. W2H: WWW interface to the GCG sequence analysis package. Bioinformatics. 1998;14:452–457. doi: 10.1093/bioinformatics/14.5.452. [DOI] [PubMed] [Google Scholar]
- Etzold T, Ulyanov A, Argos P. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Krogh A. Two methods for improving performance of an HMM and their application for gene finding. Proc Int Conf Intell Syst Mol Biol. 1997;5:179–186. [PubMed] [Google Scholar]
- Guigo R, Knudsen S, Drake N, Smith T. Prediction of gene structure. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Durbin R. Using GeneWise in the Drosophila annotation experiment. Genome Res. 2000;10:547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]