Abstract
Study Objectives:
Individual sleep timing differs and is governed partly by circadian oscillators, which may be assessed by hormonal markers, or by clock gene expression. Clock gene expression oscillates in peripheral tissues, including leukocytes. The study objective was to determine whether the endogenous phase of these rhythms, assessed in the absence of the sleep-wake and light-dark cycle, correlates with habitual sleep-wake timing.
Design:
Observational, cross-sectional.
Setting:
Home environment and Clinical Research Center.
Participants:
24 healthy subjects aged 25.0 ± 3.5 (SD) years.
Measurements:
Actigraphy and sleep diaries were used to characterize sleep timing. Circadian rhythm phase and amplitude of plasma melatonin, cortisol, and BMAL1, PER2, and PER3 expression were assessed during a constant routine.
Results:
Circadian oscillations were more robust for PER3 than for BMAL1 or PER2. Average peak timings were 6:05 for PER3, 8:06 for PER2, 15:06 for BMAL1, 4:20 for melatonin, and 10:49 for cortisol. Individual sleep-wake timing correlated with the phases of melatonin and cortisol. Individual PER3 rhythms correlated significantly with sleep-wake timing and the timing of melatonin and cortisol, but those of PER2 and BMAL1 did not reach significance. The correlation between sleep timing and PER3 expression was stronger in individuals homozygous for the variant of the PER3 polymorphism that is associated with morningness.
Conclusions:
Individual phase differences in PER3 expression during a constant routine correlate with sleep timing during entrainment. PER3 expression in leukocytes represents a useful molecular marker of the circadian processes governing sleep-wake timing.
Citation:
Archer SN; Viola AU; Kyriakopoulou V; von Schantz M; Dijk DJ. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. SLEEP 2008;31(5):608-617.
Keywords: Sleep-wake cycle, constant routine, circadian rhythms, melatonin, cortisol, leukocytes, clock genes
CIRCADIAN RHYTHMS HAVE EVOLVED AS A BIOLOGICAL ADAPTATION TO THE LIGHT-DARK CYCLE AND OTHER ENVIRONMENTAL DAILY CYCLES, WHICH currently have a period of 24 hours.1,2 These rhythms enable organisms to anticipate and to entrain to the changing physical conditions associated with the daily transition between day and night. Similarly, sleep has evolved in animals as a necessary period of restoration for the brain and the body. While they remain independent mechanisms, the circadian and sleep homeostatic systems show high levels of interaction.3 Inter-individual differences prevail in both systems.4–7 Differences in chronotype and sleep-wake timing can be studied by a simple, questionnaire-based assessment of diurnal preference.8 Laboratory-based studies have confirmed that morning or evening preference correlates with differences in the phase and period of circadian markers, such as core body temperature and melatonin rhythms.9,10 It has also been shown that diurnal preference is associated with differences in sleep structure,11,12 and survey data have highlighted large inter-individual differences in sleep duration and sleep timing.13
Almost all cells in the mammalian body possess the ability to generate circadian rhythms, which are synchronized by the central pacemaker in the SCN through mechanisms that have only in part been elucidated. These can include humoral and behavioral mechanisms, such as feeding and sleep-wake activity.14 The core molecular components of the clock are well established, although additional elements continue to be discovered, especially in output pathways where the cellular clock links to biochemical processes under circadian influence. The core mechanism is thought to consist of positive transcription factors promoting the expression of a set of proteins which feed back negatively to inhibit the transcription of their own mRNA. The whole cycle has a period of approximately 24 h. The exact period length is fine-tuned by posttranslational mechanisms such as phosphorylation, which (depending on the target site) can act to shorten or lengthen the period.15 Less is known about the molecular components of the sleep homeostat, although several genes have been linked with sleep regulation, including alleles of the human leukocyte antigen (HLA) complex, the prion protein gene (PRNP), the pre-prohypocretin gene, the gamma amino butyric acid A (GABAA) beta3 gene, the myotonic dystrophy type 1 (DM1) protein kinase gene, catecholo-methyltransferase (COMT), tumor necrosis factor alpha (TNF), adenosine deaminase (ADA), and the adenosine A2A receptor (ADORA2A), among others.16–18 Animal studies have indicated an interaction between the circadian clock and the sleep homeostat through the discovery of altered sleep structure in clock gene knockout mice.19–21 It has also been established that polymorphisms within human clock genes can affect diurnal preference,22–24 sleep timing,25,26 and sleep structure.27 The variable number tandem repeat (VNTR) polymorphism in PER3, where a 54-nucleotide coding region motif is repeated in 4 or 5 units, has been linked with multiple phenotypic parameters. The longer, 5-repeat allele has been associated with morning preference,23,28 and higher sleep propensity and worse cognitive performance in response to sleep deprivation,27 as well as being linked with an increased risk of breast cancer.29
Traditionally, the circadian marker of choice in humans has been the hormone melatonin, because its rhythm is driven by the SCN through a well-defined neural pathway, and because it can be monitored conveniently and reliably in a variety of body fluids.30 Assessment of endogenous circadian phase and period of melatonin correlates well with phenotypes of interest, such as sleep timing and diurnal preference.10 More recently, methods have been developed to specifically access molecular clock components in the periphery (for review, see Lamont et al.).31 Analysis of clock gene expression in cultured fibroblasts could, in theory, provide useful information about individual circadian period without the need for elaborate forced desynchrony protocols, but results of these approaches have been mixed so far.32 It has also been established that peripheral, nucleated blood cells show rhythmic clock gene expression in vivo, and several studies have sought to refine the technique for quantifying clock gene transcripts in a variety of leukocyte preparations, collected from subjects kept under varying conditions.33–40 While these studies have demonstrated the utility of this approach, few have shown a correlation between endogenous peripheral oscillations in clock gene expression and habitual sleep timing or hormonal markers such as melatonin or cortisol. In particular, it is not known whether variation in the phase of endogenous circadian rhythms of clock gene expression in the periphery, correlates with the entrained phase of the habitual sleep-wake cycle, even in the absence of the masking effects of the light-dark and sleep-wake cycle.41 In this study, we have assessed this relationship for the clock genes BMAL1 (ARNTL), PER2, and PER3 under constant routine conditions. We show for the first time that the phase of peripheral clock gene expression in leukocytes correlates with habitual sleep timing and that PER3 expression represents the most robust of the 3 markers of circadian gene expression.
METHODS
Participants
The protocol received a favorable review by the University of Surrey Ethics Committee, and was conducted in accordance with the principles of the Declaration of Helsinki. Participants in the study were screened for health (by history, physical examination, and standard biochemistry and hematology), and did not suffer from sleep disorders or excessive daytime sleepiness. All participants provided written, informed consent after having received a detailed explanation of the study procedures. Participants were also selected according to their genotype for the PER3 VNTR polymorphism as part of a larger study from which the data presented here have been derived (described previously in Viola et al.).27 10 participants were homozygous for the longer repeat allele (PER35/5) and 14 were homozygous for the shorter repeat allele (PER34/4) (see Table 1 for subject characteristics).
Table 1.
Subject Characteristics
| PER3 Polymorphism | PER35/5 | PER34/4 | P |
|---|---|---|---|
| N | 10 | 14 | |
| Age | 25.2 ± 1.1 | 24.8 ± 1.0 | 0.8 |
| BMI | 22.6 ± 0.6 | 21.5 ± 0.4 | 0.2 |
| Sex (M/F) | 6/4 | 8/6 | 0.9 |
| Women in luteal phase | 1 | 1 | |
| Ethnicity (European/Asian/African) | 8/2/0 | 9/3/2 | |
| Habitual bed time | 01:03 ± 1:43 | 01:03 ± 1:17 | 0.9 |
| Habitual wake time | 07:57 ± 1:29 | 08:41 ± 01:19 | 0.2 |
| Sleep duration | 06:55 ± 1:01 | 07:38 ± 0:56 | 0.1 |
Subject characteristics split by PER3 VNTR genotype. Body-mass index (BMI). Mean ± standard deviation. P values refer to comparisons between genotypes.
Field Study
The selected participants took part in a pre-laboratory field study to determine individual sleep-wake schedules. Subjects wore actigraphs (wrist-worn Actiwatch L, Cambridge Neurotechnology, Cambridge, UK) to quantify activity,42 and completed daily sleep diaries for approximately 3 weeks (on average 2.70 ± 0.17) prior to the laboratory study. The data of the first 2 weeks were analyzed to characterize the habitual sleep-wake cycle in each participant. Individual habitual sleep onset and wake times were determined from values calculated during this 2-week period based on the sleep diary data and corroborated by actiwatch data, which both showed very close agreement. Sleep onset was defined as the “lights-out” time plus the individual sleep latency time, which was estimated by the subject and recorded in the sleep diary. This time was also confirmed by inactivity in the actiwatch data. Habitual timing was defined as the median of these values. Participants were then instructed to sleep according to their habitual schedule for one week prior to the laboratory study. Compliance with this schedule was verified by analysis of the sleep diary and actigraphy prior to the start of the 5-day laboratory study. Neither meal times nor content were controlled during the field study.
Laboratory Study
On entering the laboratory phase, participants underwent habituation and baseline sleep episodes, which were timed according to each individual's habitual sleep schedule determined from the field study. Immediately following this, participants were kept awake under an approximate 40-h constant routine (CR) condition, as previously described.27 During the CR protocol, participants stayed in bed in a semi-recumbent position in constant environmental conditions with no information on clock time. Participants received hourly nutritional drinks during the CR as a substitute for main meals. These were calculated for each subject using the Harris-Benedict formula with an activity factor of 1.3.43 The participants were not allowed to leave the bed for the duration of the protocol. Wakefulness was verified from continuous electroencephalogram (EEG) and electrooculogram (EOG) recordings. During the CR period, hourly blood samples were collected from a permanent indwelling cannula to quantify melatonin, cortisol and clock gene mRNA. After the CR, volunteers went to bed in the laboratory at their habitual bedtime for a recovery sleep episode of 12 h, during which time they were free to sleep or get up as desired.
Hormone and Clock Gene Measurements
For the measurement of melatonin and cortisol, hourly blood samples (7 mL) were collected into lithium-heparin tubes. Plasma was immediately separated by centrifugation (10 min, 1,650 × g, 4°C) and stored at −20°C until analysis. Plasma melatonin and cortisol concentrations were measured by radioimmunoassay (Stockgrand Ltd, Guildford, Surrey, UK), as previously described.27
Analysis of clock gene expression followed modifications of previously published methods.37,39 Hourly blood samples (2.5 mL) were collected into PAXgene Blood RNA vacutainer tubes (PreAnalytiX, Hombrechtikon, Switzerland), mixed by inversion, incubated at room temperature for several hours, and then frozen at −20°C until extracted using the PAXgene 96 Blood RNA Kit (QIAGEN, Hilden, Germany). Extracted total RNA was analysed spectrophotometrically (ND-1000, NanoDrop Technologies, Wilmington, DE). Then 100 ng of each sample was subjected to random-primed reverse transcription by M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) in a total volume of 40 μL; 6.25% of the resulting cDNA was used in each real-time PCR reaction and amplified by the ABsolute QPCR ROX mix (ABgene, Epsom, Surrey, UK) in a 96-well plate format in an ABI Prism 7500 instrument (Applied Biosystems, Foster City, CA). Primer and probe sequences are shown in Table 2. All probes were labelled with FAM at the 5'-end and Black Hole Quencher 1 at the 3'-end. Standard curve controls were dilutions of human genomic DNA (for GAPDH) and a sequence-verified cDNA clone (for PER2, PER3, and BMAL1), respectively. The clock gene primers and probes were designed to amplify only cDNA. Negative controls included reverse-transcription reactions where RNA had been omitted, as well as blanks to which no cDNA had been added. Amplification curves were analyzed using the Prism 7500 software with log CT values being normalized against the standard curves to produce relative copy number. Clock gene expression data were normalized against levels of the housekeeping transcript GAPDH.
Table 2.
Primer and Probe Sequences (5' – 3') for Q-PCR Templates
| Gene | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| PER2 | CTGCCTTCTGCTGGCAGAGA | CCGCCCTTTCATCCACATC | TGGTTATGAAGCCCCTAGAATTCCT |
| PER3 | GGTCGGGCATAAGCCAATG | GTGTTTAAATTCTTCCGAGGTCAAA | CCCAGAGACAGCCAGGGATGCTACC |
| BMAL1 | AAACCAACTTTTCTATCAGACGATGAA | TCGGTCACATCCTACGACAAAC | ACCTCATTCTCAGGGCAGCAGATGG |
| GAPDH | caaggtcatccatgacaactttg | gggccatccacagtcttctg | accacagtccatgccatcactgcca |
Data Analysis
The analysis of the circadian markers aimed to assess the presence of circadian rhythmicity, and to quantify the amplitude and the phase of these rhythms. For the purpose of the present paper, an individual time series was considered to be rhythmic if the 95% confidence interval of the estimated amplitude of a sine wave with a circadian period of 24.2 h did not include 0. Using a nonlinear regression procedure (PROC NLIN, SAS 9.1), the function fitted to the data was:
Value(sample ti) = Mesor + Amplitude*sin((sample ti-phase)/24.2), in which the mesor, amplitude and phase were free parameters, ti represents the clock time i at which a sample was collected, and value represents the concentration or copy number of the circadian marker variable. These analyses were conducted on the raw data (clock gene relative copy number, nmol/L of cortisol, and pg/mL of melatonin), clock gene relative copy number normalized with respect to GAPDH, and z-scored data, which for the hormones was the z-scored raw data, and for the clock genes was the z-scored normalized data. Here, analyses conducted on the z-scored data are presented throughout, unless specifically stated. The analysis of the z-scored data was used to allow for a comparison of amplitudes across variables that are measured in different units. This approach first equates the total variance in each variable in each individual, and then determines how much of this variance can be attributed to a circadian signal.
The start of the recovery sleep episode after the CR was timed to occur 48 h after the habitual sleep onset time for each individual. Thus, because of differences in habitual sleep and wake timing between subjects, the individual CRs varied in duration from 37.8 to 42.5 h. Because of the individual habitual scheduling, not all individuals contributed data during the beginning and the end of the 42.5 h CR session. Data were included for analysis from a window within the CR when at least 80% of the individuals contributed data. The longest period within the CR session when hourly clock gene data were available from at least 80% of the participants was 32 h, and data analysis has been restricted to this period. Within this period, the mean number of subjects contributing data at each sampling hour was 95%. The exclusion of the initial and final hours of the CR data also helps to avoid potential masking effects on physiological variables of sleep and change in posture that may last for several hours after wake time.41
The phase of the melatonin rhythm was defined either on the basis of the onset of the nocturnal secretion, i.e. the time when melatonin concentration first exceeds the mean value, or the timing of the fitted maximum. The phase of the cortisol rhythm was defined as the fitted maximum. For both melatonin and cortisol, simple sinusoid functions were fitted to the individual time series. The phase of the clock gene rhythms was defined as the timing of the fitted maximum.
Statistical Analysis
All statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC). PROC MIXED was used to compare amplitudes between variables. P values were based on Kenward-Roger's corrected degrees of freedom.44 Comparisons of the variances (SD) of the peak timing of the clock gene rhythms were calculated using the F test. Correlations were computed with PROC CORR. Two dependent correlations measured on the same subjects (e.g. the correlation between habitual sleep timing and the rhythm of melatonin vs. the correlation between habitual sleep timing and the rhythm of BMAL1), were compared using the Fisher Z transformation, after correcting for the covariance between these correlations because one variable contributes to both correlations. Correlations obtained in 2 different groups of subjects (PER34/4 vs. PER35/5) were also compared, and for this we used the Fisher-Z transformation without correction.
RESULTS
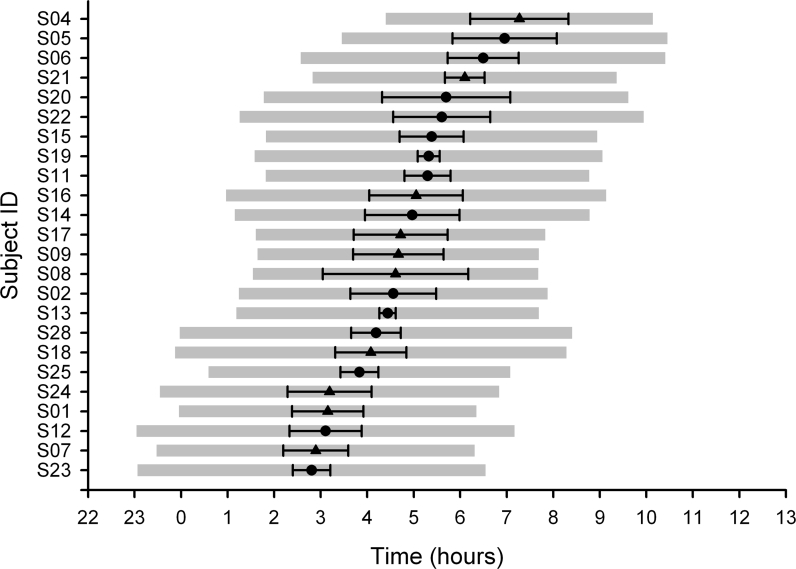
Analyses of the actigraphy and sleep diary data showed that individuals varied widely in their habitual sleep timing (Figure 1). Habitual sleep timing ranged from 22:45 to 04:40 (onset), and 06:00 to 10:40 (offset). The mean (± SD) sleep onsets, mid sleep, wake times, and sleep duration were 01:03 ± 01:27, 04:43 ± 01:20, 08:23 ± 01:25 and 07:20 ± 01:44 when the entire sample was considered. There was no significant difference between these parameters when they were analyzed according to PER3 VNTR genotype (Table 1).
Figure 1.
Habitual sleep timing. Mean sleep onset, mean sleep mid-point (± SD), and mean wake times are shown for each individual (gray bars), ordered by sleep mid-point. • = PER34/4 subjects, ▴ = PER35/5 subjects.
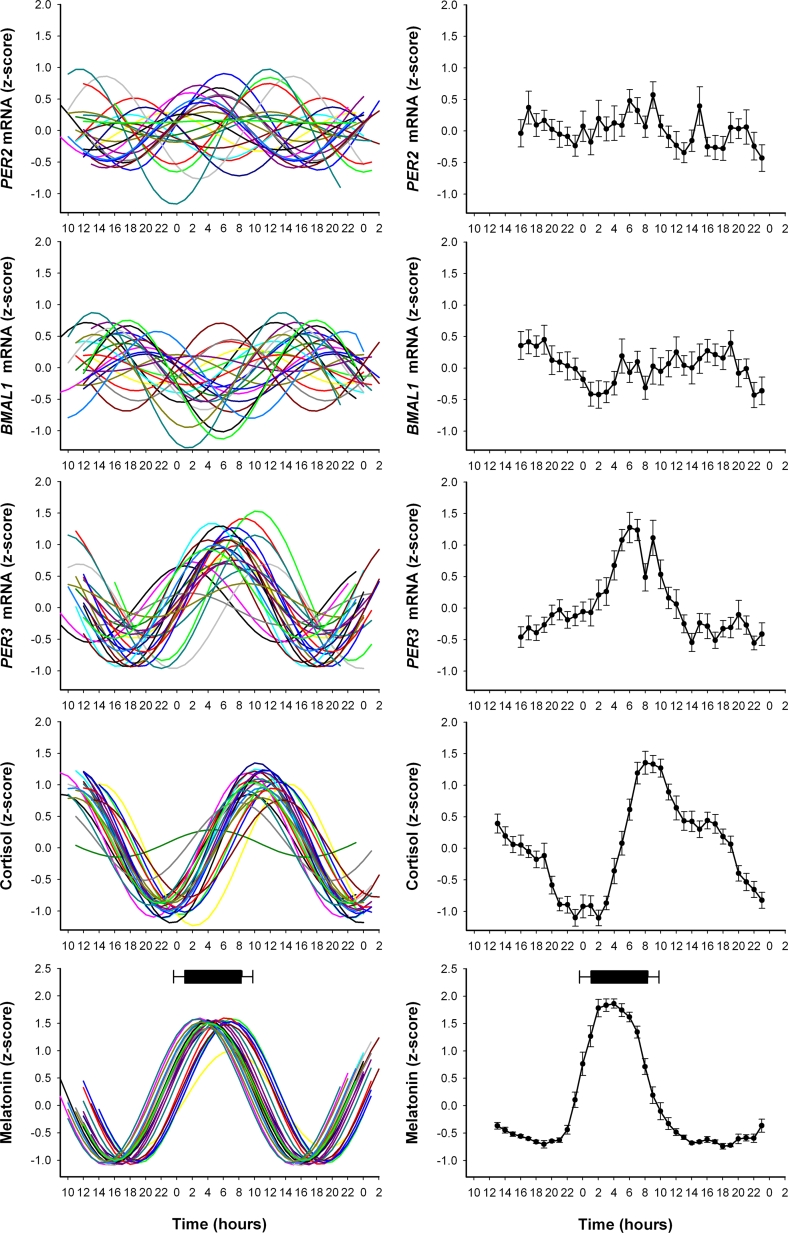
To investigate the inter-individual variation in the amplitude and phase of the five circadian markers, we plotted the curves fitted to z-scored data for each individual (Figure 2, left panel). The average profiles of the z-score values were also computed (Figure 2, right panel). As shown in Figure 2, melatonin had the least variance in amplitude and phase timing. Out of the 3 clock genes that were analyzed, PER3 had the more robust rhythm (individual data for relative copy number normalized with respect to GAPDH for each clock gene are shown in Supplementary Figure 1). To further quantify the extent of circadian rhythmicity, we determined for each circadian marker the number of individuals in whom a significant circadian amplitude was detected.
Figure 2.
Individual and average oscillations in circadian markers. Left Panel: z-score normalized rhythms from individual subjects for PER2, BMAL1, PER3, melatonin and cortisol plotted relative to clock time (data for clock genes are the relative copy number/GAPDH ratio). Right Panel: Average z-score curves for each marker for all individuals (± SD). Mean sleep onset and wake times are depicted as a bar above the melatonin profile (± SD).
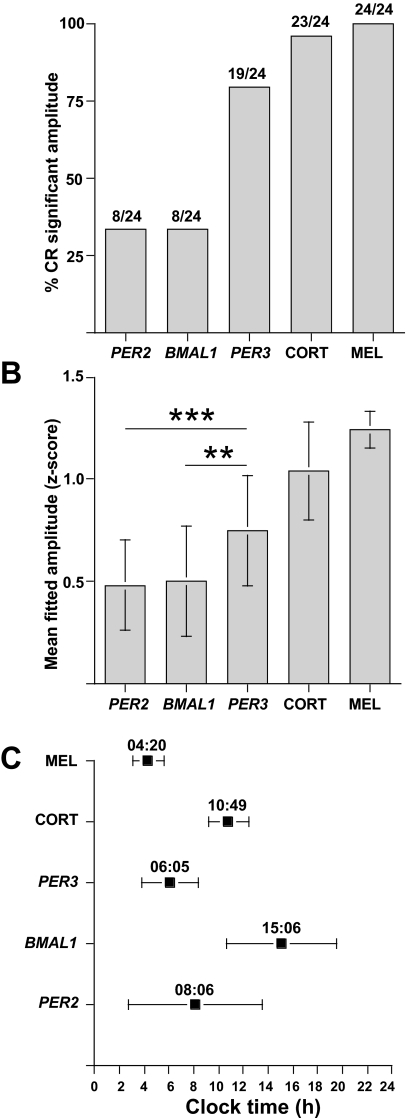
The amplitude of rhythms differed significantly from zero in all 24 individuals for melatonin (14 PER34/4, 10 PER35/5), 23 for cortisol (13 PER34/4, 10 PER35/5), 19 for PER3 (11 PER34/4, 8 PER35/5), 8 for BMAL1 (4 PER34/4, 4 PER35/5), and 8 out of 24 individuals for PER2 (4 PER34/4, 4 PER35/5) (Figure 3A). The mean z-scored amplitudes (± SD) when all 24 individuals were analyzed were 0.48 ± 0.22 for PER2, 0.50 ± 0.27 for BMAL1, 0.75 ± 0.27 for PER3, 1.04 ± 0.24 for cortisol, and 1.24 ± 0.09 for melatonin (Figure 3B). The mean fitted z-scored amplitude of the PER3 rhythm was significantly greater than the PER2 (P = 0.0004) and BMAL1 (P = 0.0027) rhythms. This was also the case when the non z-scored data for the copy number/GAPDH ratio and the relative mRNA copy number were compared (Table 3). When only significant amplitudes were included in the same analysis, the mean z-scored amplitudes were 0.72 ± 0.13 for PER2, 0.82 ± 0.16 for BMAL1, 0.85 ± 0.17 for PER3, and 1.08 ± 0.16 for cortisol (melatonin remained unchanged). With this analysis, the mean amplitude of PER3 was significantly greater than PER2 (P = 0.0004) and BMAL1 (P = 0.003).
Figure 3.
Characterization of circadian marker rhythms. (A) Percentage of individuals in whom, under constant routine conditions, a significant amplitude could be detected for each circadian marker. The actual number of individuals is shown above each bar. (B) Mean fitted z-score amplitudes (± SD). (C) Mean fitted z-score phase maxima (± SD). **P = 0.0027, ***P = 0.0004. CORT = Cortisol, MEL = Melatonin
Table 3.
Characterization of Circadian Rhythm Parameters
| A | Number of subjects with Significant amplitudes | Mean fitted amplitude | Mean fitted maxima H:min ± SD |
|---|---|---|---|
| PER2 | 11/24 (46%) | 388.1 ± 700.8 | 09:30 ± 05:39 |
| BMAL1 | 8/24 (33%) | 751.6 ± 737.8 | 14:02 ± 04:31 |
| PER3 | 20/24 (83%) | 1914.9 ± 2396.6 | 05:32 ± 02:47 |
| Cortisol | 23/24 (96%) | 111.6 ± 44.4 | 10:49 ± 01:37 |
| Melatonin | 24/24 (100%) | 31.6 ± 15.6 | 04:20 ± 01:18 |
| B | |||
| PER2 | 8/24 (33%) | 0.01 ± 0.01 | 08:06 ± 05:23 |
| BMAL1 | 8/24 (33%) | 0.03 ± 0.03 | 15:06 ± 04:28 |
| PER3 | 19/24 (79%) | 0.08 ± 0.13 | 06:05 ± 02:14 |
Percentage of subjects with significant amplitudes for circadian markers and mean fitted amplitudes (± SD) and mean fitted maxima (time ± SD) for (A) clock gene (expressed as relative copy number), cortisol (nmol/L) and melatonin rhythms (pg/mL), and (B) clock gene rhythms expressed as relative copy number normalized to GAPDH. PER3 amplitude is significantly greater than BMAL1 and PER2 amplitudes when expressed as relative copy number (P = 0.031 and 0.006, respectively) and when normalized against GAPDH (P = 0.05 and 0.01, respectively).
The timing of the mean fitted z-scored maxima (peak times), as well as the inter-individual variation, differed between the various markers. Mean peak times (± SD) were computed from only those individuals contributing significant amplitudes. They were 8:06 ± 5:23 for PER2, 15:06 ± 4:28 for BMAL1, 6:05 ± 2:14 for PER3, 10:49 ± 1:37 for cortisol, and 4:20 ± 1:18 for melatonin. Thus, the timing of the PER3 rhythm showed the least variance of the 3 clock genes (Figure 3C) and was significantly lower than the variance of PER2 (P = 0.003) and BMAL1 (P = 0.017). There was no significant difference between the peak times of PER3 and PER2 (P = 0.33), but the peak times of PER3 and PER2 were significantly different from that of BMAL1 (P = 0.0005 and P = 0.014, respectively). When z-scored data from all individuals were analyzed, the peak times were not substantially different (PER2 = 9:07 ± 6:34, BMAL1 = 14:27 ± 4:13, PER3 = 5:54 ± 2:48, Cortisol = 10:28 ± 2:18), with the largest difference being an approximate one-hour delay in PER2 peak timing. There were no significant differences between either the amplitudes or phases of any of the circadian markers when compared between PER3 VNTR genotype. However, the variance in the amplitude of the mean fitted z-scored melatonin rhythm was significantly lower in the PER35/5 subjects compared with the PER4/4 (1.26 ± 0.04 vs 1.23 ± 0.11; P = 0.003).
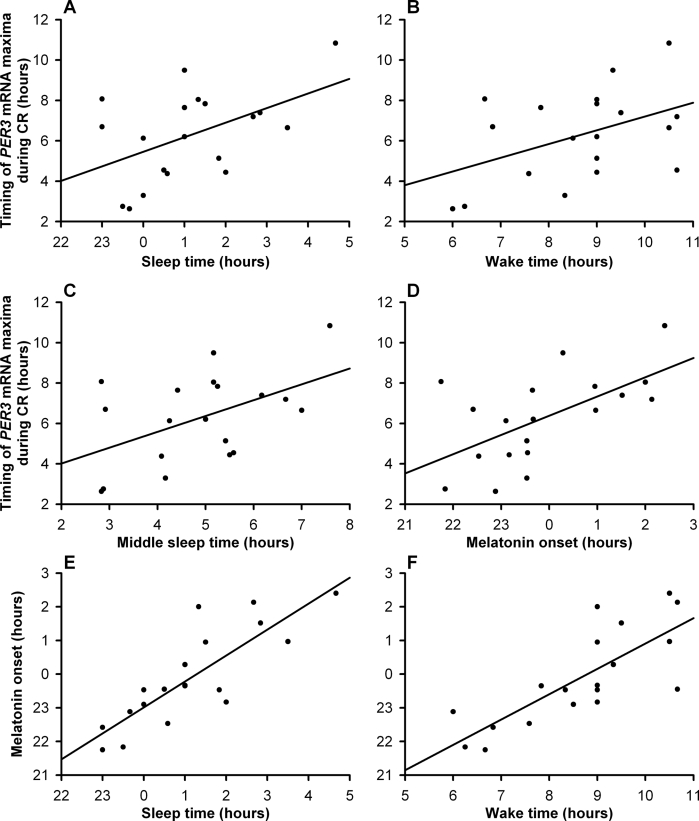
Correlation analyses were performed to assess the association between inter-individual variation in habitual sleep timing, as assessed prior to the laboratory study, and the timing of the various marker rhythms (determined from z-scored data), as assessed during the CR, in those individuals with significant rhythm amplitudes (Table 4). Correlations are presented for all subjects with significant amplitude together and also when split according to PER3 VNTR genotype. When the data from all significant subjects were analyzed irrespective of genotype, the strongest correlation was observed between individual mid-sleep time and the phase of melatonin (maximum) (r = 0.87, P < 0.0001), but both melatonin onset and peak phases also showed significant positive correlations with sleep time, mid-sleep, and wake time (Table 4, Figure 4E and 4F). The timing of the fitted maxima of the cortisol rhythms also showed significant positive correlations with these sleep-wake parameters (Table 4). Of the 3 clock genes, only the timing of the fitted maxima for PER3 showed significant correlations with sleep-wake timing and with the peak of the cortisol rhythm and the onset of melatonin, which had the strongest correlation (r = 0.59, P = 0.005) (Table 4, Figure 4A – 4D). No significant correlations were observed between PER2 or BMAL1 rhythms and sleep-wake timing or melatonin and cortisol (Table 4). However, unlike PER2 and PER3, BMAL1 showed negative correlations with the other circadian markers. None of the circadian markers correlated significantly with the overall level of expression of the clock genes when expressed as relative copy number.
Table 4.
Correlations Between Circadian Markers
| Circadian marker 1 | Circadian marker 2 | r (all subjects) | P (all subjects) | r (PER34/4) | P (PER34/4) | r (PER35/5) | P (PER35/5) |
|---|---|---|---|---|---|---|---|
| Sleep time | Melatonin (onset) | 0.78 | < 0.0001 | 0.69 | 0.0040 | 0.86 | 0.0004 |
| Mid-sleep | Melatonin (onset) | 0.84 | < 0.0001 | 0.76 | 0.0007 | 0.92 | < 0.0001 |
| Wake time | Melatonin (onset) | 0.80 | < 0.0001 | 0.72 | 0.0020 | 0.88 | 0.0002 |
| Sleep time | Melatonin (max) | 0.80 | < 0.0001 | 0.79 | 0.0006 | 0.87 | 0.0003 |
| Mid-sleep | Melatonin (max) | 0.87 | < 0.0001 | 0.83 | < 0.0001 | 0.93 | < 0.0001 |
| Wake time | Melatonin (max) | 0.83 | < 0.0001 | 0.76 | < 0.0001 | 0.90 | < 0.0001 |
| Sleep time | Cortisol (max) | 0.80 | 0.0015 | 0.74 | < 0.0001 | 0.39 | (0.26) |
| Mid-sleep | Cortisol (max) | 0.67 | 0.0002 | 0.86 | < 0.0001 | 0.48 | (0.14) |
| Wake time | Cortisol (max) | 0.65 | 0.0040 | 0.85 | 0.0020 | 0.54 | (0.09) |
| Sleep time | PER3 (max) | 0.48 | 0.030 | 0.06 | (0.86) | 0.70 | 0.0400 |
| Mid-sleep | PER3 (max) | 0.49 | 0.030 | −0.08 | (0.80) | 0.82 | 0.0070† |
| Wake time | PER3 (max) | 0.44 | (0.055) | −0.21 | (0.52) | 0.89 | 0.0009‡ |
| Melatonin (onset) | PER3 (max) | 0.59 | 0.005 | 0.30 | (0.34) | 0.80 | 0.0100 |
| Cortisol (max) | PER3 (max) | 0.48 | 0.030 | 0.26 | (0.43) | 0.70 | 0.0400 |
| Sleep time | PER2 (max) | 0.26 | (0.520) | −0.35 | (0.67) | 0.53 | (0.50) |
| Mid-sleep | PER2 (max) | 0.15 | (0.700) | −0.65 | (0.37) | 0.58 | (0.44) |
| Wake time | PER2 (max) | 0.03 | (0.930) | −0.84 | (0.16) | 0.63 | (0.40) |
| Melatonin (onset) | PER2 (max) | 0.12 | (0.770) | −0.34 | (0.70) | 0.42 | (0.60) |
| Cortisol (max) | PER2 (max) | 0.01 | (0.990) | −0.44 | (0.58) | 0.61 | (0.41) |
| Sleep time | BMAL1 (max) | −0.67 | (0.810) | 0.36 | (0.07) | −0.97 | 0.02 |
| Mid-sleep | BMAL1 (max) | −0.59 | (0.100) | 0.61 | (0.41) | −0.92 | (0.08) |
| Wake time | BMAL1 (max) | −0.43 | (0.610) | 0.86 | (0.09) | −0.76 | (0.25) |
| Melatonin (onset) | BMAL1 (max) | −0.61 | (0.770) | 0.71 | (0.32) | −0.77 | (0.25) |
| Cortisol (max) | BMAL1 (max) | −0.30 | (0.460) | 0.09 | (0.92) | −0.46 | (0.56) |
Correlations between circadian markers of sleep-wake timing, clock gene expression and hormonal rhythms. Correlations with melatonin are for timing of either the onset or maximum of the melatonin rhythm. Correlations with cortisol and clock gene rhythms are for the timing of fitted maxima. Calculations of hormone and clock gene timing are taken from z-scored data, as described in the Methods and presented in the Results. Data have been included in the analysis only from individuals contributing significant circadian amplitudes. The data are presented for all subjects and split by PER3 genotype, which included for PER34/4 and PER35/5 respectively, 14 and 10 subjects for melatonin, 13 and 10 for cortisol, 11 and 8 for PER3, 4 each for PER2, and 4 each for BMAL1. † = significant difference between PER34/4 and PER35/5 correlation (P = 0.03), ‡ = significant difference between PER34/4 and PER35/5 correlations (P = 0.004), r = correlation coefficient, nonsignificant P values are given in brackets.
Figure 4.
Scatter plots showing the correlations between PER3 expression and sleep markers for each individual (represented by each dot). The mean fitted z-score peak (time in hours) of PER3 expression is plotted against (A) sleep onset (r = 0.48, P = 0.03), (B) wake time (r = 0.44, P = 0.055), (C) mid-sleep time (r = 0.49, P = 0.03), and the mean fitted z-score melatonin onset (D) (r = 0.59, P = 0.005) (all times in hours). Also shown are the correlations for melatonin onset against sleep time (E) (r = 0.78, P < 0.0001) and wake time (F) (r = 0.80, P < 0.0001).
To investigate whether the correlations described above were affected by the PER3 polymorphism, the z-scored data for the phase timings of the circadian markers were split by genotype. The correlations between sleep-wake timing and melatonin rhythm timing remained, with the strongest correlation being between the timing of mid-sleep and the peak of melatonin in both genotypes (PER34/4 r = 0.83, P < 0.0001; PER35/5 r = 0.93, P < 0.0001) (Table 4). Whereas PER34/4 homozygotes also showed significant correlations between sleep-wake timing and the timing of cortisol rhythms, there were no such significant correlations with cortisol within the PER35/5 subjects. However, it should be noted that when these correlations are compared between genotype they are not significantly different. Conversely, PER35/5 homozygotes showed significant correlations between PER3 expression rhythms and sleep-wake timing and the timing of melatonin and cortisol, with the strongest correlation being with wake time (r = 0.89, P = 0.0009), whereas the PER34/4 homozygotes did not. In this case, the correlations with mid-sleep and wake time were stronger in PER35/5 individuals and were significantly different between the genotypes (P = 0.03 and 0.004, respectively) (Table 4). Finally, the negative correlation between sleep time and BMAL1 expression was only significant in the PER35/5 subjects.
DISCUSSION
The present data show that the phase of clock gene expression in leukocytes, assessed in the absence of the masking effects of light-dark and sleep-wake cycles, correlates with habitual sleep timing. To date, eight published studies have examined the expression of clock genes in human blood cells.33–40 Together, these studies have characterized the expression of PER1-3, CRY1, CLOCK, BMAL1, REV-ERBα and DEC1 in either mononuclear cells or unseparated leukocytes (mononuclear and polymorphonuclear cells). Apart from one study that involved 61 neonates undergoing light therapy treatment,36 these studies have been based on smaller groups of 3–12 adults, providing blood samples at either 2 time points or every 2 to 4 h over periods ranging from 20 to 39 h. Only 2 studies have attempted constant routine protocols,34,35 although one allowed limited movement and sleep episodes.35 Both studies reported correlations between the phases of melatonin and clock gene transcripts. In a recent study, the effects of a simulated shift work schedule on central and peripheral circadian markers have been studied, and blood mononuclear cell clock gene expression has been analysed over three 24-h periods during the schedule.40
This report is the first to investigate the association between habitual sleep timing and peripheral blood clock gene expression in 24 adults with sampling every hour during an approximate 40-h constant routine in the absence of a sleep-wake cycle. We chose to characterize the expression of 2 members of the negative feedback loop, PER2 and PER3, and the positive transcription factor BMAL1, which normally oscillates in opposite phase to PER.45 We used the PAXgene RNA system for blood sampling. This allows for immediate stabilisation of RNA from leukocytes (both mononuclear and polymorphonuclear cells) enabling accurate results without the need for immediate sample extraction. The fact that we found significant circadian rhythms in each of the 3 clock genes that were studied demonstrates the reliability of this method. While it is known that peripheral white blood cells show a circadian variation in the number of their composite types,46,47 it has been shown recently that there is no phase difference in the expression of PER1 between human peripheral mononuclear and polymorphonuclear cells.35 Indeed, the expression rhythms of clock genes from almost any peripheral tissue will be representative of a mixed cell population. However, the existence of significant clock gene rhythms within peripheral tissues would suggest that most cell types are oscillating in close phase agreement. As with PER1, we have no reason to expect that PER3 should show phase differences in expression between peripheral mononuclear and polymorphonuclear cells, and the well-defined PER3 rhythm we present here supports this.
In this study, we found significant mRNA oscillations for each of the 3 clock genes, with PER3 having the highest number of participants with significant oscillations. The PER3 oscillations have a mean amplitude significantly greater than that of PER2 and BMAL1, and a significantly smaller variance in peak timing. Comparison of these features shows that the robustness of PER3 as a circadian marker in peripheral leukocytes within this study is more reliable than PER2 and BMAL1.
As repeatedly reported in the published literature,10,34,40,48 we found highly significant correlations between sleep-wake timing and the timing of the rhythms of melatonin and cortisol. We also found a significant correlation between the phases of individual PER3 and melatonin rhythms, confirming previous data.34 However, no correlations were found between the rhythms in melatonin and PER2, unlike the previous study by Boivin et al.,34 or BMAL1. In addition, the current study also shows significant correlations between the phase of PER3 and the timing of sleep-wake activity and the timing of cortisol rhythms, which were absent for PER2 and BMAL1. The negative correlation trend for BMAL1 is of interest and may explain why some studies have shown the rhythm of peripheral blood BMAL1 to be both out of phase (present and 37) and in phase.38 However, the authors of the latter study did define 2 “chronotype” subject groups in which the phases of PER2 and BMAL1 were 01:44 and 21:45, and 10:33 and 12:30, respectively. In this study, the average peak times of expression for PER3, PER2 and BMAL1 were 06:05, 08:06, and 15:06, respectively. Similar expression peaks have been reported for PER2 and PER3, which are very similar to the peak time presented here for PER3, although the previous report found an approximately 6-h range across 3 subjects for both genes.34 More recently, it has been reported that after adaptation to a shift schedule, PER2 peak expression occurred shortly after the scheduled wake period and that the BMAL1 peak (during baseline) was out of phase with respect to PER2, occurring 14.6 h after wake.40 PER1, 2 and 3 were also found to have similar phase peaks around 06:00 in the study by Takimoto et al., who also presented data showing more robust rhythms in PER3.37 Another study, using the same PAXgene purification method for leukocyte RNA, reported a peak time of 07:34 for PER1, but they did not detect significant rhythms in either PER2 or BMAL1.39 Finally, when we compared these correlations for significant differences, we found that the correlation between melatonin onset and sleep time is stronger than PER3 peak timing (P = 0.006), but that a similar comparison showed that the correlation between cortisol peak timing and sleep time is no better than for PER3 (P = 0.18).
Taken together, the findings from these studies indicate that there are significant inter-individual differences in the timing of peripheral blood clock gene rhythms, especially with respect to BMAL1, and that out of all of the genes so far analyzed, PER3 demonstrates the greatest consistency. However, the data from previous studies may suffer from less frequent sampling and fewer numbers of subjects studied. It is possible that, all things being equal, other clock genes apart from the ones analysed here could also show equally robust rhythms. In addition, there will be differences in the expression of circadian markers between central and peripheral oscillators, and also between different peripheral oscillators. It is possible that the robustness of PER3 that is found in this and other studies is a specific feature of the peripheral leukocyte oscillator. Nevertheless, in this data set, and in other currently available data sets, PER3 rhythmicity compares favourably to rhythmicity in other clock genes.
Previously, we have reported that the PER3 VNTR polymorphism is associated with diurnal preference and delayed sleep phase disorder.23 In a separate laboratory study, we have also shown that the polymorphism is predictive of sleep structure and cognitive performance.27 The subjects providing blood samples for the current investigation were from the same study protocol. Consequently, all of the variables that have been reported here can be split and analyzed by PER3 VNTR genotype. When this was performed on data averaged for each group, no differences were found in the amplitudes or phases of any of the circadian markers between the PER34/4 or PER35/5 individuals. However, the melatonin amplitude variance was significantly lower in the PER35/5 subjects and this could reflect more robust melatonin rhythms in these individuals.
For this study, subjects were selected by PER3 VNTR genotype alone and not on the basis of diurnal preference or habitual sleep timing. Although there was no significant difference in diurnal preference between the 2 groups, it can be seen from Figure 1 that there is a range of inter-individual sleep-wake timing differences, with the PER35/5 subjects tending to have earlier habitual wake times than the PER34/4 subjects. It is also worth noting that within the subject group as a whole, there does exist a correlation between diurnal preference and habitual wake time (r = −0.42, P = 0.04), but not sleep onset. When the correlations between the various circadian markers were analyzed, we found that both genotypes show strong correlations between sleep-wake timing and the timing of melatonin. However, only the PER34/4 individuals show significant correlations between sleep-wake and cortisol, and this requires further investigation. When the data were split by genotype, only the PER3 rhythms in the PER35/5 individuals showed correlations with sleep-wake timing and melatonin and cortisol, with the correlations with the timing of mid-sleep and wake being significantly different from the correlations in the PER34/4 individuals. In addition, the negative correlation between sleep time and BMAL1 was only ever significant in the PER35/5 subjects. This implies that the contribution to the significant correlations between PER3 expression and other circadian markers observed in all 19 subjects is derived mainly from the PER35/5 individuals. Therefore, while amplitude and phase of the circadian markers may not differ between the genotypes, the PER3 expression is more strongly associated with sleep-wake timing in PER35/5 individuals. This finding is somewhat reminiscent of the notion that the sleep-wake cycles of morning types, with which homozygosity for the longer PER3 VNTR allele is associated,23 are more tightly controlled by circadian rhythmicity than evening types.49 Thus, while we did not observe genotype-dependent differences in sleep-wake timing or the timing of melatonin and cortisol rhythms, we did find significant differences in the correlations of these timings between PER3 VNTR genotypes. However, PER3 gene expression being a marker for entrained phase does not necessarily imply that the VNTR polymorphism should lead to differences in entrained phase. It is possible that the polymorphism may lead to differences in diurnal preference through its effect on the sleep-wake homeostat, and not through its effect on the timing of core circadian markers. Finally, while previous data clearly demonstrate associations between PER3 and sleep structure and timing in humans, animal models with a disruption to Per3 show only a modest effect on circadian phenotype, as defined by the free-running period.50 The precise functional contribution that PER3 makes to the sleep homeostat and/or the circadian clock remains to be determined.
In summary, we have performed the largest study to date measuring peripheral blood clock gene expression with the greatest temporal resolution in a constant routine in the absence of a masking sleep-wake cycle. We have demonstrated that in our study, among the clock genes measured, PER3 has the most robust circadian oscillation. This is not unusual since, in mice, PER3 shows strong circadian rhythms in brain areas such as the suprachiasmatic nucleus (SCN) and the organum vasculosum lamina terminalis (OVLT),51 but also in many peripheral tissues.45 We show that the phase of PER3 expression in leukocytes correlates with the phases of melatonin and cortisol, and with habitual sleep timing. Because the phase of leukocyte PER3 transcript levels was assessed in the absence of the sleep-wake cycle and light-dark cycle, these correlations cannot be explained by direct (masking) effects of the light-dark and sleep-wake cycle on PER3 expression in leukocytes. The mechanisms by which PER3 mRNA levels in the periphery are synchronized to the central circadian pacemaker, habitual sleep timing and melatonin timing remains to be established.
Taken together, these data show that the rhythm of PER3 expression in leukocytes represents a reliable molecular marker for the assessment of entrained phase in healthy individuals. There are PER3 VNTR genotype-dependent effects on the association between the phase of PER3 expression and sleep-wake timing, as well as other circadian markers. Assessment of the PER3 rhythm in advanced and delayed sleep phase disorder may provide further insights into the mechanism underlying these circadian sleep timing disorders.
ACKNOWLEDGMENTS
This research was funded by a grant from the BBSRC (BBS/B/08523). We thank Jayshan Carpen, Matt Cooper, Alex Garvin, and Benita Middleton for their technical support and advice.
Footnotes
Disclosure Statement
This is not an industry supported study. Dr. Scammell has received research support from Takeda and Jazz and has participated in speaking engagements for Cephalon, Jazz, Alexza, Lundbeck, and Takeda. The other authors have reported no financial conflicts of interest.
Supplementary Figure S1 for Inter-Individual Differences In Habitual Sleep Timing and Entrained Phase of Endogenous Circadian Rhythms of BMAL1, PER2 and PER3 mRNA in Human Leukocytes
Supplementary Figure S1.
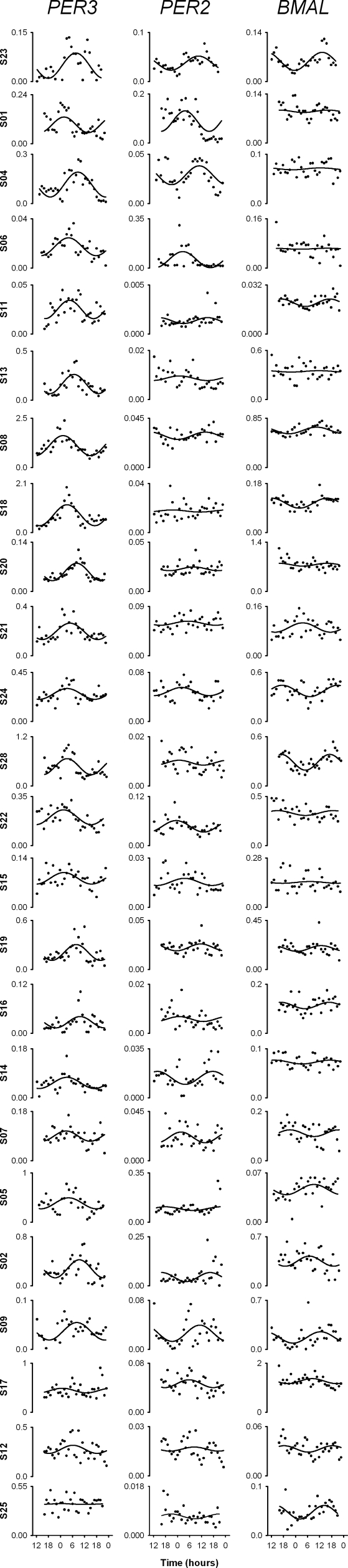
Individual clock gene oscillations. Relative copy number/GAPDH ratio data for PER3, PER2, and BMAL1 for each individual plotted against time in hours. The data have been fitted as described in the methods section.
REFERENCES
- 1.Looby P, Loudon AS. Gene duplication and complex circadian clocks in mammals. Trends Genet. 2005;21:46–53. doi: 10.1016/j.tig.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 2.von Schantz M, Jenkins A, Archer SN. Evolutionary history of the vertebrate period genes. J Mol Evol. 2006;62:701–7. doi: 10.1007/s00239-005-0185-1. [DOI] [PubMed] [Google Scholar]
- 3.Dijk DJ, von Schantz M. Timing and consolidation of human sleep, wakefulness, and performance by a symphony of oscillators. J Biol Rhythms. 2005;20:279–90. doi: 10.1177/0748730405278292. [DOI] [PubMed] [Google Scholar]
- 4.Tafti M, Franken P. Genetic dissection of sleep. J Appl Physiol. 2002;92:1339–47. doi: 10.1152/japplphysiol.00834.2001. [DOI] [PubMed] [Google Scholar]
- 5.Aeschbach D, Sher L, Postolache TT, Matthews JR, Jackson MA, Wehr TA. A longer biological night in long sleepers than in short sleepers. J Clin Endocrinol Metab. 2003;88:26–30. doi: 10.1210/jc.2002-020827. [DOI] [PubMed] [Google Scholar]
- 6.Mongrain V, Lavoie S, Selmaoui B, Paquet J, Dumont M. Phase relationships between sleep-wake cycle and underlying circadian rhythms in morningness-eveningness. J Biol Rhythms. 2004;19:248–57. doi: 10.1177/0748730404264365. [DOI] [PubMed] [Google Scholar]
- 7.Van Dongen HP, Vitellaro KM, Dinges DF. Individual differences in adult human sleep and wakefulness: Leitmotif for a research agenda. Sleep. 2005;28:479–96. doi: 10.1093/sleep/28.4.479. [DOI] [PubMed] [Google Scholar]
- 8.Horne JA, Östberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- 9.Duffy JF, Dijk DJ, Hall EF, Czeisler CA. Relationship of endogenous circadian melatonin and temperature rhythms to self-reported preference for morning or evening activity in young and older people. J Investig Med. 1999;47:141–50. [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy JF, Rimmer DW, Czeisler CA. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav Neurosci. 2001;115:895–9. doi: 10.1037//0735-7044.115.4.895. [DOI] [PubMed] [Google Scholar]
- 11.Mongrain V, Carrier J, Dumont M. Chronotype and sex effects on sleep architecture and quantitative sleep EEG in healthy young adults. Sleep. 2005;28:819–27. doi: 10.1093/sleep/28.7.819. [DOI] [PubMed] [Google Scholar]
- 12.Mongrain V, Carrier J, Dumont M. Difference in sleep regulation between morning and evening circadian types as indexed by antero-posterior analyses of the sleep EEG. Eur J Neurosci. 2006;23:497–504. doi: 10.1111/j.1460-9568.2005.04561.x. [DOI] [PubMed] [Google Scholar]
- 13.Groeger JA, Zijlstra FR, Dijk DJ. Sleep quantity, sleep difficulties and their perceived consequences in a representative sample of some 2000 British adults. J Sleep Res. 2004;13:359–71. doi: 10.1111/j.1365-2869.2004.00418.x. [DOI] [PubMed] [Google Scholar]
- 14.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 15.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 16.Tafti M, Maret S, Dauvilliers Y. Genes for normal sleep and sleep disorders. Ann Med. 2005;37:580–9. doi: 10.1080/07853890500372047. [DOI] [PubMed] [Google Scholar]
- 17.Retey JV, Adam M, Honegger E, et al. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proc Natl Acad Sci U S A. 2005;102:15676–81. doi: 10.1073/pnas.0505414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Retey JV, Adam M, Khatami R, et al. A genetic variation in the adenosine A2A receptor gene (ADORA2A) contributes to individual sensitivity to caffeine effects on sleep. Clin Pharmacol Ther. 2007;81:692–8. doi: 10.1038/sj.clpt.6100102. [DOI] [PubMed] [Google Scholar]
- 19.Wisor JP, O'Hara BF, Terao A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 21.Franken P, Dudley CA, Estill SJ, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci U S A. 2006;103:7118–23. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzenberg D, Young T, Finn L, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 23.Archer SN, Robilliard D, Skene DJ, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–5. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 24.Carpen JD, Archer SN, Skene DJ, Smits M, von Schantz M. A single-nucleotide polymorphism in the 5'-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–7. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 25.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 26.Ebisawa T. Circadian rhythms in the CNS and peripheral clock disorders: human sleep disorders and clock genes. J Pharmacol Sci. 2007;103:150–4. doi: 10.1254/jphs.fmj06003x5. [DOI] [PubMed] [Google Scholar]
- 27.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–8. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 28.Pereira DS, Tufik S, Louzada FM, et al. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- 29.Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–70. [PubMed] [Google Scholar]
- 30.Arendt J. Melatonin and the pineal gland. London: Chapman Hall; 1995. [Google Scholar]
- 31.Lamont EW, James FO, Boivin DB, Cermakian N. From circadian clock gene expression to pathologies. Sleep Med. 2007;8:547–56. doi: 10.1016/j.sleep.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Brown SA, Fleury-Olela F, Nagoshi E, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takata M, Burioka N, Ohdo S, et al. Daily expression of mRNAs for the mammalian Clock genes Per2 and clock in mouse suprachiasmatic nuclei and liver and human peripheral blood mononuclear cells. Jpn J Pharmacol. 2002;90:263–9. doi: 10.1254/jjp.90.263. [DOI] [PubMed] [Google Scholar]
- 34.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–5. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 35.Kusanagi H, Mishima K, Satoh K, Echizenya M, Katoh T, Shimizu T. Similar profiles in human period1 gene expression in peripheral mononuclear and polymorphonuclear cells. Neurosci Lett. 2004;365:124–7. doi: 10.1016/j.neulet.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 36.Chen A, Du L, Xu Y, Chen L, Wu Y. The effect of blue light exposure on the expression of circadian genes: bmal1 and cryptochrome 1 in peripheral blood mononuclear cells of jaundiced neonates. Pediatr Res. 2005;58:1180–4. doi: 10.1203/01.pdr.0000183663.98446.05. [DOI] [PubMed] [Google Scholar]
- 37.Takimoto M, Hamada A, Tomoda A, et al. Daily expression of clock genes in whole blood cells in healthy subjects and a patient with circadian rhythm sleep disorder. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1273–9. doi: 10.1152/ajpregu.00126.2005. [DOI] [PubMed] [Google Scholar]
- 38.Teboul M, Barrat-Petit MA, Li XM, et al. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med. 2005;83:693–9. doi: 10.1007/s00109-005-0697-6. [DOI] [PubMed] [Google Scholar]
- 39.Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun. 2007;354:924–8. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 40.James FO, Cermakian N, Boivin DB. Circadian rhythms of melatonin, cortisol, and clock gene expression during simulated night shift work Sleep. 2007;30:1427–36. doi: 10.1093/sleep/30.11.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duffy JF, Dijk DJ. Getting through to circadian oscillators: why use constant routines? J Biol Rhythms. 2002;17:4–13. doi: 10.1177/074873002129002294. [DOI] [PubMed] [Google Scholar]
- 42.Lockley SW, Skene DJ, Butler LJ, Arendt J. Sleep and activity rhythms are related to circadian phase in the blind. Sleep. 1999;22:616–23. doi: 10.1093/sleep/22.5.616. [DOI] [PubMed] [Google Scholar]
- 43.Harris JA, Benedict FG. A Biometric Study of Human Basal Metabolism. Proc Natl Acad Sci U S A. 1918;4:370–3. doi: 10.1073/pnas.4.12.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–97. [PubMed] [Google Scholar]
- 45.Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol Biol. 2004;5:18. doi: 10.1186/1471-2199-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abo T, Kawate T, Itoh K, Kumagai K. Studies on the bioperiodicity of the immune response. I. Circadian rhythms of human T, B, and K cell traffic in the peripheral blood. J Immunol. 1981;126:1360–3. [PubMed] [Google Scholar]
- 47.Bourin P, Mansour I, Doinel C, Roue R, Rouger P, Levi F. Circadian rhythms of circulating NK cells in healthy and human immunodeficiency virus-infected men. Chronobiol Int. 1993;10:298–305. doi: 10.1080/07420529309059712. [DOI] [PubMed] [Google Scholar]
- 48.Klerman EB, Gershengorn HB, Duffy JF, Kronauer RE. Comparisons of the variability of three markers of the human circadian pacemaker. J Biol Rhythms. 2002;17:181–93. doi: 10.1177/074873002129002474. [DOI] [PubMed] [Google Scholar]
- 49.Monk TH, Buysse DJ, Potts JM, DeGrazia JM, Kupfer DJ. Morningness-eveningness and lifestyle regularity. Chronobiol Int. 2004;21:435–43. doi: 10.1081/cbi-120038614. [DOI] [PubMed] [Google Scholar]
- 50.Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000;20:6269–75. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takumi T, Taguchi K, Miyake S, et al. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753–9. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]