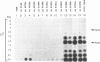
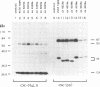
Abstract
The integration protein (IN) is required for retrovirus DNA integration into the host DNA. The function of the C terminus of the Moloney murine leukemia virus IN protein was examined. The terminal 28 amino acids were found to be nonessential. A linker insertion at position 6025, within a 36-amino-acid insertion not found in any other retrovirus, also produced viable virus.
Full text
PDF
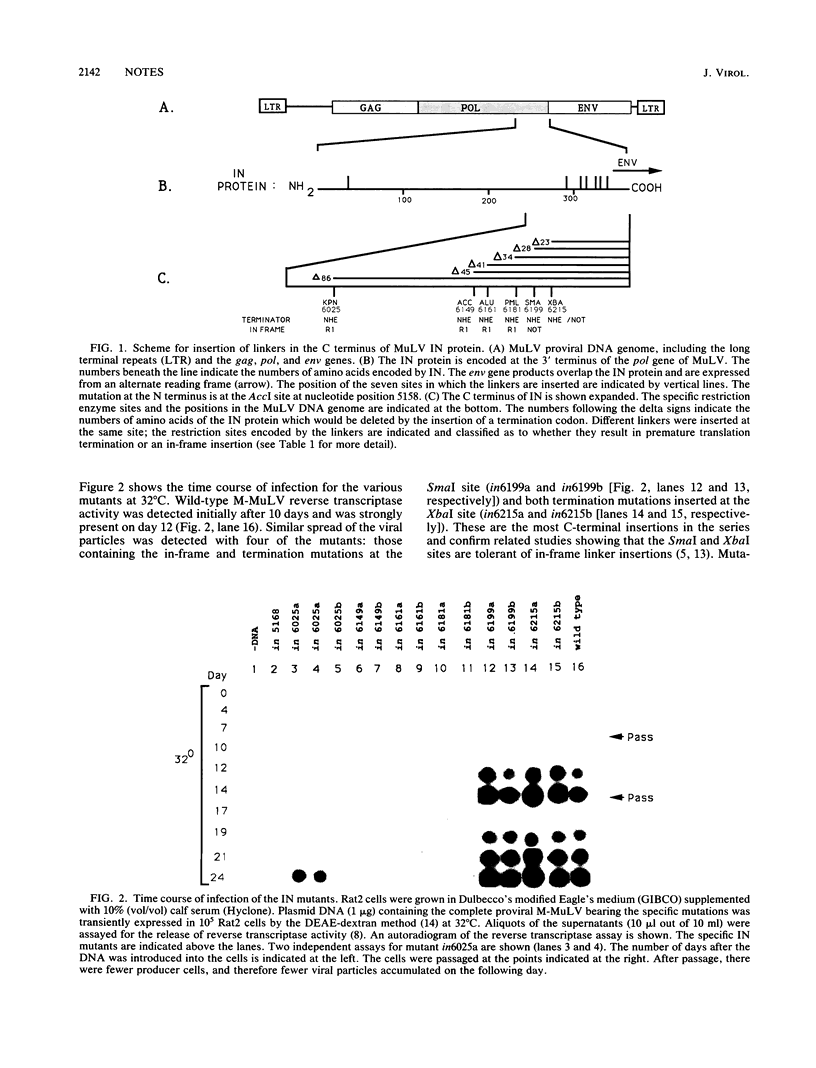



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Correct integration of retroviral DNA in vitro. Cell. 1987 May 8;49(3):347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- Brown P. O., Bowerman B., Varmus H. E., Bishop J. M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicelli J., Goff S. P. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell. 1985 Sep;42(2):573–580. doi: 10.1016/0092-8674(85)90114-x. [DOI] [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990 Aug 24;62(4):829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Donehower L. A. Analysis of mutant Moloney murine leukemia viruses containing linker insertion mutations in the 3' region of pol. J Virol. 1988 Nov;62(11):3958–3964. doi: 10.1128/jvi.62.11.3958-3964.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Varmus H. E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988 Aug 12;54(4):497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Goff S., Traktman P., Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981 Apr;38(1):239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett D. P., Mumm S. R. Unraveling retrovirus integration. Cell. 1990 Jan 12;60(1):3–4. doi: 10.1016/0092-8674(90)90707-l. [DOI] [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Swanstrom R., Olsen J. C. Nuclease mechanism of the avian retrovirus pp32 endonuclease. J Virol. 1986 Jun;58(3):970–974. doi: 10.1128/jvi.58.3.970-974.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel L. I., Goff S. P. Construction of mutants of Moloney murine leukemia virus by suppressor-linker insertional mutagenesis: positions of viable insertion mutations. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4149–4153. doi: 10.1073/pnas.81.13.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Quinn T. P., Grandgenett D. P. Genetic evidence that the avian retrovirus DNA endonuclease domain of pol is necessary for viral integration. J Virol. 1988 Jul;62(7):2307–2312. doi: 10.1128/jvi.62.7.2307-2312.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Jones R. W., Goff S. P. Insertional mutagenesis of the Abelson murine leukemia virus genome: identification of mutants with altered kinase activity and defective transformation ability. J Virol. 1988 Mar;62(3):978–986. doi: 10.1128/jvi.62.3.978-986.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdewohld H., Weiher H., Reik W., Jaenisch R., Breindl M. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J Virol. 1987 Feb;61(2):336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Schwartzberg P. L., Goff S. P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989 Jul 14;58(1):47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- Roth M. J., Schwartzberg P., Tanese N., Goff S. P. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J Virol. 1990 Oct;64(10):4709–4717. doi: 10.1128/jvi.64.10.4709-4717.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Tanese N., Goff S. P. Gene product of Moloney murine leukemia virus required for proviral integration is a DNA-binding protein. J Mol Biol. 1988 Sep 5;203(1):131–139. doi: 10.1016/0022-2836(88)90097-6. [DOI] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P., Colicelli J., Goff S. P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984 Jul;37(3):1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- Shih C. C., Stoye J. P., Coffin J. M. Highly preferred targets for retrovirus integration. Cell. 1988 May 20;53(4):531–537. doi: 10.1016/0092-8674(88)90569-7. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Tanese N., Roth M. J., Goff S. P. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J Virol. 1986 Aug;59(2):328–340. doi: 10.1128/jvi.59.2.328-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Robinson H. L. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J Virol. 1986 Nov;60(2):683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]