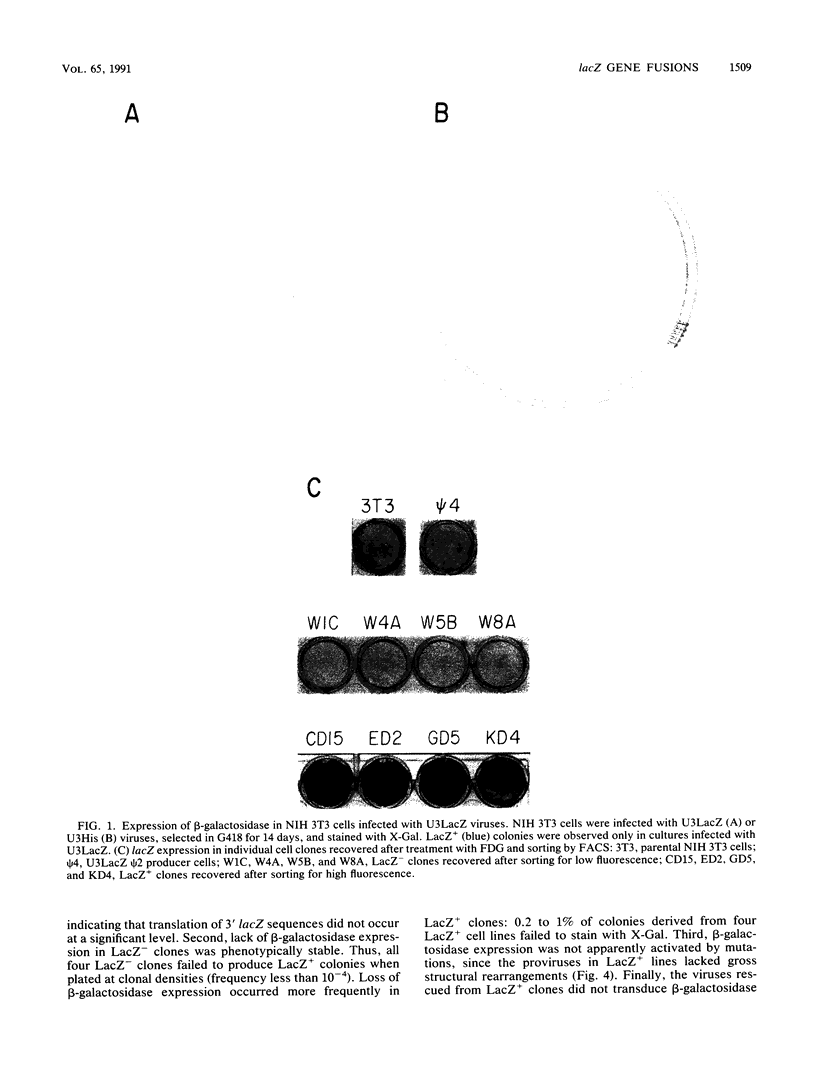
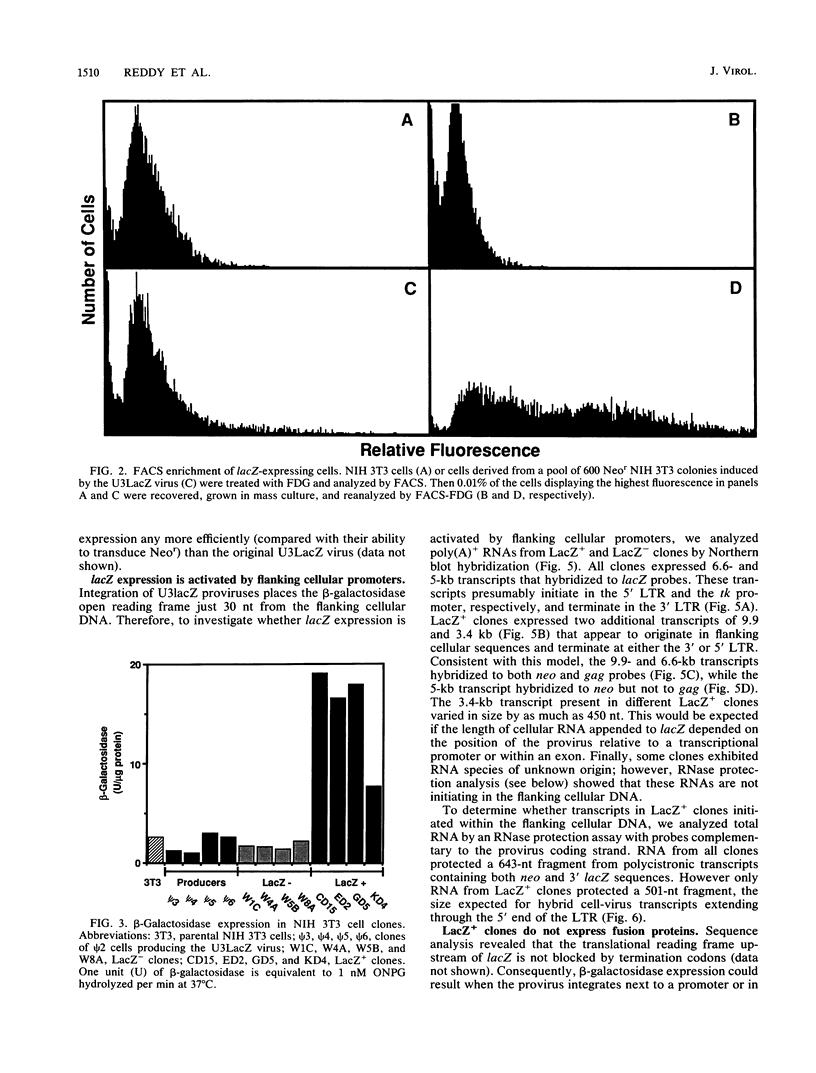
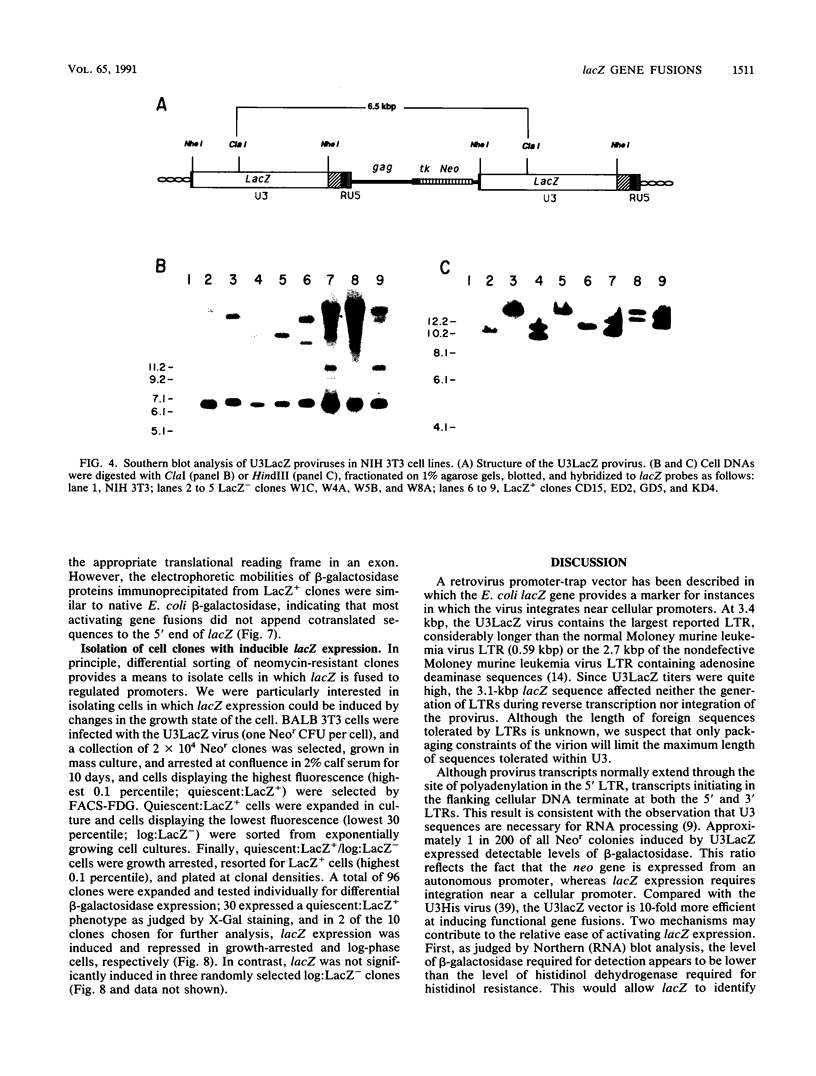
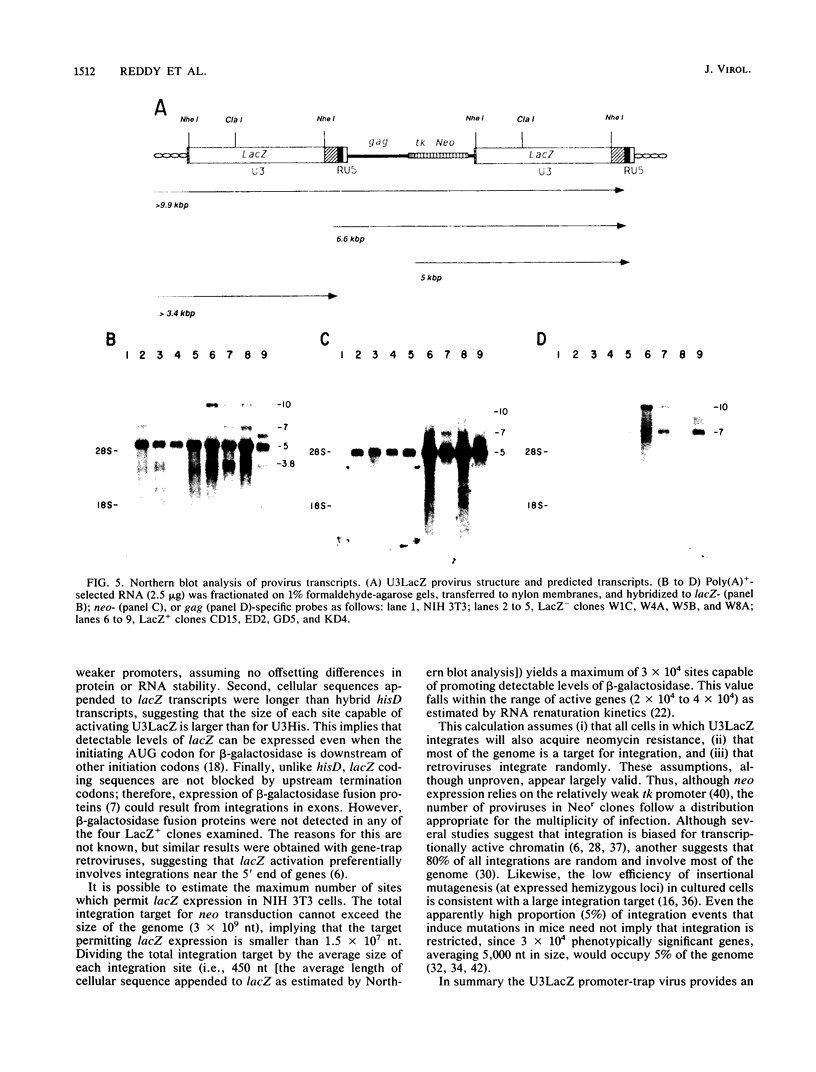
Abstract
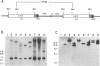
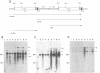
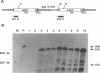
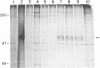
A retrovirus promoter-trap vector (U3LacZ) has been developed in which Escherichia coli lacZ coding sequences were inserted into the 3' long terminal repeat (LTR) of an enhancerless Moloney murine leukemia virus. The U3LacZ virus contains the longest reported LTR (3.4 kbp); nevertheless, lacZ sequences did not interfere with the ability of the virus to transduce a neomycin resistance gene expressed from an internal promoter. Duplication of the LTR placed lacZ sequences in the 5' LTR just 30 nucleotides from the flanking cellular DNA. Approximately 0.4% of integrated proviruses expressed beta-galactosidase as judged by 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-Gal) staining, and individual clones expressing lacZ were isolated by fluorescence-activated cell sorting. In all clones examined, beta-galactosidase expression resulted from the fusion of lacZ sequences to transcriptional promoters located in the flanking cellular DNA. Furthermore, by differential sorting of neomycin-resistant cell populations, clones were isolated in which lacZ expression was induced and repressed in growth-arrested and log phase cells, respectively.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen N. D., Cran D. G., Barton S. C., Hettle S., Reik W., Surani M. A. Transgenes as probes for active chromosomal domains in mouse development. Nature. 1988 Jun 30;333(6176):852–855. doi: 10.1038/333852a0. [DOI] [PubMed] [Google Scholar]
- Barklis E., Mulligan R. C., Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986 Nov 7;47(3):391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- Bellofatto V., Shapiro L., Hodgson D. A. Generation of a Tn5 promoter probe and its use in the study of gene expression in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1035–1039. doi: 10.1073/pnas.81.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Shepherd S., Lee K., McCall K., Barbel S., Ackerman L., Carretto R., Uemura T., Grell E. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989 Sep;3(9):1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Brenner D. G., Lin-Chao S., Cohen S. N. Analysis of mammalian cell genetic regulation in situ by using retrovirus-derived "portable exons" carrying the Escherichia coli lacZ gene. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5517–5521. doi: 10.1073/pnas.86.14.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. A promoterless retroviral vector indicates that there are sequences in U3 required for 3' RNA processing. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1197–1201. doi: 10.1073/pnas.84.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Frederiksen K., Jat P. S., Valtz N., Levy D., McKay R. Immortalization of precursor cells from the mammalian CNS. Neuron. 1988 Aug;1(6):439–448. doi: 10.1016/0896-6273(88)90175-4. [DOI] [PubMed] [Google Scholar]
- Gossler A., Joyner A. L., Rossant J., Skarnes W. C. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989 Apr 28;244(4903):463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- Hantzopoulos P. A., Sullenger B. A., Ungers G., Gilboa E. Improved gene expression upon transfer of the adenosine deaminase minigene outside the transcriptional unit of a retroviral vector. Proc Natl Acad Sci U S A. 1989 May;86(10):3519–3523. doi: 10.1073/pnas.86.10.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr W. G., Nolan G. P., Serafini A. T., Herzenberg L. A. Transcriptionally defective retroviruses containing lacZ for the in situ detection of endogenous genes and developmentally regulated chromatin. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 2):767–776. doi: 10.1101/sqb.1989.054.01.090. [DOI] [PubMed] [Google Scholar]
- King W., Patel M. D., Lobel L. I., Goff S. P., Nguyen-Huu M. C. Insertion mutagenesis of embryonal carcinoma cells by retroviruses. Science. 1985 May 3;228(4699):554–558. doi: 10.1126/science.3838595. [DOI] [PubMed] [Google Scholar]
- Koncz C., Martini N., Mayerhofer R., Koncz-Kalman Z., Körber H., Redei G. P., Schell J. High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Mouellic H., Lallemand Y., Brûlet P. Targeted replacement of the homeobox gene Hox-3.1 by the Escherichia coli lacZ in mouse chimeric embryos. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4712–4716. doi: 10.1073/pnas.87.12.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Mulligan R. C., Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983 May;33(1):153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- Miller A. D., Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986 Aug;6(8):2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan G. P., Fiering S., Nicolas J. F., Herzenberg L. A. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton P. A., Coffin J. M. Bacterial beta-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol Cell Biol. 1985 Feb;5(2):281–290. doi: 10.1128/mcb.5.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S., Ozgur K., Lu M., Chang W., Mohan S. R., Kumar C. C., Ruley H. E. Structure of the human smooth muscle alpha-actin gene. Analysis of a cDNA and 5' upstream region. J Biol Chem. 1990 Jan 25;265(3):1683–1687. [PubMed] [Google Scholar]
- Rohdewohld H., Weiher H., Reik W., Jaenisch R., Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987 Feb;61(2):336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J. R., Rubenstein J. L., Nicolas J. F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986 Dec 1;5(12):3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence S. E., Gilbert D. J., Swing D. A., Copeland N. G., Jenkins N. A. Spontaneous germ line virus infection and retroviral insertional mutagenesis in eighteen transgenic Srev lines of mice. Mol Cell Biol. 1989 Jan;9(1):177–184. doi: 10.1128/mcb.9.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. P., Fenner S., Greenoak G. E., Moran C., Coffin J. M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988 Jul 29;54(3):383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Quintrell N., Ortiz S. Retroviruses as mutagens: insertion and excision of a nontransforming provirus alter expression of a resident transforming provirus. Cell. 1981 Jul;25(1):23–36. doi: 10.1016/0092-8674(81)90228-2. [DOI] [PubMed] [Google Scholar]
- Varmus H. Retroviruses. Science. 1988 Jun 10;240(4858):1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Robinson H. L. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986 Nov;60(2):683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Sharp J. A., Summers W. C. Nucleotide sequence of the thymidine kinase gene of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., Noda T., Gray D. A., Sharpe A. H., Jaenisch R. Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell. 1990 Aug 10;62(3):425–434. doi: 10.1016/0092-8674(90)90008-3. [DOI] [PubMed] [Google Scholar]
- von Melchner H., Reddy S., Ruley H. E. Isolation of cellular promoters by using a retrovirus promoter trap. Proc Natl Acad Sci U S A. 1990 May;87(10):3733–3737. doi: 10.1073/pnas.87.10.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Melchner H., Ruley H. E. Identification of cellular promoters by using a retrovirus promoter trap. J Virol. 1989 Aug;63(8):3227–3233. doi: 10.1128/jvi.63.8.3227-3233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]