Abstract
The crystal structure of the RNA dodecamer 5′-GGCC(GAAA)GGCC-3′ has been determined from x-ray diffraction data to 2.3-Å resolution. In the crystal, these oligomers form double helices around twofold symmetry axes. Four consecutive non-Watson–Crick base pairs make up an internal loop in the middle of the duplex, including sheared G·A pairs and novel asymmetric A·A pairs. This internal loop sequence produces a significant curvature and narrowing of the double helix. The helix is curved by 34° from end to end and the diameter is narrowed by 24% in the internal loop. A Mn2+ ion is bound directly to the N7 of the first guanine in the Watson–Crick region following the internal loop and the phosphate of the preceding residue. This Mn2+ location corresponds to a metal binding site observed in the hammerhead catalytic RNA.
The study of RNA structure by NMR and x-ray crystallographic methods is currently flourishing due to both improvements in methods of synthesis and purification and the impetus provided by discoveries of new biological functions of RNA. A common element of RNA secondary structure is the internal loop, an interruption in double helical RNA by a series of bases that cannot form standard Watson–Crick pairs. Internal loops are found, for example, in ribosomal RNA, ribozymes, viroids, protein regulatory sites, and SELEX-evolved RNAs. Characterization of the three-dimensional structure of internal loops and their effect on the helices that bracket them is still in an early stage. The crystal structures of several RNA oligomers incorporating symmetric internal loops have been previously determined (1–4) and shown to have continuous base pairing with formation of U·G, U·C, and U·U non-Watson–Crick pairs. The helices containing these internal loops generally retain an A-form geometry; however, the presence of tandem U·C base pairs in one structure (1) induced a dramatic widening of the major groove from about 4 Å to about 8 Å. Perturbations in regular RNA helices by internal loops may be utilized by regulatory proteins to recognize specific RNA structures such as the rev-responsive element (RRE) (5) and the iron regulatory element (IRE) (6). Recently, it has been shown that the G·U mispair responsible for recognition and aminoacylation of tRNAAla by its synthetase can be substituted by other non-Watson–Crick base pairs, implying that distortion in the helix induced by mispairing and not a particular sequence may be responsible for recognition (7).
MATERIALS AND METHODS
Synthesis, Crystallization, and Data Collection.
The dodecaribonucleotide rGGCCGAAAGGCC was synthesized by in vitro transcription with T7 RNA polymerase as described (9, 10). Crystals were grown at room temperature by the hanging drop vapor diffusion method from a solution of 2 mM RNA mixed with an equal volume of the reservoir solution consisting of 20 mM NaCl, 5 mM MnCl2, 50 mM Tris·HCl (pH 7.5), 30% PEG 400. Data were collected from plate-like crystals at the Stanford Synchrotron Radiation Laboratory, Palo Alto, CA, to 2.27-Å resolution. These crystals belong to space group P6522 with cell dimensions of a = b = 37.71 Å and c = 88.30 Å. The 1869 reflections collected to 2.27-Å resolution represented 92.4% of a complete dataset. The Rmerge for this data was 7.45% and the average F2/σ was 2.8 for all data to 2.27 Å and 3.3 for the data to 2.5 Å.
Structure Solution and Refinement.
The structure was determined by molecular replacement using the amore program package (11). A variety of search models were tested including canonical RNA helices, NMR structures, crystal structures, and models constructed from combinations of these sources. A successful molecular replacement solution was finally found using a standard A-form duplex for the four Watson–Crick base pairs and a head-to-head G·A base pair (12) as determined in a previous crystallographic study. This model was subjected to rigid body and positional refinement followed by simulated annealing with an initial temperature of 3000 K by using the program x-plor (13). The simulated annealing refinement shifted the G·A base pairing geometry to that found in this structure, resulting in a significant drop in the R factor. Simulated annealing tests at either higher or lower starting temperature did not lead to a correct solution. Electron density maps based on phases calculated from this 5-base-pair model showed weak density for the missing A·A pair, which was then added to the model. Repeated cycles of positional refinement, simulated annealing, and thermal parameter refinement lowered the crystallographic R factor to 18.6% (Rfree = 21.8%) for 1502 data between 2.3- and 8.0-Å resolution excluding reflections less than 3.0σ. The rms deviation in restrained bond distances was 0.02 Å and in bond angles was 1.1°. The refined model consists of a single-stranded RNA dodecamer, 20 water molecules, and one Mn2+ ion in the asymmetric unit.
RESULTS
We report herein the crystal structure of the RNA dodecamer GGCCGAAAGGCC at 2.3-Å resolution. Although in solution this sequence exists primarily as a “tetraloop” hairpin structure (10), in the crystal the RNA fragment forms a double helix with the base-paired strands related by a crystallographic twofold axis. Other sequences that exist mainly as single-stranded tetraloops in solution have also formed duplexes in the crystal (1–3), presumably due to the high “concentration” in the crystal that favors formation of the bimolecular structure. The GGCCGAAAGGCC duplex (GAAA Duplex) consists of Watson–Crick base-paired ends flanking an internal loop (indicated by the underlined nucleotides) of four consecutive nonstandard base pairs: G·A, A·A, A·A, and A·G. Because of the crystal symmetry, only one strand (or 6 base pairs) is unique. A schematic diagram of the double helix incorporating the internal loop as found in the crystal is shown in Fig. 1 and a stereodiagram is given in Fig. 2.
Figure 1.
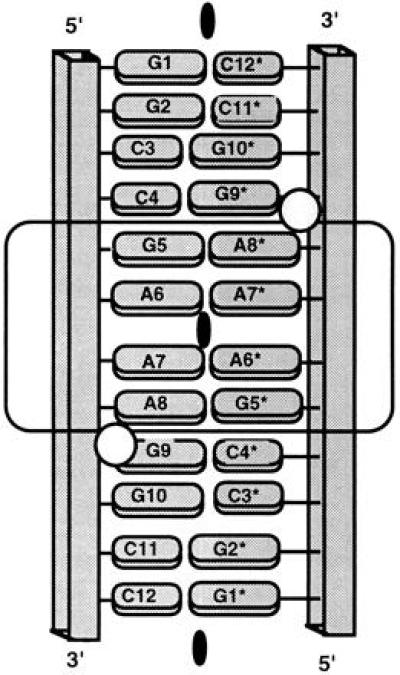
Schematic diagram of the GAAA duplex, as characterized in crystals of the dodecamer ribonucleotide r(GGCCGAAAGGCC). The phosphodiester backbone is depicted as a rectangular tube and the bases as ovals (purines, large; pyrimidines, small). The crystallographic twofold axis relating the first strand (residues 1–12) to the second strand (residues 1*–12*) is indicated as a blackened oval. Because of this exact symmetry only 6 base pairs are unique, i.e., base pairs 1–6·7*–12* are the same as pairs 1*–6*·7–12. The 4 terminal base pairs on either end are formed by standard Watson–Crick pairing, while the middle 4 bases form an internal loop of nonstandard pairs as indicated by the boxing. Bound Mn2+ ions are represented by circles.
Figure 2.
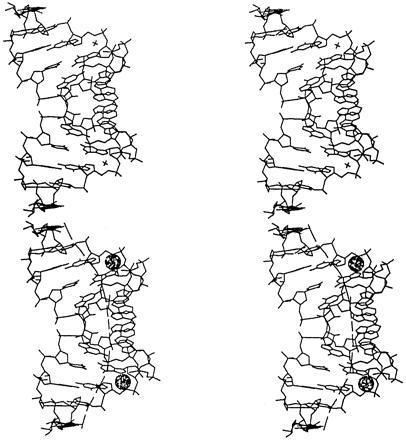
Stereoview of the GAAA duplex illustrating the twofold symmetry of the double helix and the curvature of the axis. A crystallographically related helix stacks coaxially as described in the text. Repetition of this interaction generates pseudoinfinite helices throughout the crystal. The local helix axes are shown as thick lines through the helix. The bound Mn2+ ions are shown as spheres. Bound waters are not shown in this illustration.
Another crystallographic twofold axis relates two dodecamer duplexes such that they stack in a head-to-tail fashion, with the 5′ and 3′ ends of one molecule abutting the 3′ and 5′ termini of the other (Fig. 2). The result is a pseudoinfinite helix throughout the crystal with continuous base stacking and a break in the backbone connectivity only at the 5′ and 3′ ends of each dodecamer. The observation of coaxial interhelical stacking is virtually universal in RNA oligonucleotide crystal structures and an important determinant of RNA tertiary structure (14).
The GAAA duplex is compared with a standard A-form RNA helix in Fig. 3. The distortion of the dodecamer helix introduced by the internal loop is apparent in this figure. Fitting of the GAAA duplex to a standard RNA-A helix results in an overall standard deviation of more than 4.0 Å, which is very large when compared with other RNA duplexes containing internal loops [1.5 Å (1) and 1.0 Å (2)]. The bowing of the helix apparent in Fig. 3a causes a compression of the major groove to 2.0 Å in the internal loop and expansion of the minor groove to 13.5 Å at its maximum (the corresponding groove widths for an RNA-A helix are 4.0 Å and 11.1 Å, respectively). The helix is curved by 33.8° from end to end so that at the internal loop the helix axis is offset at the maximum by 6.2 Å from a linear helix (15). The greatest perturbation in the helix axis occurs at the junction between the C·G pairs at the end of the Watson–Crick segments and the G·A pairs beginning and ending the internal loop. The kink angles at these junctions are both 10.9° and the translational offsets are about 1.0 Å (15). Fig. 3a clearly shows the narrowing of the helix in the internal loop where the diameter is reduced to 13.3 Å from 19.6 Å in the Watson–Crick paired ends (compared with 17.4 Å in a standard RNA-A duplex).
Figure 3.

Comparison of the GAAA duplex (a) and a canonical RNA-A dodecamer double helix (b) viewed from three orthogonal directions. The mini-helices are viewed perpendicular to and down the helix axes in exactly the same orientation. A ribbon passing through the phosphates emphasizes backbone distortion.
Drawings of the G·A and A·A base pairs observed in the crystal structure are shown with hydrogen bonds indicated in Fig. 4. The G·A pairs in the GAAA duplex are of the sheared (reverse Hoogsteen) variety observed in the single-stranded tetraloop structure adopted by this sequence in solution (10) and the hammerhead catalytic RNA (16, 17). The formation of sheared G·A base pairs versus head-to-head G·A pairs (12) has been inferred to be present in internal loops found in ribosomal RNA (18) and favored by certain flanking sequences (19, 20). In addition to the two base–base hydrogen bonds (N3G–N6A and N2G–N7A) stabilizing the sheared G·A pairing, the adenine amino group makes a hydrogen bond to the 2′-hydroxyl of the guanine ribose and the guanine N1 makes an interstrand hydrogen bond to a phosphate oxygen of the preceding residue (pro-Rp oxygen to A7*) as shown in Fig. 4. This last base-backbone hydrogen bond has not been observed in previous G·A pairs and may be dependent on the neighboring A·A noncanonical base pair. The thermodynamic stability of G·A pairs (21, 22) may be related to the formation of these four hydrogen bonds. This structure also includes a novel type of A·A base pairing not previously observed crystallographically. The asymmetric A·A base pairs make only two base–base hydrogen bonds (N6–N1 and N7–N6) compared with the total of four H-bonds in the G·A pairs. This may contribute to the lower thermodynamic stability of A·A base pairs in internal loops (21, 23).
Figure 4.
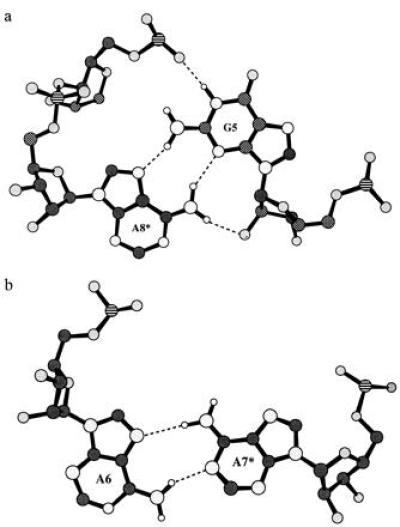
Structures of the G·A and the A·A base pairs found in this duplex. In these diagrams phosphorous is indicated by a lined pattern, the carbons are shaded dark gray, the oxygens are lighter gray, and the nitrogens are shaded the lightest gray. The hydrogens are shown as unshaded circles in their calculated positions. Dashed lines are used to indicate hydrogen bonding. (a) A G·A base pair viewed perpendicular to the plane formed by the bases. Four interstrand hydrogen bonds are shown, two base–base and two base–backbone, as described in the test. (b) An A·A base pair. Only two base–base hydrogen bonds are formed by this asymmetric pair.
Fig. 5 shows the stepwise stacking of the noncanonical base pairs in the internal loop. Interestingly, the guanine of the G·A pair (G5/G5*) is unstacked on the 3′ side, while the adenine stacks over the following A·A pair and one of the adenines in the A·A pair (A6/A6*) is unstacked on the 5′ side. This unstacking leaves the base planes available for potential interaction.
Figure 5.
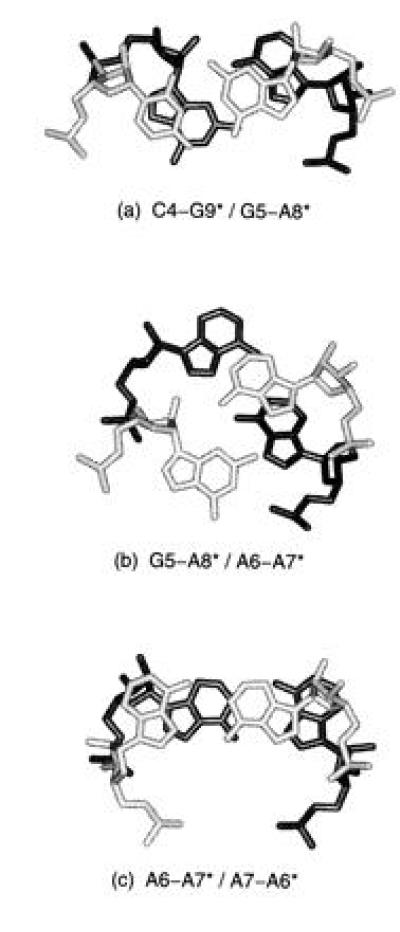
Stepwise stacking of noncanonical base pairs found in the GAAA internal loop. Base pairs shown in light gray stack above those shown in black. The upper base pair is indicated before the slash in the title under each illustration. Symmetry related stacking is not shown but is inferred. In addition to stacking interactions, helical twist angles are also apparent. (a) Stacking of the final base pair of Watson–Crick region, C4·G9*, on the first base pair of the internal loop, G5·A8*. G5 is seen to be unstacked on the 3′ side and A6 is unstacked on its 5′ side. The helical twist angle between the C1′–C1′ vectors of two successive base pairs is 16°. (b) Stacking of the G5·A8* base pair on A6·A7* pair. The twist angle between these base pairs is 59°. (c) Stacking of the consecutive A·A base pairs (A6·A7* above A7·A6*). The helical twist angle between these pairs is 34°.
Finally, an additional feature of this crystal structure is the identification of a tightly bound hydrated manganese ion in each strand of the duplex. The hydrated Mn2+ is directly coordinated to the N7 of G9 (of the Watson–Crick G9·C4* pair) and the pro-Rp phosphate oxygen of A8, which is the previous A·G base pair. Another Mn2+ hydrate is bound to the symmetrically related A8* and G9* residues.
DISCUSSION
Noncanonical G·A and A·A base pairs are extremely common in internal loops of biological RNAs including ribosomal RNAs, ribozymes, small nuclear (sn) RNAs, and others. The possible functional significance of the GAAA internal loop sequence is emphasized by the conservation and high frequency of occurrence of tandem G·A, A·A (5′-GAAA), as well as G·A, A·G base pairs in chloroplast and eubacterial 23S ribosomal RNA (18). The characteristic base-backbone interstrand hydrogen bonding involving the guanine of the G·A pair and the conserved metal binding site utilizing the adenine may contribute to the high frequency of occurrence of these motifs in ribosomal RNA. Also, the unusual stacking dislocation between the G·A and A·A base pairs, in which the 3′ side of G5 and the 5′ side of A6 are totally unstacked (see Fig. 5), presents a unique surface for recognition and/or a potential intercalation site. The depiction of tandem G·A, A·A base pairs at high resolution in the GAAA duplex furthers our understanding of the role of these base pairs and enables more accurate models to be constructed for a variety of RNA molecules.
The recently determined crystal structures of two hammerhead ribozymes (16, 17) contain a conserved G·A, A·G motif that may be compared with the G·A, A·A motif in the GAAA duplex. Fig. 6 compares a region in domain II of the hammerhead ribozyme (16) consisting of a Watson–Crick C·G base pair followed by consecutive G·A, A·G noncanonical base pairs, with the C·G, G·A, A·A motif observed in the GAAA duplex. As shown, the conformation of the C·G and G·A base pairs in the GAAA duplex is similar to that of the analogous base pairs in the hammerhead. However, the conformation of the adjacent base pairs (A6·A7* in the GAAA duplex and G8·A13 in the hammerhead) are extremely different. For example, as shown in Fig. 4a for the GAAA duplex, the G5 N1 forms an interstrand hydrogen bond with the pro-Rp phosphate oxygen of A7*, whereas in the hammerhead ribozyme the guanine N1 forms an interstrand hydrogen bond with the 2′-hydroxyl of the following residue. Thus, an interstrand hydrogen bond involving the guanine N1 and the sugar phosphate backbone may be important for stabilization of sheared G·A pairs even though it may be accomplished in very different ways depending on the neighboring base pair.
Figure 6.

Stereoviews comparing the structures of the analogous metal binding sites in the GAAA duplex and the hammerhead ribozyme. (a) Base pairs C11.1·G10.1, G12·A9, and A13·G8 of domain II in the DNA–RNA hybrid hammerhead ribozyme (15). (b) Base pairs C4·G9*, G5·A8*, and A6·A7* are shown for the GAAA duplex The positions of the Mn2+ ions are indicated by crosses. The carbon and nitrogen atoms in the C4·G9* and G5·A8* base pairs of the GAAA duplex were superimposed on the corresponding atoms in the hammerhead ribozyme to obtain this common orientation.
It is also apparent from Fig. 6 that the base stacking interactions between the G·A and A·A base pairs in the GAAA duplex are very different from the corresponding stacking observed between the G·A and A·G base pairs in domain II of the hammerhead ribozyme. In the hammerhead, the tandem G·A base pairs lead to interstrand stacking of the two guanine bases, whereas, as seen in Figs. 5 and 6, there is no interstrand stacking in the internal loop of the GAAA duplex and in fact, G5 is not stacked at all on its 3′ side. In addition, the other unusual features of the internal loop in the GAAA duplex, the bowed and narrowed helix, are not observed in the internal loop of domain II in the hammerhead ribozyme.
A specific metal binding site is found in a pocket between C·G and G·A base pairs in both the GAAA duplex and two forms of hammerhead catalytic RNA (16, 17). This site binds Mn2+ in the GAAA duplex, either Mn2+ or Cd+2 in the DNA/RNA hammerhead ribozyme (16), and Mg2+ in the all RNA hammerhead structure (17). Both Mn2+ and Cd+2 are directly coordinated to the guanine N7 of the C·G pair and the adenine pro-Rp oxygen of the G·A pair. Biochemical studies support the preference for direct coordination of Mn2+ to the N7 position of guanine (24). In the all RNA hammerhead ribozyme structure (17), a Mg2+ hydrate is bound directly to the pro-Sp oxygen of the adenine in the G·A pair and through its water shell to the O6 and N7 positions of the guanine in the C·G pair. Based on these observations, the base-pair sequence C·G, G·A appears to form a module that provides and is stabilized by a metal binding pocket when the G·A pair is in the sheared conformation.
In conclusion, the direct observation of bending and narrowing in the internal loop of this RNA helix suggests two potential modes for specific recognition of helical distortion by RNA binding proteins. The distortion of the helical backbone in the internal loop, characterized by the dramatic narrowing of the helical diameter and width of the major groove, may represent a unique local recognition site. On the other hand, the curvature of the helix propagates a potential global recognition signal that may function far away from the internal loop itself. Future studies of protein–RNA complexes are needed to show whether one or both of these types of distortion is actually utilized for recognition.
Acknowledgments
Special credit is due to Jaru Jancarik who conducted the initial crystallization experiments. This work was supported by National Institutes of Health Grants GM 4921501 (S.R.H.) and AI33098 (A.P.) and Research Career Development Award AI01051 (A.P.). Facilities and equipment were provided through support of the Office of Energy Research, Office of Health and Environmental Research, Health Effects Research Division of the U.S. Department of Energy. Diffraction data was collected at the synchrotron at the Stanford Synchrotron Radiation Laboratory.
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 283D). The refined coordinates of the GAAA duplex have been deposited in the Nucleic Acids Database (NDB) (8) (URL051).
References
- 1.Holbrook S R, Cheong C, Tinoco I, Jr, Kim S-H. Nature (London) 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 2.Baeyens K J, DeBondt H L, Holbrook S R. Nat Struct Biol. 1995;2:56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- 3.Cruse W B T, Saludijan P, Biala E, Strazewski P, Prange T, Kennard O. Proc Natl Acad Sci USA. 1994;91:4160–4164. doi: 10.1073/pnas.91.10.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lietzke S E, Barnes C L, Berglund J A, Kundrot C E. Structure (London) 1996;4:917–930. doi: 10.1016/s0969-2126(96)00099-8. [DOI] [PubMed] [Google Scholar]
- 5.Bartel D P, Zapp M L, Green M R, Szostak J W. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 6.Theil E C. Biofactors. 1993;4:87–93. [PubMed] [Google Scholar]
- 7.Gabriel K, Schneider J, McClain W H. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 8.Berman H M, Olson W K, Beveridge D L, Westbrook J, Gelbin A, Demeny T, Hsieh S-H, Srinivasan A R, Schneider B. Biophys J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milligan J, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heus H A, Pardi A. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 11.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 12.Leonard G A, McAuley-Hecht K E, Ebel S, Lough D M, Brown T, Hunter W N. Curr Biol. 1994;2:483–494. doi: 10.1016/S0969-2126(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 13.Brünger A T. x-plor, A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. Version 3.1. [Google Scholar]
- 14.Murphy F L, Wang Y H, Griffith J D, Cech T R. Science. 1994;265:1709–1712. doi: 10.1126/science.8085157. [DOI] [PubMed] [Google Scholar]
- 15.Lavery R, Sklenar H. J Biomol Struct Dynam. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 16.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 17.Scott W G, Finch J T, Klug A. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 18.Gautheret D, Konings D, Gutell R R. J Mol Biol. 1994;242:1–8. doi: 10.1006/jmbi.1994.1552. [DOI] [PubMed] [Google Scholar]
- 19.Cheng J W, Chou S H, Reid B R. J Mol Biol. 1992;228:1037–1041. doi: 10.1016/0022-2836(92)90312-8. [DOI] [PubMed] [Google Scholar]
- 20.Katahira M, Sato H, Mishima K, Uesugi S, Fujii S. Nucleic Acids Res. 1993;21:5418–5424. doi: 10.1093/nar/21.23.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SantaLucia J J, Kierzek R, Turner D H. Biochemistry. 1990;29:8813–8819. doi: 10.1021/bi00489a044. [DOI] [PubMed] [Google Scholar]
- 22.SantaLucia J, Kierzek R, Turner D H. J Am Chem Soc. 1991;113:4313–4322. [Google Scholar]
- 23.SantaLucia J, Jr, Kierzek R, Turner D H. Biochemistry. 1991;30:8243–8250. doi: 10.1021/bi00247a021. [DOI] [PubMed] [Google Scholar]
- 24.Steinkopf S, Sletten E. Acta Chem Scand. 1994;48:388–392. doi: 10.3891/acta.chem.scand.48-0388. [DOI] [PubMed] [Google Scholar]


