Abstract
The Arabidopsis PAD4 gene previously was found to be required for expression of multiple defense responses including camalexin synthesis and PR-1 gene expression in response to infection by the bacterial pathogen Pseudomonas syringae pv. maculicola. This report describes the isolation of PAD4. The predicted PAD4 protein sequence displays similarity to triacyl glycerol lipases and other esterases. The PAD4 transcript was found to accumulate after P. syringae infection or treatment with salicylic acid (SA). PAD4 transcript levels were very low in infected pad4 mutants. Treatment with SA induced expression of PAD4 mRNA in pad4–1, pad4–3, and pad4–4 plants but not in pad4–2 plants. Induction of PAD4 expression by P. syringae was independent of the regulatory factor NPR1 but induction by SA was NPR1-dependent. Taken together with the previous observation that pad4 mutants have a defect in accumulation of SA upon pathogen infection, these results suggest that PAD4 participates in a positive regulatory loop that increases SA levels, thereby activating SA-dependent defense responses.
Plants respond to pathogen attack by activation of an array of inducible defense responses (1). If a potential pathogen triggers a strong form of resistance called gene-for-gene resistance, plant defense responses are activated rapidly, preventing the pathogen from causing disease. Gene-for-gene resistance occurs when the product of a pathogen gene, called an avirulence (avr) gene, is recognized by a corresponding specific resistance (R) gene in the plant. R-avr interactions are thought to be receptor–ligand-binding events that trigger a form of programmed cell death called the hypersensitive response (HR) and rapid expression of defense responses (2). Infection by virulent pathogens also causes activation of defense responses, but this occurs more slowly than it does in gene-for-gene resistance.
Salicylic acid (SA) plays a central role in signaling during gene-for-gene resistance and responses to virulent pathogens, indicating that similar signal transduction mechanisms can be involved in both of these responses. Plants that are unable to accumulate SA because of the presence of a transgene encoding salicylate hydroxylase (nahG) fail to express PR genes during gene-for-gene resistance or infection by virulent pathogens. They also display greatly enhanced susceptibility to avirulent and virulent pathogens (3, 4).
Arabidopsis thaliana mutants are being used to study SA-dependent regulation of defense responses. A large collection of mutants that are compromised in disease resistance was obtained by screening for enhanced disease susceptibility (eds) to infection by the virulent Pseudomonas syringae strain P. syringae pv. maculicola ES4326 (Psm ES4326) (5–7). These eds mutations include alleles of npr1 and pad4, as well as alleles of a large number of other genes that are less well characterized (5, 8).
Plants carrying npr1 (also called nim1) mutations fail to express the defense genes PR-1, BGL2, and PR-5 when treated with SA (9–11). They also show enhanced susceptibility to virulent P. syringae strains (9–11). Not all SA-dependent responses are NPR1-dependent, because synthesis of the antimicrobial compound camalexin requires SA but not NPR1 (12, 13). NPR1 interacts with transcription factors that bind to essential elements of the PR-1 promoter, suggesting that it may function by altering the activities of transcription factors required for defense gene expression (14).
Plants carrying pad4 mutations display reduced camalexin synthesis, PR-1 expression, and SA levels when infected with Psm ES4326 (15). The SA accumulation defect appears to be the cause of the other defects, because SA treatment before infection restores camalexin synthesis and PR-1 expression (15). Defense response defects are not observed in pad4 plants infected with an isogenic avirulent strain carrying the avirulence gene avrRpt2, demonstrating that PAD4 is not required in this case of gene-for-gene resistance. The phenotypes of pad4 mutants are consistent with the idea that PAD4 is required for amplification of weak signals, such as those resulting from infection by a virulent pathogen, to a level sufficient for activation of SA signaling. If this is true, then the SA-generating signal produced by recognition of avrRpt2 must be sufficiently strong that PAD4-dependent amplification is not required.
In this report, we describe isolation of PAD4 by positional cloning. The predicted PAD4 amino acid sequence has regions of similarity to eukaryotic triacyl glycerol lipases and esterases. The patterns of PAD4 expression in response to SA treatment or pathogen infection suggest that PAD4 and SA act in a positive signal-amplification loop required for activation of defense responses.
Methods
Plants and Growth Conditions, DNA and RNA Analysis, Inoculation with Bacteria, Treatment with Salicylic Acid, and Camalexin Quantification.
Plants were grown as described (15). DNA and RNA analyses were carried out as described (15), except that for the RNA blots, a single-stranded antisense PAD4 probe was made from the plasmid pDJ5.1 (ATCC strain DH5α/AtcPAD4) by using antisense primer 5′-CGTGAAATTGAGGTGGAGAGAGATTGGTTTCCG-3′. Inoculations with bacteria, SA treatments, and camalexin quantitation were carried out as described (15).
Isolation of pad4–2.
The pad4–2 mutant was isolated from fast neutron-mutagenized Landsberg erecta (Ler) seed (Lehle Seeds, Round Rock, TX) in a screen for suppressors of RPP5-mediated resistance to Peronospora parasitica (16). Allelism with pad4–1 was determined in F1 and F2 complementation tests.
Isolation of pad4–3 and 4–4.
The pad4–3 and pad4–4 mutants were isolated in the Columbia (Col) ecotype from a screen for Arabidopsis mutants with enhanced susceptibility to the fungal pathogen Erysiphe orontii. The screen was carried out by inoculating 4.5-week-old M2 Arabidopsis plants grown from fast neutron-mutagenized seed pools (Lehle Seeds) with E. orontii conidia as described (17). Plants were scored at 2–3 weeks after infection, and heavily infected plants were allowed to set seed. Progeny of the putative mutants were retested to confirm the enhanced-susceptibility phenotype. Complementation testing with the pad4–1 allele revealed that two of the mutations, now called pad4–3 and pad4–4, were pad4 alleles.
Markers Used for Mapping PAD4.
We made cleaved amplified polymorphic sequence (CAPS) markers corresponding to markers m409, m457, and AtEm1 (GenBank accession no. Z11158) and to the ends of YACs (yeast artificial chromosomes) CIC7A4 (right end [R] and left end [L]), CIC9D9 (L), and yUP1E3 (L). The YAC ends were cloned by using a modified version of the adapter-ligation protocol (18) and partially sequenced. The sequence was used to design primers for the PCR. PCR then was performed on Col and Ksk genomic DNA, and the products were digested with a battery of restriction enzymes to detect polymorphisms. BAC (bacterial artificial chromosome) ends were also cloned by adapter ligation. BAC T8N21 (R) and T5I22 (L) ends and cosmid inserts 8 and 23 were converted into restriction fragment length polymorphism (RFLP) markers by using them to probe Southern blots containing genomic DNA from Col and Ksk digested with a battery of restriction enzymes. Detailed information about the CAPS will be available at http://genome-www.stanford.edu/Arabidopsis/aboutcaps.html.
Construction of the Cosmid Contig Spanning PAD4.
The YAC and BAC clones used in this study were obtained from the Arabidopsis Biological Research Center at Ohio State University. BAC DNA was purified on a CsCl gradient (19) and partially digested with TaqI. The fragments were cloned into the ClaI site of the binary vector pCLD04541 (20). The cosmid clones were packaged into bacteriophage λ particles by using the Gigapack XL kit from Stratagene. Thirty-six randomly chosen cosmids from the library were aligned into a contig by using BAC end probes T8N21R and T5I22L and inserts from cosmids that hybridized to these two probes. DNA preparations from cosmids that complemented the camalexin-deficient phenotype of pad4–1 were analyzed by EcoRI, HindIII, and BamHI digestion followed by Southern hybridization with probes made from various fragments of cosmid 8. A restriction map of the cosmids then was constructed.
Isolation of the PAD4 cDNA Clone.
A cDNA library was constructed by using poly(A) RNA purified from wild-type Columbia leaves infected with Psm ES4326. The 5′ and 3′ random amplified cDNA ends (RACE) of the PAD4 cDNA were isolated by using the Marathon cDNA isolation kit (CLONTECH). The gene-specific primers used were 5′-CGTGAAATTGAGGTGGAGAGAGATTGGTTTCCG-3′ and 5′-GAATTGTTAGGTAAAAAGCTGGTGGTGATAACCGG-3′ for the 5′ and 3′ RACE products, respectively. A longer cDNA (no. 2) was isolated by using primers 5′-ATGGACGATTGTCGATTCGAG-3′ and 5′-AGAATATATAGTAACATTCATCAGAAAGTC-3′, corresponding to the ends of the cDNA sequence, and cloned into plasmid pCR2.1 (Invitrogen).
DNA Sequence Analysis.
Fragments obtained from EcoRI and HindIII digests of cosmid 8 were subcloned into pBluescript SK(+) (Stratagene). These subcloned fragments then were sequenced by using an Applied Biosystems automated sequencer. To analyze mutant alleles, PAD4 sequences were amplified from wild-type and pad4 mutant plants and sequenced directly. The sequence data were analyzed by using the software lasergene (DNAstar, Madison, WI). Sequence data also were submitted for a blast (21) search of GenBank. Multiple sequence alignment of the predicted protein sequences was performed by using the clustalw 1.73 program at http://transfac.gbf-braunschweig.de/dbsearch/clustalw.html.
Results
Positional Cloning of PAD4.
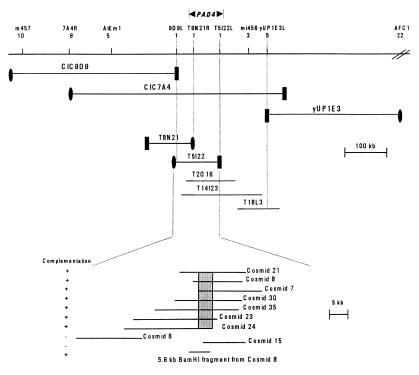
We used a map-based cloning strategy to isolate PAD4 in an effort to gain insight into the function of PAD4 in controlling defense responses. As reported previously, PAD4 is located on chromosome 3 between GL1 and BGL2 (15). We carried out further mapping of PAD4 with 312 pad− F2 plants from a cross between pad4–1 plants (Col accession) and wild-type Keswick (Ksk) plants by using CAPS markers (22). PAD4 was found to lie between markers m457 and AFC1 (Fig. 1). The physical map of this region showed that most of it was covered by overlapping YAC clones (23). Mapping with markers generated from YAC ends revealed that PAD4 lies between the left end of YAC CIC9D9 (9D9L) and the left end of YAC yUP1E3 (1E3L). Hybridization of the Arabidopsis Biological Resource Center BAC library filters with CIC9D9L and yUP1E3L probes and searching of the Arabidopsis BAC fingerprint database at http://genome.wustl.edu/gsc/arab/arabidopsis allowed us to identify and align BACs in this region (Fig. 1). Mapping with RFLP markers derived from the right end of BAC T8N21 (T8N21R) and the left end of T5I22 (T5I22L) revealed that PAD4 lies on BAC T5I22.
Figure 1.
Positional cloning and structure of the PAD4 gene. A 5-cM region between CAPS markers m457 and AFC1 was partially spanned with YAC, BAC, and cosmid clones. The number of recombination events between PAD4 and a particular marker among 620 chromosomes tested is shown below the marker. ●, Right ends, and ■, left ends of YAC and BAC clones. Cosmids 7, 8, 21, 23, 24, 30, and 35 and the indicated 5.6-kb BamHI fragment from cosmid 8 complemented the pad4–1 mutation. Shading indicates the region common to all these cosmids. Only 2 of the 13 noncomplementing cosmids are shown here.
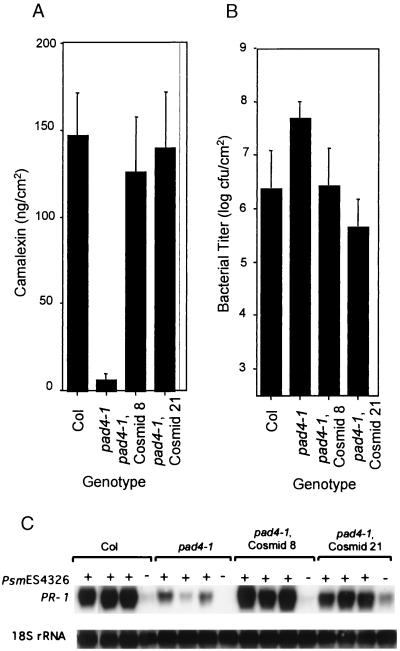
Complementation testing was used to identify the PAD4 gene within BAC T5I22. A cosmid library was constructed by subcloning DNA from BAC T5I22 into the binary vector pCLD04541 (20). DNA blot hybridization was used to assemble the cosmids into a contig covering BAC T5I22. Twenty cosmids that collectively contained all of the BAC DNA were used to transform pad4–1 plants, and the transformants were tested for complementation of the camalexin-deficient phenotype of pad4–1. Seven cosmids (numbers 7, 8, 21, 23, 24, 30, and 35; Fig. 1) complemented the camalexin-accumulation phenotype of pad4–1 plants whereas 13 other cosmids failed to complement. Fig. 2 shows that cosmids 8 and 21 complemented the camalexin-accumulation, PR-1 expression, and bacterial growth phenotypes, respectively, caused by the pad4–1 mutation. Furthermore, a 5.6-kb BamHI fragment from cosmid 8 (Fig. 1) also complemented the camalexin-deficient phenotype of pad4–1 (data not shown), demonstrating that this fragment contains PAD4.
Figure 2.
Complementation of the camalexin-deficient phenotype (A), enhanced bacterial growth phenotype (B), and the PR-1 transcript accumulation phenotype (C) of pad4–1 by cosmids 8 and 21. Wild-type (Col), pad4–1, and transgenic pad4–1 containing cosmid 8 or cosmid 21 were infected with Psm ES4326. Camalexin levels in infected leaves were determined 48 hr after infection. Bacterial titer was determined 3 days after infection, and PR-1 mRNA levels were determined 36 hr after infection. For A and B, each bar represents the mean and SD of six replicate samples. In C, the 18S rRNA probe was used to evaluate uniform loading. Similar results were obtained in another independent experiment.
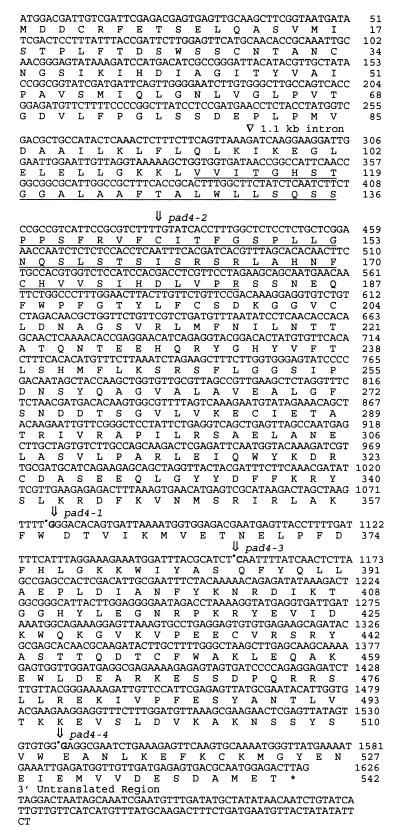
The DNA sequence of this 5.6-kb region of cosmid 8 was determined and used to perform a blast search (21). The predicted protein sequence showed similarity to lipases and other esterases. To identify the PAD4 gene within this region, we first isolated and sequenced a cDNA clone (cDNA 1) corresponding to the lipase-like gene. Examination of the genomic sequence revealed the existence of an in-frame ATG 7 bases upstream from the 5′ end of cDNA 1 (Fig. 3). Using a PCR primer beginning with this upstream ATG, we were able to amplify a cDNA product (cDNA 2), suggesting that cDNA 2 represents the full-length protein. We amplified the 5.6-kb region of the genomic DNA from wild-type Col, Landsberg erecta (Ler), and the four pad4 mutant alleles and determined the DNA sequence of the amplified products. Fig. 3 shows that each mutant allele had a single mutation in the 5.6-kb region. All of these mutations lie in the predicted ORF of the lipase-like gene, demonstrating that it is PAD4.
Figure 3.
Structure of the PAD4 gene showing the position of the intron and all four mutations in the coding sequence and the 3′ untranslated region. Insertion of an extra T at nucleotide position 430 occurs in pad4–2, codon TGG at position 359 is changed to TAG in pad4–1, codon CAA at position 386 is changed to TAA in pad4–3, and a G is missing from codon 513 in pad4–4. The underlined region displays sequence similarity to triacylglycerol lipases and esterases as shown in Fig. 4. cDNA 1 starts at nucleotide 46. PAD4 is located on the sequenced BAC clone F2206 (GenBank accession no. AL050300.1).
PAD4 Displays Sequence Similarity to Triacylglycerol Lipases.
Fig. 4 shows an alignment of the N-terminal region (amino acids 111–181) of the PAD4 sequence with those of other lipases and an esterase. Although the level of amino acid identity between PAD4 and the lipases is relatively low (27–35% over these 70 aa), PAD4 is as similar to any of these known lipases as they are to each other (Fig. 4). The region similar to lipases includes three conserved amino acid residues that form a catalytic triad: a serine, an aspartate, and a histidine (Fig. 4) (24). Interestingly, the lipase similarity also is present in the product of EDS1, another Arabidopsis gene with a crucial role in activation of defense responses (25). The COOH-terminal 360-aa of PAD4 did not show significant sequence similarity to any known protein. However, because pad4–1, pad4–3, and pad4–4 all cause truncation of this region of the protein (Fig. 3), the C-terminal region must be essential for PAD4 function.
Figure 4.
Amino acid sequence comparison of the predicted PAD4 protein with other lipase and lipase-like genes. The putative lipase catalytic triad consisting of a serine, histidine, and aspartate is indicated by arrows. RhizoTGL, triacylglycerol lipase precursor 1 from Rhizomucor miehei; FusaTGL, triacylglycerol lipase from Fusarium heterosporum; Rhizolip, triacylglycerol lipase precursor 1 from Rhizomucor niveus; Thermolip, lipase from Thermomyces lanuginosus; AspFAE, ferulic acid esterase A from Aspergillus niger; AtEDS1, A. thaliana EDS1; AtPAD4, A. thaliana PAD4. Invariant residues are indicated in bold letters, and conserved amino acids are underlined.
PAD4 Expression Is Induced by Pathogen Infection and SA.
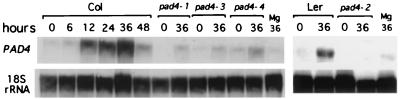
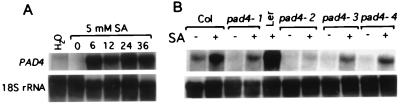
To examine the effect of pathogen infection on PAD4 transcript levels, we performed RNA blot analysis on wild-type and pad4 leaves infected with Psm ES4326. Fig. 5 shows that PAD4 mRNA levels increased beginning at 12 hr and reached a maximum by 36 hr after infection. Curiously, we observed that PAD4 transcript levels were very low in all of the four pad4 mutants even after infection with Psm ES4326 (Fig. 5). A possible explanation for this is that PAD4 function is required for activation of PAD4 expression. The previous observation that pad4 mutants are deficient in SA accumulation after Psm ES4326 infection (15) suggests a possible mechanism. If activation of PAD4 expression requires SA, then PAD4 could be required to produce the SA in response to Psm ES4326 infection. To test these ideas, we treated wild-type Columbia, Landsberg erecta, and pad4 mutant plants with SA and examined levels of the PAD4 transcript. Fig. 6 shows that PAD4 mRNA levels in wild-type, pad4–1, pad4–3, and pad4–4, but not pad4–2 plants, increased rapidly after SA treatment. These results suggested that SA is sufficient for PAD4 mRNA induction. The mutation in pad4–2 plants causes a translation stop early in the protein (amino acid position 181). mRNAs containing premature chain termination mutations (“nonsense mRNAs”) often are unstable because they are subject to nonsense-mediated mRNA decay (NMD) (26). Chain termination mutations near the 5′ end of an ORF tend to cause a stronger NMD effect than those near the 3′ end (27). This could be the reason why we could detect strong SA induction of the PAD4 transcript in pad4–1, pad-3, and pad4–4, but not pad4–2 plants.
Figure 5.
After infection by Psm ES4326, PAD4 transcript levels are very high in wild-type plants and greatly reduced all pad4 mutant alleles. Leaves from wild-type (Col) and all four pad4 mutants were excised 0, 6, 12, 24, 36, or 48 hr after infection. Mg indicates leaves mock-inoculated with 10 mM MgSO4 and harvested after 36 hr. Similar results were obtained in another independent experiment.
Figure 6.
PAD4 mRNA is induced by SA in wild-type, pad4–1, pad4–3, and pad4–4 but not in pad4–2. Wild-type (Col and Ler) and pad4 plants were treated with 5 mM SA in 0.02% Silwet L-77 (vol/vol) until uniformly wet. Control samples were treated with 0.02% Silwet L-77 (H2O). (A) Wild-type (Col) plants were sprayed with 5 mM SA, and PAD4 mRNA levels were determined 0, 6, 12, 24, and 36 hr after treatment. (B) Wild-type (Col and Ler) and pad4 plants were treated with 5 mM SA, and PAD4 mRNA levels were determined 0 and 6 hr after treatment. Similar results were obtained in another independent experiment.
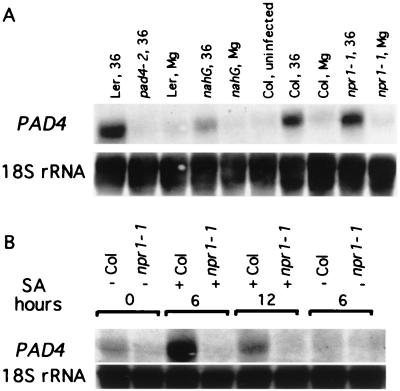
To confirm the requirement of SA for induction of PAD4 by pathogen infection, we examined PAD4 transcript levels in the SA-deficient nahG plants. Fig. 7A shows that in plants infected with Psm ES4326, PAD4 transcript levels were much lower in nahG plants than in wild type. This shows that Psm ES4326 induction of PAD4 expression is strongly SA-dependent.
Figure 7.
PAD4 mRNA induction by Psm ES4326 is SA-dependent but NPR1-independent whereas induction by SA is NPR1-dependent. (A) Wild-type (Col and Ler), nahG, and npr1–1 plants were infected with Psm ES4326. Samples were analyzed for PAD4 mRNA 36 hr after infection. Mg indicates leaves mock-inoculated with 10 mM MgSO4 and harvested at 36 hr. (B) Wild-type (Col) and npr1–1 plants were treated with 5 mM SA, and PAD4 mRNA levels were determined at 0, 6, and 12 hr. Control samples were treated with 0.02% Silwet L-77 (H2O). Similar results were obtained in another independent experiment.
Psm ES4326 Induction of PAD4 Expression Is NPR1-Independent Whereas SA Inducibility of PAD4 Is NPR1-Dependent.
To test whether pathogen induction of PAD4 mRNA expression requires NPR1, we examined the levels of PAD4 mRNA in wild-type and npr1–1 plants after pathogen infection. Fig. 7A shows that PAD4 transcript levels were comparable to wild type in npr1–1 plants infected with Psm ES4326. To determine whether induction of PAD4 by exogenous SA requires NPR1, we treated wild-type and npr1–1 plants with 5 mM SA and examined PAD4 expression. We found that PAD4 transcript levels were undetectable in npr1–1 plants after SA treatment (Fig. 7B). These results demonstrate that Psm ES4326-induced PAD4 expression is NPR1-independent, but SA-induced PAD4 expression is NPR1-dependent.
Discussion
PAD4 is required for expression of multiple defense responses after pathogen infection. The predicted sequence of PAD4 is similar to those of triacylglycerol lipases and an esterase. Lipases are hydrolytic enzymes that break down triacylglycerols into fatty acids and glycerol. There is evidence for the involvement of lipids and lipases in cellular signaling. For example, it has been shown that diacylglycerol is capable of activating protein kinase C in vitro and in vivo (28). The activation of protein kinase C is required to modulate many Ca2+-dependent cellular processes (29). It is possible that the lipolytic activity of PAD4 leads to the synthesis or degradation of a molecule involved in signal transduction pathways, leading to disease resistance. However, PAD4 is also similar to a ferulic acid esterase from Aspergillus niger (Fig. 4), and so it is possible that its substrate is not a lipid.
EDS1, another Arabidopsis gene involved in defense responses, was cloned recently (25). The predicted EDS1 sequence shows similarity to the same class of eukaryotic lipases as PAD4 (Fig. 4). EDS1 is a key component of disease-resistance pathways activated by the TIR-NBS-LRR class of R genes in response to bacterial and oomycete pathogens (30, 31). Like mutations in PAD4, mutations in EDS1 cause increased susceptibility to the virulent pathogen P. syringae pv. tomato (Pst DC3000) and some compatible and incompatible Peronospora isolates (30, 31). Comparison of the spectrum of pathogens affected by pad4 and eds1 has been complicated by the fact that the well characterized alleles are in different ecotypes. The PR-1 expression phenotypes of both eds1 (25) and pad4 (15) suggest that both genes act upstream from SA. The observation that two genes required for regulation of defense responses share a triacylglycerol lipase motif suggests that this motif is relevant to the function of these genes.
The pattern of PAD4 expression is consistent with the idea that PAD4 and SA form part of a signal-amplification loop that is required for expression of PR-1 and other defense responses. In this model, pathogen infection causes some signal, possibly a low level of SA, which induces PAD4 expression. PAD4 activity stimulates SA accumulation, which further induces PAD4 expression. Previous characterization of pad4–1 showed that PAD4 is required for SA accumulation after Psm ES4326 infection (15). In this work, we found that SA treatment is sufficient to activate PAD4 expression and that SA is necessary for full activation of PAD4 expression in response to infection. The pattern of PAD4 expression in pad4 mutants is also consistent with a role for PAD4 in an SA amplification loop. In pad4 mutants, PAD4 was not induced significantly by infection, but it was induced by SA. This result could be due to the requirement of PAD4 for SA accumulation and activation of PAD4 expression by SA. Alternatively, the apparent increase in PAD4 mRNA in pad4 mutants treated with SA could be due to stronger activation of PAD4 expression by SA than by infection.
There is other evidence supporting the idea that SA acts in a positive autoregulatory fashion. SA treatment increased expression of EDS1, even though EDS1 was shown to function upstream of SA-inducible PR-1 expression (25). In the lsd6 lesion-mimic mutant, lesion formation is associated with elevated SA levels and PR gene expression, and SA is required for lesion formation (32). Small amounts of SA potentiate H2O2 production, cell death, and expression of defense genes including phenylalanine ammonia lyase (PAL) in response to infection (33). H2O2 production and cell death both lead to increased SA concentrations, and PAL activity is required for SA synthesis (34).
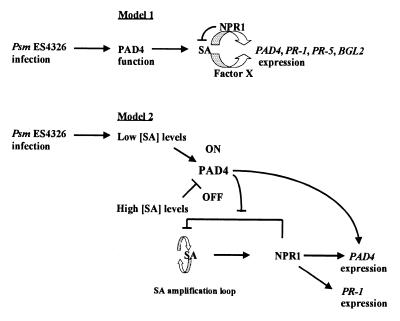
Curiously, PAD4 expression in response to Psm ES4326 infection did not require NPR1, whereas PAD4 expression in response to SA did require NPR1. Fig. 8 shows two models that may explain this observation. Model 1 postulates that SA is required for PAD4 expression, and its effect may be mediated either by NPR1 or by a pathogen-inducible factor that has not yet been identified. NPR1 is proposed to have a negative effect on SA levels, because infected npr1 plants exhibit higher SA levels than infected wild-type plants (10). We have proposed a similar model previously to explain why expression of PR-5 and BGL2 in response to SA is NPR1-dependent, but expression in response to Psm ES4326 infection is NPR1-independent (5). There are other examples of SA-dependent, NPR1-independent responses. These include camalexin synthesis in response to Psm ES4326 infection (13) and expression of PR-1, PR-5, and BGL2 in cpr6 mutants (35).
Figure 8.
Proposed models for the roles of PAD4, SA, and NPR1 in defense gene expression. (Model 1) SA is necessary but not sufficient for activation of expression of defense genes including PAD4. Another component is required—either NPR1 or some unknown factor X from the pathogen. NPR1 also inhibits SA accumulation. (Model 2) Different SA levels modulate PAD4 activity differently. Low SA levels activate and very high SA levels inactivate PAD4. Activated PAD4, in turn, stimulates expression of defense genes and inhibits the repressing activity of NPR1 on the SA amplification loop. Very high SA levels turn PAD4 off. In this situation, NPR1 activity is required for defense gene expression.
Model 2 postulates that there are two ways to induce PAD4 expression: one that requires NPR1 and SA, and another that requires PAD4 and SA. In addition to its effect on activation of gene expression, NPR1 inhibits an SA amplification loop. PAD4 counters this inhibition. The activities of PAD4 are promoted by low levels of SA but inhibited by high levels of SA. Consequently, when plants are infected with Psm ES4326 (leading to a presumed low initial level of SA), PAD4 is activated and induces PAD4 expression independently of NPR1. However, when plants are sprayed with SA, SA levels are high, PAD4 activity is repressed, and NPR1 is required to induce PAD4 expression. PAD4 increases SA levels by reducing the NPR1-dependent inhibition of SA amplification. Future experiments will be designed to test these models.
Acknowledgments
We thank Philip Howard for technical assistance, Xinnian Dong for seeds of Ler plants carrying the nahG transgene, Eric Holub for Keswick seeds, the ABRC for clones, and Suwei Zhao and Kongyi Jiang of the DNA Sequencing Facility at Center for Agricultural Biotechnology, University of Maryland Biotechnology Institute. We are grateful to David Bouchez for providing data before publication and Bart Feys for pointing out the in-frame ATG upstream from the end of PAD4 cDNA 1. We thank Fumiaki Katagiri for useful discussions and critical reading of the manuscript. This research was supported in part by National Science Foundation Grant MCB 9723493 to J.G. and National Institutes of Health Grant GM 48707 to F.M.A.
Abbreviations
- SA
salicylic acid
- YAC
yeast artificial chromosome
- CAPS
cleaved amplified polymorphic sequence
- BAC
bacterial artificial chromosome
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF188329).
References
- 1.Hammond-Kosack K E, Jones J D G. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 3.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 4.Delaney T P, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessman H, Ward E, et al. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 5.Glazebrook J, Rogers E E, Ausubel F M. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers E E, Ausubel F M. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volko S M, Boller T, Ausubel F M. Genetics. 1998;149:537–548. doi: 10.1093/genetics/149.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao H, Glazebrook J, Clarke J D, Volko S, Dong X. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 9.Cao H, Bowling S A, Gordon S, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney T P, Friedrich L, Ryals J A. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah J, Tsui F, Klessig D F. Mol Plant–Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- 12.Glazebrook J, Ausubel F M. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Last R L. Plant Cell. 1996;8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Fan W, Kinkema M, Li X, Dong X. Proc Natl Acad Sci USA. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou N, Tootle T L, Tsui F, Klessig D F, Glazebrook J. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker J E, Coleman M J, Szabo V, Frost L N, Schmidt R, van der Biezen E A, Moores T, Dean C, Daniels M J, Jones J D. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reuber T L, Plotnikova J M, Dewdney J, Rogers E E, Wood W, Ausubel F M. Plant J. 1998;16:473–485. doi: 10.1046/j.1365-313x.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- 18.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1995. [Google Scholar]
- 20.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 23.Camilleri C, Lafleuriel J, Macadre C, Varoquaux F, Parmentier Y, Picard G, Caboche M, Bouchez D. Plant J. 1998;14:633–642. doi: 10.1046/j.1365-313x.1998.00159.x. [DOI] [PubMed] [Google Scholar]
- 24.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley S, Turkenburg J P, Christiansen L, Huge-Jensen B, Norskov L, et al. Nature (London) 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 25.Falk A, Feys B J, Frost L N, Jones J D G, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Culbertson M, R. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 27.Peltz S W, Brown A H, Jacobsen A. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 28.Go M, Sekiguchi K, Hideaki N, Kikkawa U, Nishizuka Y. Biochem Biophys Res Commun. 1987;144:598–605. doi: 10.1016/s0006-291x(87)80008-6. [DOI] [PubMed] [Google Scholar]
- 29.Nishizuka Y. Science. 1986;233:305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- 30.Aarts N, Metz M, Holub E, Staskawicz B J, Daniels M J, Parker J E. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker J E, Holub E B, Frost L N, Falk A, Gunn N D, Daniels M J. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, Steiner H Y, Ryals J. Plant Cell. 1995;7:2013–2022. doi: 10.1105/tpc.7.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mauch-Mani B, Slusarenko A J. Plant Cell. 1996;8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirasu K, Nakajima H, Rajasekhar V K, Dixon R A, Lamb C. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clarke J D, Liu Y, Klessig D F, Dong X. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]