Abstract
Eukaryotic Cu,Zn superoxide dismutases (CuZnSODs) are antioxidant enzymes remarkable for their unusually stable β-barrel fold and dimer assembly, diffusion-limited catalysis, and electrostatic guidance of their free radical substrate. Point mutations of CuZnSOD cause the fatal human neurodegenerative disease amyotrophic lateral sclerosis. We determined and analyzed the first crystallographic structure (to our knowledge) for CuZnSOD from a prokaryote, Photobacterium leiognathi, a luminescent symbiont of Leiognathid fish. This structure, exemplifying prokaryotic CuZnSODs, shares the active-site ligand geometry and the topology of the Greek key β-barrel common to the eukaryotic CuZnSODs. However, the β-barrel elements recruited to form the dimer interface, the strategy used to forge the channel for electrostatic recognition of superoxide radical, and the connectivity of the intrasubunit disulfide bond in P. leiognathi CuZnSOD are discrete and strikingly dissimilar from those highly conserved in eukaryotic CuZnSODs. This new CuZnSOD structure broadens our understanding of structural features necessary and sufficient for CuZnSOD activity, highlights a hitherto unrecognized adaptability of the Greek key β-barrel building block in evolution, and reveals that prokaryotic and eukaryotic enzymes diverged from one primordial CuZnSOD and then converged to distinct dimeric enzymes with electrostatic substrate guidance.
Keywords: β-barrel, evolution, metalloenzyme, x-ray crystallography, macromolecular assembly
Superoxide dismutases (SODs) have a major role in defense against oxygen toxicity and regulation of reactive oxygen levels (1, 2). The manganese- and iron-containing enzymes, which share a common α/β fold, are the major SODs characterized in prokaryotes and mitochondria (1, 3). In contrast, the structurally unrelated Cu,Zn SODs (CuZnSODs) are widespread among eukaryotes but until recently (4) known in only a small number of Gram-negative bacterial species, including human and animal pathogens (2, 5). The eukaryotic CuZnSODs are cytoplasmic, whereas the bacterial CuZnSODs are secreted to the periplasm (4, 6). Single-site mutants of human CuZnSOD (HSOD) are associated with the fatal neurodegenerative disease, familial amyotrophic lateral sclerosis (FALS) (7, 8). The localization of FALS mutations to residues contributing to the exceptionally stable β-barrel fold and dimer interface in HSOD highlights their importance for the enzyme’s function (7). Thus, we expected that the CuZnSOD from the luminescent symbiont of Leiognathid fish (9) would share the β-barrel fold and dimer interface of the eukaryotic CuZnSODs (1, 10).
Here, we present the atomic structure of Photobacterium leiognathi (PhCuZnSOD) at 1.9-Å resolution, refined to an R factor of 19.3%. The PhCuZnSOD structure reveals a new type of dimeric assembly, distinct from that conserved among eukaryotic CuZnSODs. Comparative analyses of the dimeric interfaces and electrostatic guidance systems between P. leiognathi and eukaryotic CuZnSOD structures supports independent functional evolution in prokaryotic (P class) and eukaryotic (E class) CuZnSODs.
METHODS
Expression and Purification.
Five liters of Luria–Bertani (LB) medium with sodium ampicillin (100 mg/ml) was inoculated with Escherichia coli MC1061 containing pBR322-derived PhCuZnSOD expression plasmid, grown to midlogarithmic phase at 37°C, then induced with 1 mM isopropyl β-d-thiogalactose, and grown to stationary phase. PhCuZnSOD was expressed to high levels and was found in the periplasm and within cells. Cells were broken with a French press, DNA was precipitated in 50 mM MnCl2, and a 40–85% ammonium sulfate cut was used to precipitate PhCuZnSOD. Finally, dialysis into ice-cold 20 mM Tris·HCl, pH 8.4/50 mM NaCl/1 mM CuSO4 buffer caused PhCuZnSOD to form an isoelectric precipitate, resulting in ≈300 mg of highly purified (>99%) enzyme.
Crystallization and Phase Determination.
For crystallization experiments, PhCuZnSOD was dialyzed into 60 mM potassium phosphate (pH 6.5) and concentrated to 20 mg/ml over a 6–8000 Da cutoff membrane. Crystals of PhCuZnSOD (space group C2 with cell dimensions a = 120.7 Å, b = 87.0 Å, and c = 43.5 Å and β = 90.6°) were obtained by vapor diffusion at 20°C with 42% 2-methyl-2,4-pentanediol/60 mM potassium phosphate, pH 6.5, and improved by macroseeding (11). Initial low-resolution electron density maps calculated with diffraction data from three heavy atom derivatives [1 mM K2IrCl6, 1 mM platinum(ethylenediamine)dichloride, and 10 mM K2OsCl6] showed the subunit and dimer boundaries for three subunits (1 and 1/2 dimers) in the asymmetric unit. A 1.9-Å resolution diffraction data set, which consisted of 116,490 observations for 30,277 unique reflections (83% complete, Rsym = 4.9%), was collected from one crystal on a Xentronics area detector mounted on a Rigaku rotating anode operating at 40 kV × 80 mA. Initial phases were obtained by molecular replacement using an entire subunit with metal ions of the Cys6Ala mutant bovine CuZnSOD (BSOD) coordinates (PDB code 3SOD) (12) as a search model with the amore program package (13). The highest peak in the translation function of one PhCuZnSOD subunit was automatically used to position the two other subunits with the phased translation function by screening all other rotation peaks. This solution matched the clear noncrystallographic axes observed in a self-rotation function (κ section 120° and 180°) and correctly located the three heavy atom positions in an iridium derivative (K2IrCl6, 80% complete at 2.4 Å, Rsym = 5.7%, and Riso = 20%).
Crystallographic Refinement and Model Fitting.
Rigid-body refinement of the three subunits gave a correlation coefficient of 22% and an R factor of 48% for diffraction data from 15-Å to 4-Å resolution. Refinement of the molecular replacement model with x-plor (14) against native diffraction data from 6-Å to 2.8-Å resolution gave an R factor of 33% and an R-free of 53% (5% of the reflections). The model was then manually rebuilt into both 2Fo − Fc and Fo − Fc electron density maps with the graphics program turbo-frodo (15). Addition of water molecules during subsequent refitting and refinement cycles without noncrystallographic symmetry restraints gave a final R factor of 19.3% for 29,110 reflections (all data) between 6-Å and 1.9-Å resolution, and a free R factor of 25%. The final model contains 150–151 residues for each subunit with overall deviations from ideal geometry of 0.005 Å for bond distances and 1.4° for bond angles. For the three PhCuZnSOD subunits (3314 protein atoms), the rms deviation is 0.27 Å for all atoms and 0.2 Å for backbone atoms. Backbone dihedral angles all lie in allowed regions of the Ramachandran diagram. Pro-135, which is the only cis-Pro out of nine, ends the shortened loop 7,8. Temperature factors average 18 Å2 for main chain, 21 Å2 for side chain, 22 Å2 for three copper ions, 15 Å2 for three zinc ions, and 31 Å2 for 361 solvent molecules.
Structural Analysis.
The programs pqms and ms (16) were used to calculate solvent-accessible molecular surfaces with a 1.4-Å probe and buried molecular surfaces with a 1.6-Å probe. Electrostatic potential, calculated with the program delphi (17) using parse parameters for radii and atomic charges and dielectric constants of 2 for solute and 80 for solvent, was mapped to the solvent-accessible molecular surface from positions 1.4 Å out along surface normals. Each PhCuZnSOD subunit has a net charge of +1.
Differential Scanning Calorimetry and Gel Filtration Chromatography.
Temperature scans at 1°/min were obtained on a Microcal-2 with PhCuZnSOD at 3 mg/ml in 100 mM potassium phosphate (pH 7.8). The profile consisted of a major peak at 71°C with a small peak at Tm 62–67°C, which is probably from damaged protein. The denaturation is completely irreversible as shown by the rescan after heating to 100°C. Gel filtration chromatography on a Superose 12 HR 10/30 (Pharmacia) column, equilibrated with 60 mM potassium phosphate, pH 6.5/150 mM NaCl, gave a single peak corresponding to a PhCuZnSOD dimer with an apparent molecular mass of 31 kDa.
RESULTS AND DISCUSSION
Novel P-Class CuZnSOD Dimer Interface.
PhCuZnSOD shares the eight-stranded Greek key β-barrel fold characteristic of the E-class CuZnSODs (18). Both P-class and E-class CuZnSODs, furthermore, form homodimers that have a twofold symmetry axis roughly parallel to the β-barrel axis, preserve the opposing orientation of the two active sites within the dimer, and have similar overall dimensions (PhCuZnSOD ≈70 Å × 30 Å × 30 Å versus bovine CuZnSOD (BSOD) ≈60 Å × 30 Å × 30 Å) (Fig. 1). Despite these overall similarities between P-class and E-class enzymes, the dimer interface in PhCuZnSOD is formed from β-strands that are diametrically opposite those used in the E-class CuZnSODs. The PhCuZnSOD dimer juxtaposes β-strands 5e and 4f across the dimer interface (Fig. 1a), whereas the classic E-class CuZnSODs dimer interface juxtaposes N- and C-terminal β-strands 1a and 8h (Fig. 1b). The different packing arrangements are reminiscent of the front-to-front versus back-to-back dimerization of the immunoglobulin variable versus constant domains (20). However, the immunoglobulin variable domain packing is needed to create the antigen-binding site, while both P-class and E-class dimer packing produce essentially the same orientation of CuZnSOD active sites. In the P-class CuZnSODs, conserved hydrophobic residues of the classic E-type interface have been substituted with charged or hydrophilic residues or deleted (Fig. 2). Likewise, residues of the PhCuZnSOD interface that are sequence-conserved among P-class CuZnSODs are different in E-class CuZnSODs, suggesting these two distinct dimer assemblies are mutually exclusive. Distinct homodimeric assemblies from like subunits have also been reported within the hemoglobin family (clam versus vertebrate hemoglobins; ref. 26) and between the α- and β-chemokine families (27). Documenting this unexpected variance in dimer assembly for the CuZnSOD β-barrel supports the hypothesis that this phenomenon may provide biologically important evolutionary variation among other highly conserved dimeric assemblies.
Figure 1.
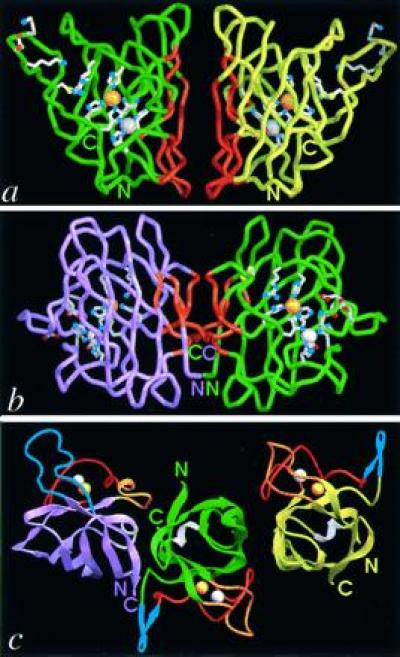
Comparison of the folds and dimer assemblies of P-class PhCuZnSOD with E-class bovine CuZnSOD (BSOD). PhCuZnSOD dimer Cα trace (green and yellow subunits) (a) and BSOD dimer (18) Cα trace (purple and green subunits) (b) viewed perpendicular to the dimer twofold axis and down into the active-site Cu for one subunit. Functionally important side chains for the active site, disulfide cysteines, and electrostatic recognition residues are displayed with white bonds and atom-colored spheres (red oxygen, blue nitrogen, and yellow sulfur atoms). Dimer interface regions in both P- and E-class CuZnSOD are highlighted (orange), and active-site metal ions are shown as spheres for the copper (orange) and zinc (silver). (c) PhCuZnSOD dimer (Right) and BSOD dimer (18) (Left) shown following superposition of one subunit (Cα atoms) from each. The central subunit (green) is displayed for PhCuZnSOD only and the subunit color code matches a and b. Loops are color-coded by function: blue for contributing to electrostatic attraction of substrate (loop 7,8 in BSOD, Upper Left, and SS loop in PhCuZnSOD, Upper Right), orange for forming the dimer interface (SS loop in BSOD and loop 7,8 in PhCuZnSOD), red for binding Zn ion, and white for the second Greek key connection across the end of the β-barrel. For loop nomenclature, see Fig. 2 legend. The view is down the dimer twofold axis and approximately 90° away from that in a and b. Figs. 1, 3a, 4, and 5 were generated with the Application Visualization System (AVS) (Advanced Visual Systems, Waltham, MA) and Figs. 1c and 3a were made with ribbons (19) implemented in AVS by Alexandre Shah.
Figure 2.

Sequence and secondary structure of PhCuZnSOD compared with other P- and E-class CuZnSODs. Sequences of two bacterial CuZnSODs, PhCuZnSOD (21, 22), and HpCuZnSOD (Haemophilus parainfluenzae) (23), are aligned with sequences of eukaryotic bovine (BSOD, ref. 24) and human (HSOD, ref. 25) CuZnSODs, based on our structural comparison. Bacterial enzyme sequences share little homology with eukaryotic sequences [e.g., 28% identity between PhCuZnSOD and BSOD (10)] but display mutual sequence similarities of about 70% and share a common pattern of insertions/deletions (2, 23). For both the PhCuZnSOD and BSOD sequences, residues in the dimer interface are color-coded and underlined in orange, and those involved in the long-range electrostatic attraction of the substrate anion are in blue. The common CuZnSOD secondary structural elements are represented between the two pairs of sequences, with bounds for PhCuZnSOD. β-strands are named by letter in sequence order and by number clockwise around the β-barrel (18), and those specific to PhCuZnSOD are shown above the sequence. Cu and Zn ligands are marked with an asterisk, the buried Asp with ▪, and the catalytically important Arg with ▾. Fig. 2 was generated with framemaker (Frame Technology Corporation, San Jose, CA).
In the extensive symmetric PhCuZnSOD dimer interface (Fig. 3a), which has overall dimensions of 25 × 19 Å, side-chain contributions predominate (≈90%) over main-chain contacts (≈10%), and hydrophobic interactions (≈70%) over hydrophilic (≈30%), resulting in 740 Å2 per subunit buried to a sphere of 1.6-Å radius. Trp-83 in the Zn loop is central to the largest hydrophobic cluster at the dimer interface, which also includes large contributions from Pro-106, Leu-108, and Pro-110 from β-strand 4f and the following Greek key loop, as well as smaller contributions from Val-29 and Phe-81 (Fig. 3b). The top and bottom of the interface are sealed by a smaller hydrophobic cluster formed between Met-41 and Phe-96, and a hydrophilic cluster in which the Asp-85 side chain forms a hydrogen bond to Tyr-26 and a salt bridge to Lys-25 across the interface, and the Asn-24 side chain hydrogen bonds to the Trp-83 main chain. The dimer interface is completed by an unusual ring of 11 hydrogen-bonded solvent molecules buried within a cavity filling the dimer interface between the two hydrophobic clusters (Fig. 3a).
Figure 3.
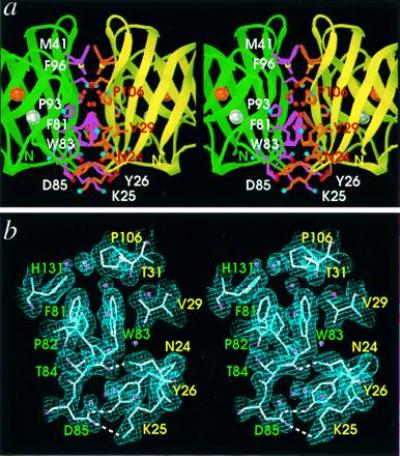
PhCuZnSOD dimer interface structure and electron density. Stereo pairs are oriented with the twofold axis vertical to match Fig. 1a. Labels indicate one-letter amino acid code and residue number. (a) Close-up displaying key side chains of the dimer interface with light purple bonds and white labels (for green subunit) or orange bonds and red labels (for yellow subunit) and atom-colored spheres (see Fig. 1a). The Asn-105 side chain and main-chain atoms of Ala-94, Asn-105, Pro-106, and Leu-108 hydrogen bond with the internal ring of buried water molecules, shown as red spheres. Active-site metal ions are shown as spheres for the copper (orange) and zinc (silver). (b) Refined model (atom-colored), solvent water molecules (pink spheres), and 2Fo − Fc electron density map (contoured at 1.0 SD above the mean) at the dimer interface. Hydrogen bonds spanning the interface are shown as white dashed lines and residue labels are color-coded by subunit. b was generated with turbo-frodo (15).
In contrast to the PhCuZnSOD interface, the BSOD dimer interface (20), which has dimensions of 18 × 15 Å and 540 Å2 of buried surface area per subunit, has more main-chain contributions (≈35%), two pairs of main-chain hydrogen bonds, and only two internal solvent molecules. Although gel-filtration experiments unambiguously revealed the presence of a PhCuZnSOD dimer in solution (data not shown), both the major contributions of side-chain interactions and the buried water ring within the PhCuZnSOD dimer interface suggest more flexibility and less stability than for the classic E-class dimer. This presumption is supported by differential scanning calorimetry data (data not shown) demonstrating a thermal unfolding transition of 71°C for PhCuZnSOD, in contrast to 88°C for bovine CuZnSOD (28). An extreme form of this decreased dimer stability of the P-class enzymes may be evident in E. coli CuZnSOD (29), which can apparently be purified as a monomer (30).
Distinct Electrostatic Guidance of Substrate to Conserved Active Site.
The catalytic Cu ion of PhCuZnSOD is solvent-exposed at the molecular surface and liganded by His-45, -47, -70, and -125, whereas the Zn is buried and liganded by His-70, -79, and -88 and Asp-91 (Figs. 1a and 2). In both P- and E-class CuZnSODs the ligands of Cu and Zn are identical, occur in the same order within sequence, and generate similar ligand geometry as well as the same intra (6 Å) and inter (30 Å) subunit metal ion distances. Like E-class CuZnSODs, PhCuZnSOD conserves the active-site Arg (Arg-144) and the buried aspartic acid (Asp-129) that hydrogen bonds His ligands of both the Cu (His-45) and Zn (His-79) ions (Fig. 2).
In contrast to the conserved ligand sequence order, active-site geometry, and adjacent positive side chain of Arg-144, the structure and character of the channel responsible for long-range electrostatic recognition are dramatically different between P- and E-class CuZnSODs (Fig. 4 a and b). The sequence basis for this difference is the 4-residue deletion starting after amino acid 137 (Fig. 2). The diffusion-limited rate of CuZnSOD-catalyzed superoxide dismutation (31) is attributable to two highly conserved features of the E-class enzymes: direct electrostatic stabilization by the conserved active-site Arg (32–35) and long-range electrostatic recognition and guidance (34, 35). PhCuZnSOD lacks homologues to BSOD Glu-130, Glu-131, and Lys-134 in E-class CuZnSOD loop 7,8, which form the ordered hydrogen-bonding network critical to long-range guidance of superoxide (34, 36). Consequently, the strategy for electrostatic attraction of the free radical anion must differ in P- and E-class CuZnSODs. In PhCuZnSOD, the active-site architecture (Fig. 4 a and b) and electrostatic potential (Fig. 4 c and d) suggest that Lys-57, Asp-58, and Lys-60, which are present in the insert forming an extension of the SS loop (Fig. 4a), are major players in the electrostatic recognition and guidance of substrate in P-class CuZnSODs. This 8-residue loop insertion is absent in E-class CuZnSODs and faces loop 7,8 from across the active site channel (Fig. 4 a and b). Lys-57, in particular, is a candidate for site-directed mutagenesis studies testing for a role in electrostatic guidance, analogous to those performed on similar residues in E-class CuZnSODs (36). These alternative recognition strategies in P-class and E-class CuZnSODs conserve the highly positive electrostatic potential around the Cu active sites (Fig. 4 c and d), which are located similar distances apart on opposing sides of the dimers.
Figure 4.
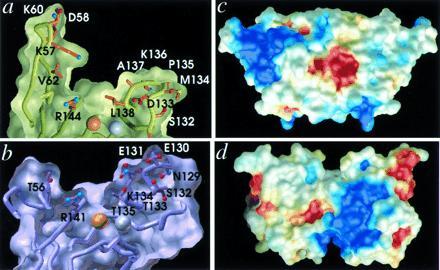
Conservation and variation of the active channel shape and electrostatic potential between P- and E-class CuZnSODs. (a) Active-site channel cross-section for P-class PhCuZnSOD and (b) E-class BSOD. Whereas the shape and dimensions are conserved between the two classes (at left, PhCuZnSOD Val-62 coincides with BSOD Thr-56 in the SS loop, and at right, PhCuZnSOD Leu-138 coincides with BSOD Thr-135 in loop 7,8), the structural location, conformation, and identity of the residues producing the long-range electrostatic attraction of the substrate anion are dramatically different, suggesting separate divergent evolution followed by convergence. Electrostatic potential mapped onto the molecular surface of the PhCuZnSOD dimer (c) (oriented to match Fig. 1a) and the BSOD dimer (d) (oriented to match Fig. 1b) and colored as blue positive potential (>3kT/q) and red negative potential (<−3kT/q). The network of charged residues Lys-57, Asp-58, and Lys-60, in the extended PhCuZnSOD SS loop (upper left) forms the positive potential for substrate attraction, whereas Glu-131, Glu-132, and Lys-134 in the extended loop 7,8 (lower right) provide the equivalent function in BSOD.
β-Barrel Flexibility.
The β-barrel is a common and versatile super secondary structure found in proteins as diverse as enzymes, immunoglobulins, and viral capsids (20, 37). The inherent flexibility of the CuZnSOD β-barrel can accommodate numerous local changes in structure through concerted shifts in surrounding residues (10, 12). The overall rms deviation from superposition of 132 Cα atoms between the PhCuZnSOD and BSOD structures (Fig. 5a) is only 1.45 Å with larger differences (up to 2.5 Å) in the first two β-strands, where sequence homology between the two CuZnSODs is weak (Fig. 2). In the PhCuZnSOD structure, however, we find three novel examples of the adaptability of the CuZnSOD β-barrel fold: the essential intrasubunit disulfide bond, the Zn loop stabilization, and the second Greek key connection (Fig. 5).
Figure 5.
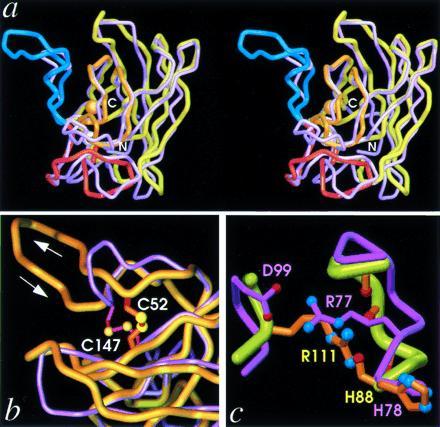
β-Barrel rearrangements. (a) Stereo pair superposition of the Cα traces of PhCuZnSOD (yellow) and BSOD (purple) subunits viewed perpendicular to the β-barrel axis (approximately 90° from Fig. 1a), showing the large insertion in PhCuZnSOD SS loop (blue, left) as well as distortions in the bottom β-sheet (center and right). The functional elements and metal ions of PhCuZnSOD are color-coded as in Fig. 1c. (b) Close-up view of SS loop in superimposed subunits, as for a. PhCuZnSOD disulfide (orange bonds with yellow spheres) Cys-52 (aligned beneath BSOD Asn-51) is located on the other side of the SS loop relative to BSOD disulfide (magenta bonds with yellow spheres) Cys-55 (both at top), while the position of PhCuZnSOD Cys-147 (BSOD Cys-144) is conserved (both below). (c) Close-up view of Zn loop in superimposed subunits, as for a, showing the functional equivalence of PhCuZnSOD Arg-111 (orange bonds and atom-colored spheres) and BSOD Arg-77 (light purple bonds and atom-colored spheres) in stabilizing the Zn loop. BSOD Arg-77 forms a salt bridge with Asp-99 (left), while PhCuZnSOD Arg-111 hydrogen bonds with four main-chain carbonyl oxygen atoms (right).
In the extended SS loop sequence in PhCuZnSOD, relative to E-class SODs, disulfide Cys-52 of PhCuZnSOD aligns positionally with BSOD Asn-51 but substitutes structurally for BSOD Cys-55 on the other side of the SS loop (Fig. 5b). This reveals a danger in the common practice of using disulfide-bonded cysteines as anchor points for homology modeling experiments, regardless of their evolutionary relatedness. Stabilization of the Zn ligand loop in both P and E classes involves a critical Arg side chain conserved within, but not, between the two classes (PhCuZnSOD Arg-111 versus BSOD Arg-77) and represents another example of loop rearrangements within a β-barrel fold. PhCuZnSOD Arg-111 is hydrogen-bonded to carbonyl oxygen atoms of Zn loop residues Phe-81, Thr-84, Asp-85, Asn-87, and His-88, whereas a distinct buried salt bridge forms between E-class CuZnSOD Arg-77 and Asp-99 side chains to accomplish similar Zn loop stabilization (Fig. 5c). Finally, the Greek key loop, which joins β-strand 4f and 7g and is closed by main-chain β-strand hydrogen bonds in E-class CuZnSODs, is deleted in PhCuZnSOD and replaced by a single α-helical turn, capped by Thr-113 (see Fig. 1c).
Structural Implications for Evolution, Function, and Stability.
Our structural results on the dimer interface and electrostatic guidance system, like the results from phylogenetic analysis (38), are not consistent with the previously proposed gene transfer origin for PhCuZnSOD, in which the symbiotic bacterium acquired the CuZnSOD gene from its eukaryotic host, the Leiognathid fish (39). Furthermore, the independently evolved dimer and electrostatic loop in PhCuZnSOD date the existence of the primordial CuZnSOD to more than 1.5 billion years ago and prior to the origin of eukaryotes as also suggested by phylogenetic analysis (2). Thus, the origin of CuZnSOD likely coincides with the era when significant oxygen began to accumulate in the atmosphere, which supports the hypothesis that CuZnSODs play a major biological role in defenses against reactive oxygen species.
We hypothesize that the putative SOD precursor first evolved divergently to form the distinct P- and E-class CuZnSODs, which then convergently optimized to distinct enzyme dimers with symmetry-equivalent loops responsible for electrostatic recognition of the superoxide anion substrate. As both SOD classes share conserved ligand stereochemistry and sequence order, the two metal ion active site, but not the dimer interface or loops providing electrostatic guidance, would have preexisted in the original SOD precursor. These structural predictions for SOD evolution suggest CuZnSOD should occur in many prokaryotes, as recently suggested from biochemical (29) and molecular biological (4) studies. These new results are consistent with our previous hypothesis (10) that a six-stranded β-barrel with twofold internal symmetry was the evolutionary precursor to present day eight-stranded β-barrels found in CuZnSODs. In particular, the new P-class structure suggests an unexpected versatility in the internally symmetric precursor: possible independent functional evolution of each half of the six-stranded β-barrel. Primordial SOD gene duplication and fusion allowed separate optimal diversification of the two halves, which is impossible in homodimers coded by a single gene. These new structural results may, therefore, explain the extreme adaptability of folds, such as the Greek key β-barrel, that occur in many different types of proteins and can be generated by gene duplication-fusion events due to their internal twofold symmetry. Such adaptability may also explain the choice of folds favored in biology out of the total large set of possible folds.
The existence of distinct structural pathways to an active CuZnSOD dimer expands our knowledge of the structural features necessary and sufficient for CuZnSOD activity. Moreover, our structural characterization of two modern enzyme classes reveals both an unexpected flexibility in the optimization of SOD function and the evident biological importance of the enzyme’s dimer assembly and electrostatic recognition, as these evolved twice in distinct but structurally related forms. However, the P-class interface, which emphasizes side-chain interactions and includes a buried water ring, is expected to be more flexible and less stable, as also predicted for FALS mutants (7). If the greater stability of the E-class enzymes reflects requirements from the higher oxygen stress in eukaryotes, then comparisons of these differences may aid our understanding of the defect in FALS SOD mutants. The discovery of such novel insights in an enzyme as thoroughly studied as CuZnSOD argues convincingly for analyzing a diversity of species in structure–function studies of any protein family.
Acknowledgments
We thank Robert A. Hallewell for construction of the expression plasmid, Michael J. Hickey for gel filtration experiments, Jean-Luc Pellequer for help in electrostatic potential calculations, and Duncan E. McRee and Katrina T. Forest for discussions. This work was in part supported by grants from the National Science Foundation (MCB-9220055 to H.M.S) and the National Institutes of Health (GM-37684 to E.D.G. and GM-39345 to J.A.T.).
Footnotes
Abbreviations: SOD, superoxide dismutase; CuZnSOD, Cu,Zn superoxide dismutase; PhCuZnSOD, Photobacterium leiognathi CuZnSOD; BSOD, bovine CuZnSOD; P class, prokaryotic CuZn SOD; E class, eukaryotic CuZnSOD; FALS, familial amyotrophic lateral sclerosis.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Brookhaven National Laboratory, Upton, NY 11973 (reference 1YAI).
References
- 1.Beyer W, Imlay J, Fridovich I. Prog Nucleic Acid Res Mol Biol. 1991;40:221–253. doi: 10.1016/s0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- 2.Bordo D, Djinovic K, Bolognesi M. J Mol Biol. 1994;238:366–386. doi: 10.1006/jmbi.1994.1298. [DOI] [PubMed] [Google Scholar]
- 3.Grace S C. Life Sci. 1990;47:1875–1886. doi: 10.1016/0024-3205(90)90399-c. [DOI] [PubMed] [Google Scholar]
- 4.Kroll J S, Langford P R, Wilks K E, Keil A D. Microbiology. 1995;141:2271–2279. doi: 10.1099/13500872-141-9-2271. [DOI] [PubMed] [Google Scholar]
- 5.St. John G, Steinman H M. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinman H M, Ely B. J Bacteriol. 1990;175:1198–1202. doi: 10.1128/jb.175.4.1198-1202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H-X, Hentati A, Tainer J A, Iqbal Z, Cayabyab A, et al. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 8.Brown R H., Jr Cell. 1995;80:687–692. doi: 10.1016/0092-8674(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 9.Puget K, Michelson A M. Biochem Biophys Res Commun. 1974;58:830–838. doi: 10.1016/s0006-291x(74)80492-4. [DOI] [PubMed] [Google Scholar]
- 10.Getzoff E D, Tainer J A, Stempien M M, Bell G I, Hallewell R A. Proteins. 1989;5:322–336. doi: 10.1002/prot.340050408. [DOI] [PubMed] [Google Scholar]
- 11.Redford S M, McRee D E, Getzoff E D, Steinman H M, Tainer J A. J Mol Biol. 1990;212:449–451. doi: 10.1016/0022-2836(90)90323-E. [DOI] [PubMed] [Google Scholar]
- 12.McRee D E, Redford S M, Getzoff E D, Lepock J R, Hallewell R A, Tainer J A. J Biol Chem. 1990;265:14234–14241. doi: 10.2210/pdb3sod/pdb. [DOI] [PubMed] [Google Scholar]
- 13.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 14.Brünger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 15.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1989. pp. 77–78. [Google Scholar]
- 16.Connolly M L. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 17.Sitkoff D, Sharp K A, Honig B. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
- 18.Tainer J A, Getzoff E D, Beem K M, Richardson J S, Richardson D C. J Mol Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- 19.Carson P. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 20.Getzoff E D, Tainer J A, Olson A J. Biophys J. 1986;49:191–206. doi: 10.1016/S0006-3495(86)83634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffens G-J, Bannister J V, Bannister W H, Flohé L, Gunzler W A, Kim S-M, Otting F. Hoppe-Seyler’s Z Physiol Chem. 1983;364:675–690. doi: 10.1515/bchm2.1983.364.1.675. [DOI] [PubMed] [Google Scholar]
- 22.Steinman H M. J Biol Chem. 1987;262:1882–1887. [PubMed] [Google Scholar]
- 23.Kroll J S, Langford P R, Loynds B M. J Bacteriol. 1991;173:7449–7457. doi: 10.1128/jb.173.23.7449-7457.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinman H M, Naik V R, Abernethy J L, Hill R L. J Biol Chem. 1974;249:7326–7338. [PubMed] [Google Scholar]
- 25.Sherman L, Dafni N, Lieman-Hurwitz J, Groner Y. Proc Natl Acad Sci USA. 1983;80:5465–5469. doi: 10.1073/pnas.80.18.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royer W E, Love W E, Fenderson F F. Nature (London) 1985;316:277–280. doi: 10.1038/316277a0. [DOI] [PubMed] [Google Scholar]
- 27.Covell D G, Smythers G W, Gronenborn A M, Clore G M. Protein Sci. 1994;3:2064–2072. doi: 10.1002/pro.5560031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepock J R, Frey H E, Hallewell R A. J Biol Chem. 1990;265:21612–21618. [PubMed] [Google Scholar]
- 29.Benov L T, Fridovich I. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 30.Battistoni A, Rotilio G. FEBS Lett. 1995;374:199–202. doi: 10.1016/0014-5793(95)01106-o. [DOI] [PubMed] [Google Scholar]
- 31.Klug D, Rabani J, Fridovich I. J Biol Chem. 1972;247:4839–4842. [PubMed] [Google Scholar]
- 32.Tainer J A, Getzoff E D, Richardson J S, Richardson D C. Nature (London) 1983;306:284–287. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- 33.Beyer W F, Fridovich I, Mullenbach G T, Hallewell R A. J Biol Chem. 1987;262:11182–11187. [PubMed] [Google Scholar]
- 34.Getzoff E D, Tainer J A, Weiner P K, Kollman P A, Richardson J S, Richardson D C. Nature (London) 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- 35.Fisher C L, Cabelli D E, Tainer J A, Hallewell R A, Getzoff E D. Proteins. 1994;19:24–34. doi: 10.1002/prot.340190105. [DOI] [PubMed] [Google Scholar]
- 36.Getzoff E D, Cabelli D E, Fisher C L, Parge H E, Viezzoli M S, Banci L, Hallewell R A. Nature (London) 1992;358:347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- 37.Woolfson D N, Evans P A, Hutchinson E G, Thornton J M. Protein Eng. 1993;6:233–245. doi: 10.1093/protein/6.5.461. [DOI] [PubMed] [Google Scholar]
- 38.Smith M W, Doolittle R F. J Mol Evol. 1992;34:175–184. doi: 10.1007/BF00182394. [DOI] [PubMed] [Google Scholar]
- 39.Martin J P, Fridovich I. J Biol Chem. 1981;256:6080–6089. [PubMed] [Google Scholar]


