Abstract
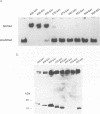
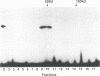
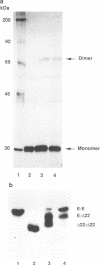
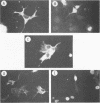
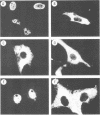
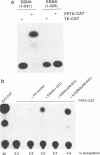
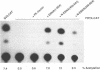
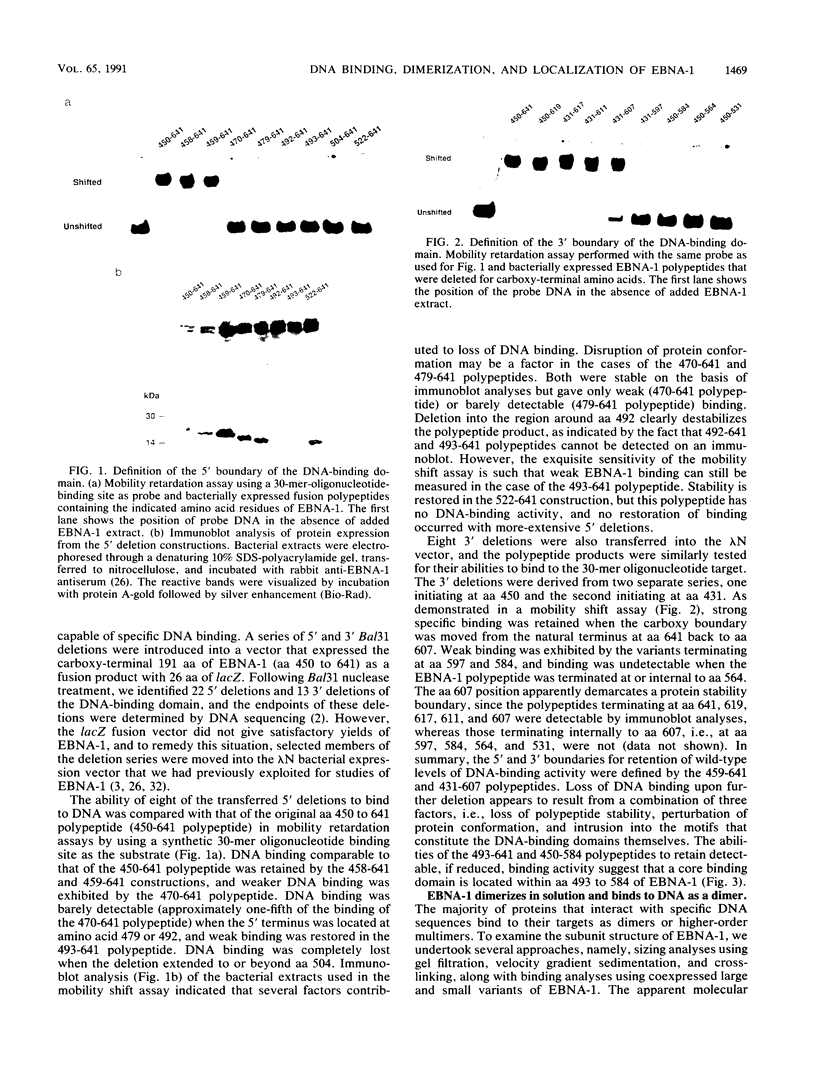
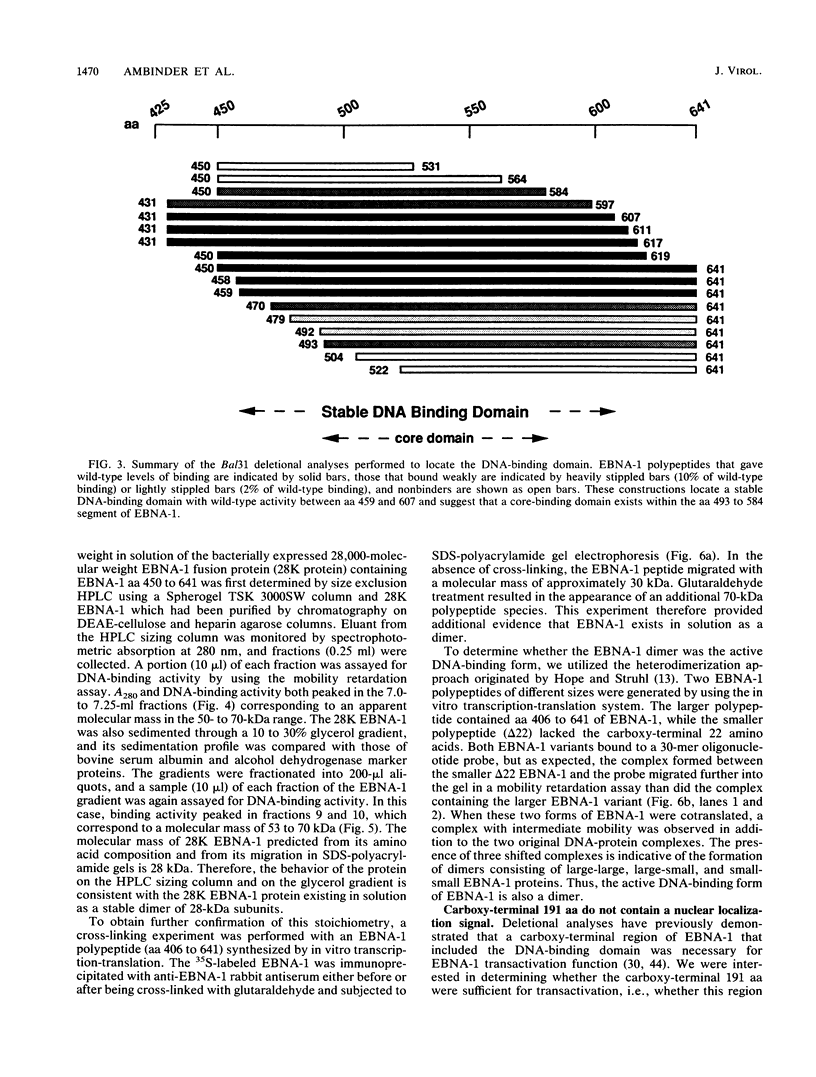
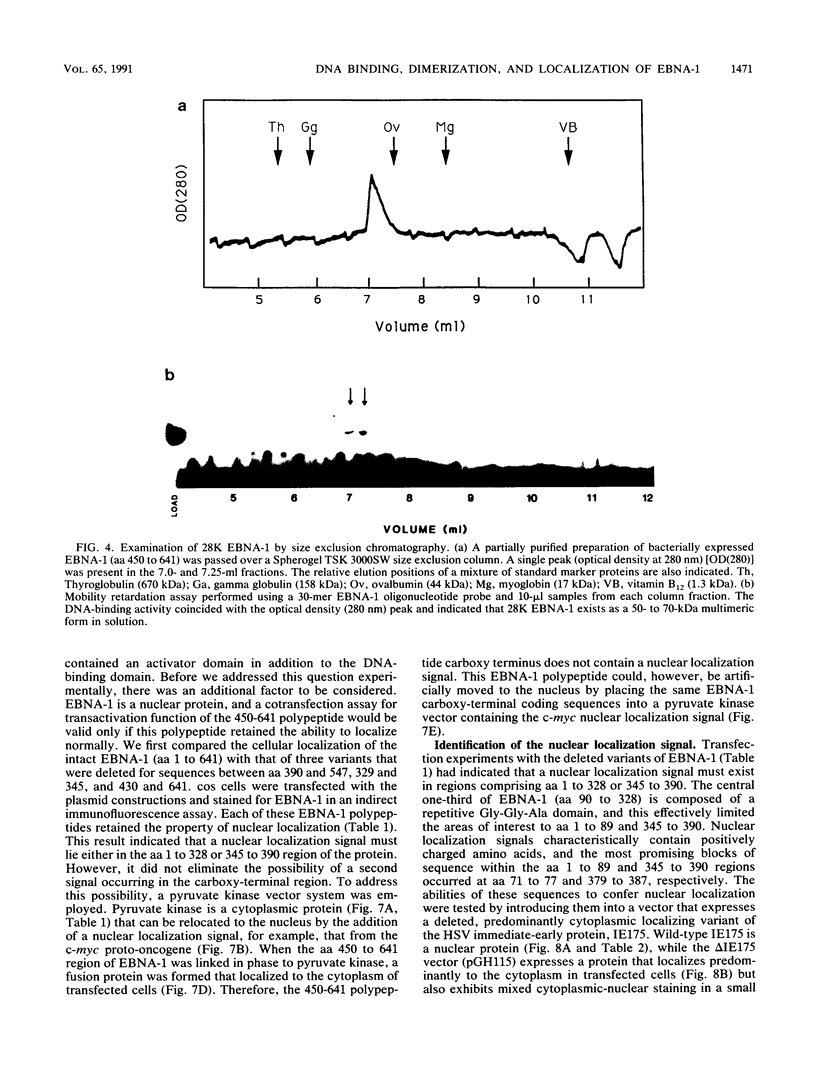
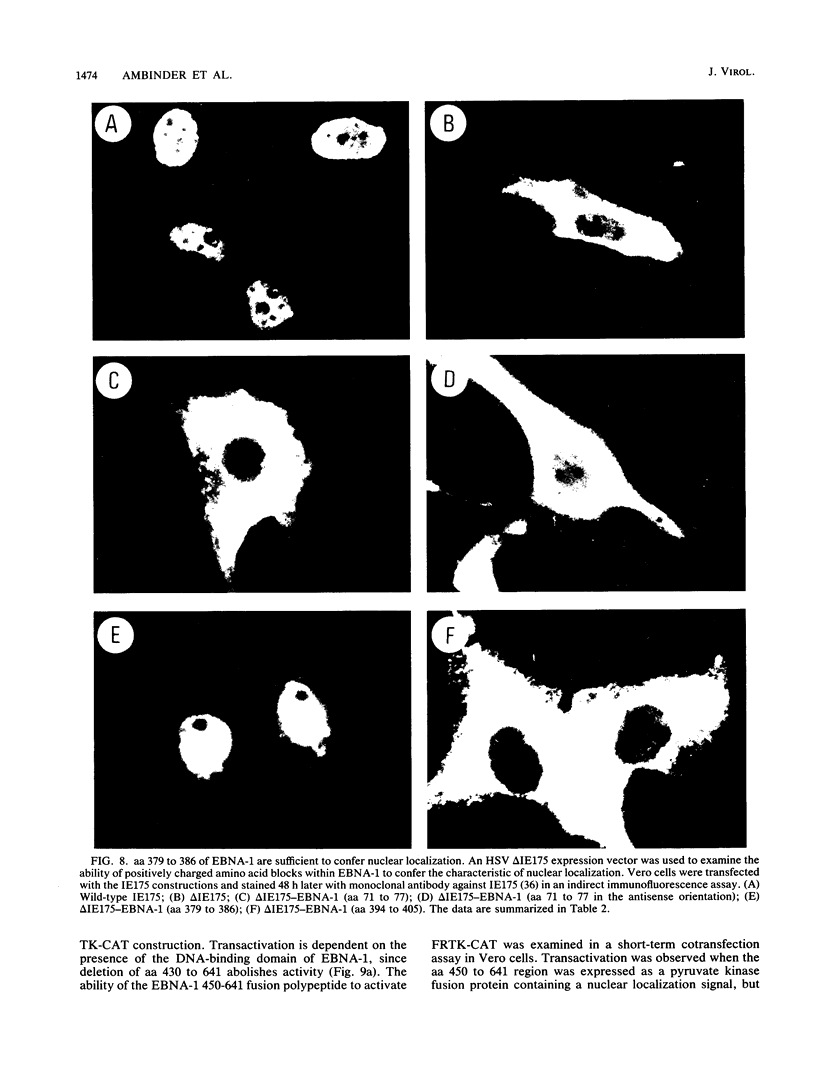
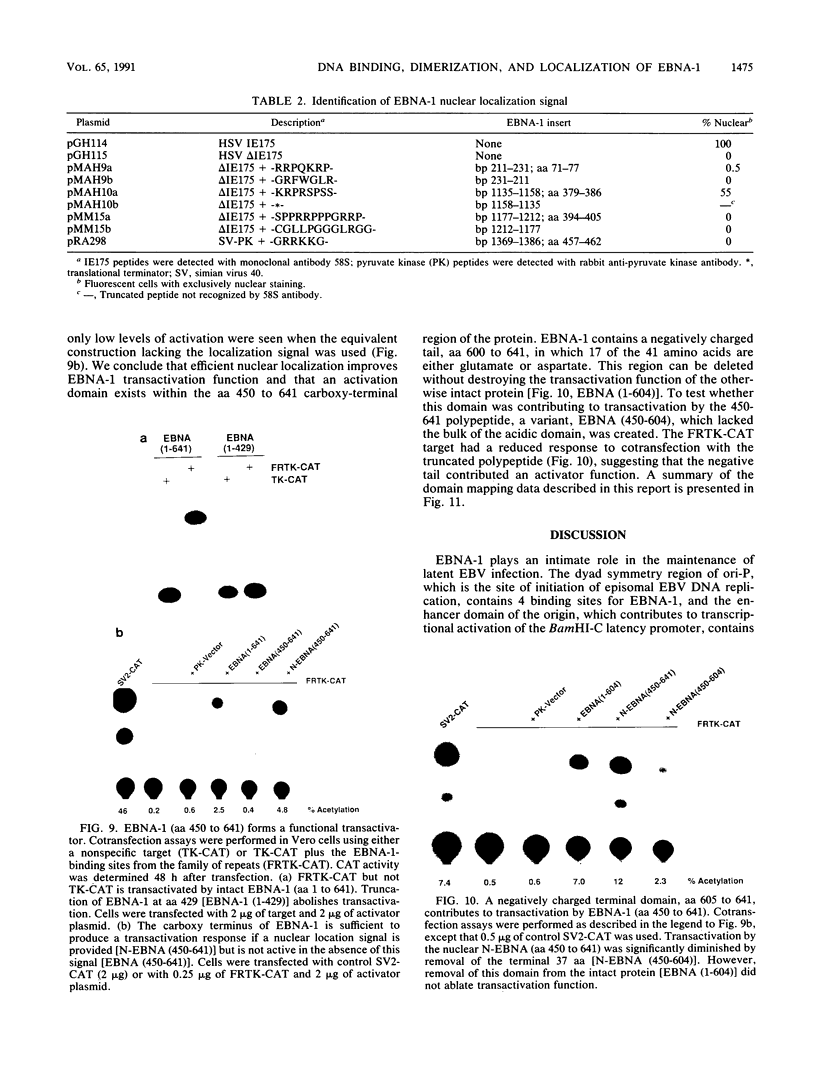
The Epstein-Barr virus (EBV)-encoded latency product EBNA-1 is functionally pleiotropic, being required for replication of the episomal form of the EBV genome and having a role in the regulation of latency transcription. EBNA-1 is a direct DNA-binding protein, and both replication and transactivation are dependent on the interaction of EBNA-1 with its cognate DNA recognition sequences. To better understand EBNA-1 function, we have further characterized the DNA-binding domain of EBNA-1 and have examined the contributions of other domains of the protein to EBNA-1 transactivation activity. A Bal31 deletional analysis of the carboxy-terminal region of EBNA-1 identified a core DNA-binding domain located between amino acids 493 and 584. Column chromatographic, sedimentation, and cross-linking studies indicated that EBNA-1 exists in solution as a dimer. Mobility retardation assays using in vitro-translated variants of EBNA-1 showed that the active DNA-binding form of EBNA-1 is also a dimer. In short-term cotransfections, a pFRTK-CAT target containing EBNA-1-binding sites from the EBV origin of plasmid replication, ori-P, was transactivated by a carboxy-terminal EBNA-1 construction (amino acids 450 to 641) that also carried a c-myc nuclear localization signal. These reconstruction experiments demonstrated that a transactivation domain exists within the carboxy-terminal region of EBNA-1, that transactivation is more efficient when a nuclear localization signal is present, and that the natural karyophilic signal lies outside of the carboxy-terminal 191 amino acids. To identify the EBNA-1 nuclear localization signal, small oligonucleotides representing EBNA-1 sequences that encode clusters of basic peptides were transferred into two different vectors expressing cytoplasmic proteins (pyruvate kinase and herpes simplex virus delta IE175 protein) and the cellular locations of the fusion constructions were determined by immunofluorescence staining of transfected cells. In this way we identified a functional nuclear localization signal, Leu-Lys-Arg-Pro-Arg-Ser-Pro-Ser-Ser, encompassing amino acids 379 to 386 of the EBNA-1 protein.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Ambinder R. F., Shah W. A., Rawlins D. R., Hayward G. S., Hayward S. D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990 May;64(5):2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. N., Dong D. L., Hayward G. S., Hayward S. D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990 Jul;64(7):3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden T., Lupton S., Levine A. J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989 Jul;63(7):3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Lee W. M. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem. 1989 Oct 25;264(30):18019–18023. [PubMed] [Google Scholar]
- Dasmahapatra B., Rozhon E. J., Schwartz J. pBD7, a novel cell-free expression vector with efficient translation initiation signal. Nucleic Acids Res. 1987 May 11;15(9):3933–3933. doi: 10.1093/nar/15.9.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. D., Triezenberg S. J., McKnight S. L. Expression of a truncated viral trans-activator selectively impedes lytic infection by its cognate virus. Nature. 1988 Sep 29;335(6189):452–454. doi: 10.1038/335452a0. [DOI] [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang K. T., Rawlins D. R., Rosenfeld P. J., Shero J. H., Kelly T. J., Hayward G. S. Multiple tandemly repeated binding sites for cellular nuclear factor 1 that surround the major immediate-early promoters of simian and human cytomegalovirus. J Virol. 1987 May;61(5):1559–1570. doi: 10.1128/jvi.61.5.1559-1570.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. H., Hayward S. D., Rawlins D. R. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J Virol. 1989 Jan;63(1):101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kimball A. S., Milman G., Tullius T. D. High-resolution footprints of the DNA-binding domain of Epstein-Barr virus nuclear antigen 1. Mol Cell Biol. 1989 Jun;9(6):2738–2742. doi: 10.1128/mcb.9.6.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lawrence J. B., Villnave C. A., Singer R. H. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988 Jan 15;52(1):51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Adams A., Bjursell G., Bornkamm G. W., Kaschka-Dierich C., Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976 Apr 15;102(3):511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Lupton S., Levine A. J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985 Oct;5(10):2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Heller M., Petti L., O'Shiro E., Kieff E. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science. 1984 Dec 14;226(4680):1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- Milman G., Hwang E. S. Epstein-Barr virus nuclear antigen forms a complex that binds with high concentration dependence to a single DNA-binding site. J Virol. 1987 Feb;61(2):465–471. doi: 10.1128/jvi.61.2.465-471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman G., Scott A. L., Cho M. S., Hartman S. C., Ades D. K., Hayward G. S., Ki P. F., August J. T., Hayward S. D. Carboxyl-terminal domain of the Epstein-Barr virus nuclear antigen is highly immunogenic in man. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6300–6304. doi: 10.1073/pnas.82.18.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare P., Hayward G. S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985 Mar;53(3):751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Polvino-Bodnar M., Kiso J., Schaffer P. A. Mutational analysis of Epstein-Barr virus nuclear antigen 1 (EBNA 1). Nucleic Acids Res. 1988 Apr 25;16(8):3415–3435. doi: 10.1093/nar/16.8.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rawlins D. R., Milman G., Hayward S. D., Hayward G. S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985 Oct;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Reisman D., Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986 Nov;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. L., Richardson W. D., Smith A. E. The effect of protein context on nuclear location signal function. Cell. 1987 Jul 31;50(3):465–475. doi: 10.1016/0092-8674(87)90500-9. [DOI] [PubMed] [Google Scholar]
- Sample J., Hummel M., Braun D., Birkenbach M., Kieff E. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5096–5100. doi: 10.1073/pnas.83.14.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter S. D., Zweig M., Hampar B. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect Immun. 1981 Dec;34(3):684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. E., Kalderon D., Roberts B. L., Colledge W. H., Edge M., Gillett P., Markham A., Paucha E., Richardson W. D. The nuclear location signal. Proc R Soc Lond B Biol Sci. 1985 Oct 22;226(1242):43–58. doi: 10.1098/rspb.1985.0078. [DOI] [PubMed] [Google Scholar]
- Speck S. H., Strominger J. L. Analysis of the transcript encoding the latent Epstein-Barr virus nuclear antigen I: a potentially polycistronic message generated by long-range splicing of several exons. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8305–8309. doi: 10.1073/pnas.82.24.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Marsh K., Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985 Feb;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989 Jun;63(6):2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers W. P., Grogan E. A., Shedd D., Robert M., Liu C. R., Miller G. Stable expression in mouse cells of nuclear neoantigen after transfer of a 3.4-megadalton cloned fragment of Epstein-Barr virus DNA. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5688–5692. doi: 10.1073/pnas.79.18.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokenski D. A., Yates J. L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989 Jun;63(6):2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. 1985 Feb 28-Mar 6Nature. 313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]