Abstract
Chemical cross-linking and high resolution MS have been integrated successfully to capture protein interactions and provide low resolution structural data for proteins that are refractive to analyses by NMR or crystallography. Despite the versatility of these combined techniques, the array of products that is generated from the cross-linking and proteolytic digestion of proteins is immense and generally requires the use of labeling strategies and/or data base search algorithms to distinguish actual cross-linked peptides from the many side products of cross-linking. Most strategies reported to date have focused on the analysis of small cross-linked protein complexes (<60 kDa) because the number of potential forms of covalently modified peptides increases dramatically with the number of peptides generated from the digestion of such complexes. We report herein the development of a user-friendly search engine, CrossSearch, that provides the foundation for an overarching strategy to detect cross-linked peptides from the digests of large (≥170-kDa) cross-linked proteins, i.e. conjugates. Our strategy combines the use of a low excess of cross-linker, data base searching, and Fourier transform ion cyclotron resonance MS to experimentally minimize and theoretically cull the side products of cross-linking. Using this strategy, the (αβγδ)4 phosphorylase kinase model complex was cross-linked to form with high specificity a 170-kDa βγ conjugate in which we identified residues involved in the intramolecular cross-linking of the 125-kDa β subunit between its regulatory N terminus and its C terminus. This finding provides an explanation for previously published homodimeric two-hybrid interactions of the β subunit and suggests a dynamic structural role for the regulatory N terminus of that subunit. The results offer proof of concept for the CrossSearch strategy for analyzing conjugates and are the first to reveal a tertiary structural element of either homologous α or β regulatory subunit of phosphorylase kinase.
Chemical cross-linking of proteins is a versatile technique with uses ranging from screening to obtaining relative or absolute structural information for interacting proteins (1). For large proteins that are refractive to techniques such as crystallography or NMR, cross-linking provides a means of gaining moderate to low resolution structural data by generating distance measurements between specific regions of interacting proteins (intermolecular cross-linking) or individual proteins (intramolecular cross-linking). Advances in modern MS methods have promoted a resurgence in the use of cross-linking by eliminating the need for large quantities of conjugates (term used herein to denote either proteins or peptides that are covalently cross-linked) to isolate and identify distinct regions of inter- or intramolecular cross-linked peptides (2, 3). Nanomolar amounts of conjugates are now routinely digested in gel to provide sufficient quantities of peptides for detection by MS methods such as LC ESI MS. Despite these advances, however, cross-linked peptides are still often difficult to detect because the signal intensity for these peptides is generally far lower than that for non-modified peptides, the latter of which have the potential to co-elute and mask the former by ion suppression mechanisms inherent in the use of LC ESI MS (4). A low yield of specific cross-linked peptides from dimeric conjugates, which are the most easily analyzed, often results from continuing cross-linking reactions that form multiple high mass conjugates, a phenomenon termed nonspecific cross-linking (1). This phenomenon is particularly problematic in the analysis of large hetero-oligomeric complexes, such as the (αβγδ)4 phosphorylase kinase (PhK)1 complex used as a model in this study, because the yield of conjugates comprising just two subunits invariably decreases markedly with time as additional subunits become cross-linked to previously formed dimeric conjugates (5).
Arguably the greatest obstacle in the identification of cross-linked peptides is the extensive array of products that are formed after cross-linking and digestion of cross-linked proteins. This is particularly true for large conjugates (>100 kDa) for which the number of potential products is estimated to increase exponentially with the number of peptides generated by proteolysis (6). As reactants, proteins add considerable complexity to a cross-linking reaction because they contain many reactive amino acid side chains that may be targeted by the cross-linker, leading to their modification by either cross-linking or monoderivatization (covalent modification without cross-linking) (7). In nearly all cases, the number of monoderivatization products will greatly exceed those of cross-linking, particularly for heterobifunctional cross-linkers such as the one used in this study, namely N-[γ-maleimidobutyryloxy]succinimide ester (GMBS). Because cross-linkers are usually selective as opposed to specific (8, 9), GMBS not only may react with a variety of side chains, but the resultant monoderivatization products may in turn form a variety of alternative stable secondary products as will be described herein. Further product complexity results from the fact that modifications may alter the enzyme-catalyzed hydrolysis of each protein in a conjugate by inhibiting proteolysis of amide bonds adjacent to the residue modified; this is particularly problematic for trypsin as lysine is the residue most commonly targeted by conventional chemical cross-linkers (8). The end result is that the heterogeneity of modifications possible in the formation of a conjugate and of its digestion products renders useless the direct comparison of native and cross-linked forms of proteins as a method for detecting cross-linked peptides. Labeling techniques have been developed to reduce the number of potential candidates for analysis of digests of conjugates. These include end labeling peptides with 18O (10, 11), enrichment with affinity-tagged cross-linkers (12, 13), combined isotope and affinity tagging (14), cross-linking non-labeled and 14N-labeled protein pairs (15), isotope-labeled cross-linkers (16–18), and MS-cleavable reagents (19–22). All have been used successfully in the analysis of low mass conjugates; however, the success of any labeling strategy is dependent upon many factors, especially the size of the conjugate being analyzed (6). To minimize the number of candidate ions observed in the analysis of large proteins by chemical cross-linking, we have used an alternative approach to the labeling strategies by simply using a minimal stoichiometric excess of cross-linker, which for convenience we term MSEC. The MSEC approach works particularly well with the PhK complex because it binds small organic reagents (a descriptor for most cross-linkers) with high affinity (23–25) as do many other proteins (for a review, see Ref. 26). We have shown previously that PhK specifically binds small hydrophobic cross-linkers, such as the geometric isomers of phenylenedimaleimide (25) and GMBS (23), and that non-active structural analogs of those cross-linkers inhibit its cross-linking by the parent compound (25). Using the MSEC approach with GMBS, we demonstrate herein that this reagent cross-links the hexadecameric PhK complex to form sufficient amounts of a βγ dimer for analysis and that the formation of this conjugate occurs with a tolerable amount of monoderivatized products.
Although the methods described above streamline somewhat the analysis of cross-linked proteins, predicting the chemical composition of any specific candidate ion requires considerable computational power given the large pool of products possible from the cross-linking and digestion of conjugates. A number of programs have been developed to predict the identity of cross-linked peptides; however, they either require the use of labeled reagents (6, 14–19, 27, 28) or significant user input (29–33). We developed a Web-based, user-friendly search engine, CrossSearch, that both provides a statistical approach to culling masses corresponding to side products of cross-linking and predicts the composition of cross-linked peptides from the mass output obtained for digests of cross-linked proteins. CrossSearch provides the underpinning for an overall strategy for predicting and verifying the composition of cross-linked peptides from both small and large conjugates, especially when used in conjunction with MSEC. Using this strategy in combination with FT-ICR MS, we identified residues involved in the intramolecular cross-linking of the 125-kDa regulatory β subunit of PhK in the context of the entire hexadecameric complex. This cross-linking result represents the first report of a tertiary structural element for either of the two large, homologous α and β subunits of PhK.
EXPERIMENTAL PROCEDURES
Proteins—
PhK was purified from the psoas muscle of New Zealand White rabbits (34); dialyzed against 50 mm Hepes (pH 6.8), 0.2 mm EDTA, 10% sucrose; and stored at −80 °C. Its concentration was determined by methods described previously (34). The mAbs against the α, β, and γ subunits of PhK were described previously (35, 36), and the anti-calmodulin mAb was from Zymed Laboratories Inc. All other secondary conjugates were from Southern Biotechnology.
Cross-linking—
PhK was cross-linked with GMBS essentially as described previously (23) with cross-linking initiated by addition of GMBS and carried out at 30 °C for 2.5 min at pH 8.2 in 50 mm Hepes, 0.2 mm EDTA. Final concentrations of PhK (αβγδ protomer) and GMBS in the reaction were 0.47 and 4.7 μm, respectively. The reaction was terminated by adding an equal volume of SDS buffer (0.125 M Tris (pH 6.8), 20% glycerol, 5% β-mercaptoethanol, 4% SDS) followed by brief vortexing. The PhK subunits were separated on 6–18% linear gradient polyacrylamide gels and stained with Coomassie Blue. Western blotting of the proteins was performed on PVDF membranes with subunit-specific mAbs as described previously (23). All cross-linking reactions were performed at least twice using different preparations of PhK.
To determine regions of cross-linking in the βγ conjugate, the cross-linked PhK complex was resolved by preparative SDS-PAGE and stained with Coomassie Blue. The bands corresponding to the βγ conjugate and the remaining monomeric β and γ subunits were excised from the gel, sectioned, and exchanged with three aliquots (each ∼5× the volume of the gel slice) of 50 mm ammonium bicarbonate, 50% acetonitrile to remove SDS. The proteins were then reduced in 10 mm dithiothreitol for 1 h at 55 °C and carboxymethylated with 50 mm iodoacetic acid for 1 h in the dark. The gel pieces were washed as described above with 50 mm ammonium bicarbonate followed by several exchanges with 50 mm ammonium bicarbonate, 50% acetonitrile. After removing the last wash, the gels were dried in a SpeedVac (Savant) and treated with trypsin (Promega; 12.5 ng/μl) for 24 h at 30 °C. Peptides were extracted from the gel pieces with 50% acetonitrile, 5% formic acid.
MS Analyses—
All samples were concentrated on a Centrivac concentrator (Labconco) to a final volume of 20 μl and pressure-loaded onto a C18 reverse phase nanocolumn (75-μm inner diameter fused silica packed in house with 9 cm of 100-Å, 5-μm, Magic C18 particles, Michrom Bioresources). Following a wash with 0.1% formic acid for 15 min at 0.5 μl/min, the column was mounted on the electrospray stage of an FT-ICR mass spectrometer (LTQ FT, ThermoFinnigan, San Jose, CA), and peptides were eluted at an approximate flow rate of 0.3 μl/min over a 120-min period using a gradient of 0–90% acetonitrile (Buffer A = 0.1% formic acid; Buffer B = acetonitrile, 0.1% formic acid). The source was operated at 1.9 kV with the ion transfer temperature set to 350 °C. LC MS data were obtained in a hybrid linear ion trap FT-ICR mass spectrometer equipped with a 7-tesla magnet. The mass spectrometer was controlled using an Xcalibur software package to continuously perform mass scan analysis on the FT instrument followed by MS/MS scans on the ion trap for the six most intense ions with a dynamic exclusion of two repeat scans (30-s repeat duration and 90-s exclusion duration) of the same ion. Normalized collision energy for MS/MS was set to 35%.
Data Analyses—
For data analyses, dta files were created on Bioworks Browser version 3.2. The corresponding log file was used to generate a list of parent ions for which the corresponding charges and tandem mass spectra were obtained. The resulting mass list ((M + H)+) was submitted to CrossSearch, the cross-link search engine, to generate a list of potential conjugates. The precursor ion and corresponding tandem mass spectrum matched to each cross-link assignment were visually examined and were further analyzed only if the parent ion was clearly above the noise base line (∼6×) and had an isotopic envelope that was characteristic of a peptide. An additional requirement was that the tandem mass spectrum be performed only on the monoisotopic peak with at least nine fragment ions required for assignment. The remaining tandem mass spectra were then searched by the Sequest algorithm (37) included in Bioworks 3.2 using a modified version of a contaminant protein data base supplied by the vendor (ThermoFinnigan). Data base modification included addition of the polypeptide sequences for the α (P18688), β (P12798), γ (P00518), and δ (P02593) Swiss-Prot/TrEMBL subunits of rabbit muscle PhK. Each search was performed using a 50-ppm error window for the mass of the parent ion with a fragment ion tolerance of 1.00 for peptides arising from both complete tryptic or random hydrolysis. The results of the searches were filtered using the following set of criteria for low confidence: minimum cross-correlation scores (Xcorr) of 1.5, 2.0, and 2.5 for plus 1, 2, and 3 charged ions, respectively, and a delta correlation score (Δcorr) greater than 0.08. Potential conjugates that could be matched to a protein sequence in this data base were rejected. The fragmentation patterns of the remaining candidates were analyzed for consistency with the predicted chemistry of cross-linking.
The tandem mass spectrum of each conjugate was analyzed using a combination of programs that were subsequently verified by an “in-house” spreadsheet. A list of masses from the tandem mass spectrum of each candidate parent ion was uploaded to MS2Links, part of a suite of programs (Collaboratory for MS3D) developed by Dr. Malin Young and co-workers (31) at Sandia National Laboratories, Livermore, CA. Theoretical fragmentation of the conjugate in the positive mode was accomplished using the sequences for each peptide in the conjugate and the masses for each intervening chemical cross-link and its possible fragmentation products using a mass error tolerance not exceeding 0.7 Da for each fragment ion. A modification table was generated for the GMBS cross-linker based on both predicted and observed amide fragmentations of the reagent (23). Potential matching ion assignments for each fragment mass generated by MS2Links were verified using a spreadsheet containing an array of masses predicted for the cross-linker (and fragments thereof) and also those generated for each peptide in the conjugate using MS Product, a fragmentation tool in Protein Prospector developed at the University of California, San Francisco (38). Final assignments were also checked by hand for verification and accuracy.
Cross-link Search Engine—
The CrossSearch engine was constructed with an intuitive XHTML/PHP-based “front end” that allows users to enter a peptide sample, information on sample preparation techniques, and masses identified by MS. The “back end” comprises an MySQL data base containing enzyme splicing information, tags for reactive amino acids, cross-linker mass data, and rule sets defining possible peptide reactants from digests of cross-linked proteins, the chemistry of cross-linking for the reagent chosen, and potential products and side products of the cross-linking reaction. PHP (scripting language) is used to interface the Web front end with the MySQL back end, and a combination of XHTML and cascading style sheets (CSS) languages are used to ensure compatibility with a wide range of browsers and overall ease of use.
RESULTS
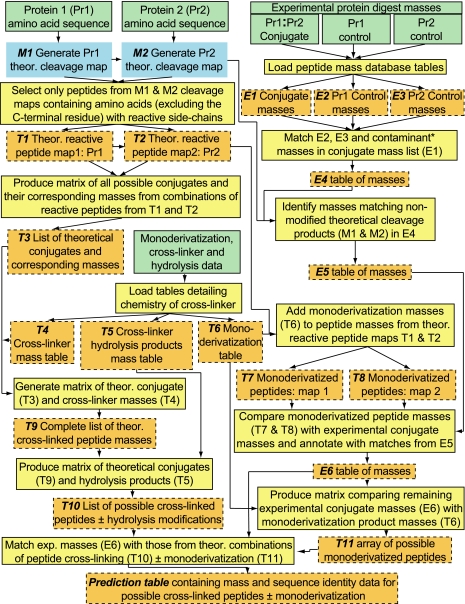
We developed a cross-link search engine, CrossSearch, that works in combination with high resolution mass spectrometry to predict chemically cross-linked regions between, or within, proteins by generating a list of hypothetical conjugates and their corresponding masses from digests of the cross-linked proteins. A schematic of the search engine showing its basic operating principles is presented in Fig. 1. The boxes in the left half show the flow of theoretical data generated from user-entered components of the reaction (green), including the identity of the chemical cross-linker and the amino acid sequences of the proteins that are covalently linked (Pr1 and Pr2). The theoretical data are cross-referenced against the experimental data (right half), comprising mass lists obtained from MS analyses of digests of the cross-linked proteins (i.e. Pr1·Pr2 conjugate) and Pr1 and Pr2 monomers remaining after termination of the cross-linking reaction that serve as controls. Digests of the monomers help to identify, and thus eliminate, side products of the cross-linking reaction in that they are assumed to contain many of the products (and contaminants) observed in the digest of the Pr1·Pr2 conjugate that are not related directly to the cross-linking of Pr1 and Pr2.
Fig. 1.
Flow diagram of the CrossSearch engine. Green rectangles indicate the search engine data input sites that allow entry of the amino acid sequences of the protein (Pr) conjugate pair, chemical cross-linker, protease (e.g. trypsin), and mass lists generated by MS analyses of digests of the conjugate and corresponding non-cross-linked Pr components. Theoretical (Theor.) digests of Pr1 and Pr2 (blue rectangles) may be carried out either by the search engine or uploaded directly from other processing tools, such as PeptideMass from the ExPASy website. Perl programming and MySQL data base iterations are shown in yellow. Orange rectangles denote MySQL tables and matrices that can be manipulated and accessed multiple times throughout the analysis without reiteration of the generating step. *, after contaminant in the experimental column of the search engine designates polyacrylamide and other non-peptide components.
The primary function of the search engine is to separate potential candidate cross-linked peptides from all possible side products of the cross-linking reaction. This is carried out in a series of steps, the first of which is to generate temporary data base tables (orange) of experimental masses (E2 and E3) from Pr1 and Pr2 controls and theoretical masses from an array of all possible products for comparison against the experimental mass list for the protein conjugate (E1). To calculate the array of theoretical conjugates and side products, potential reaction sites on Pr1 and Pr2 are determined by first generating corresponding cleavage maps (blue) for each protein (M1 and M2) and then selecting only those peptides that are potentially reactive through use of a rule set defined in the data base (computational and iterative steps shown in yellow). The protein rule set defines reactive peptides (T1 and T2) as only those containing amino acids with nucleophilic side chains (His, Lys, Arg, Cys, and Tyr) with the C-terminal residue of each peptide being excluded because its modification would be assumed to inhibit cleavage by the protease (1). The user can choose other combinations of reactive amino acids to accommodate the side-chain specificity of different types of cross-linking reagents.
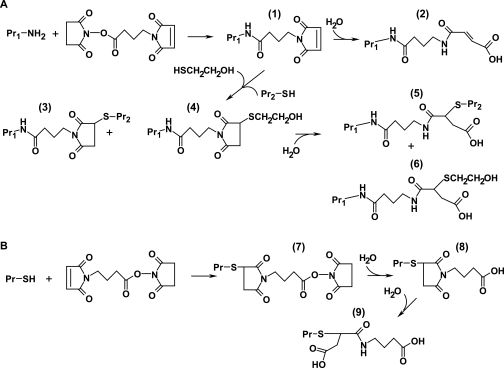
A cross-linker rule set is derived from the chemistries reported for the cross-linker used. As an example, possible products of the reaction between proteins and the heterobifunctional cross-linker GMBS are shown in Fig. 2, which illustrates the large number of products that may be formed. The formation of a single intermolecular cross-link between the identical residues of two proteins can actually result in conjugates having different masses based on the susceptibility of the maleimide ring to hydrolysis after cross-linking (Fig. 2A, products 3 and 5). Potentially competing side reactions are also contained in the cross-linker rule set. The preferential reactions of Cys and Lys with the succinimidyl and maleimido groups, respectively, of GMBS can form at least five relatively stable side products (products 2, 4, 6, 8, and 9) as a direct result of competing reactions between protein side chains and exogenous nucleophiles, including water, for the remaining functional group of the cross-linker. The number of both these possible side products, or monoderivatized peptides, and cross-linked conjugates is far greater than shown if one takes into account the fact that other protein side chains may react with the cross-linker to form stable products, i.e. these reagents are selective, not specific (8). The masses corresponding to all these chemical modifications are loaded into tables (T4–T6) for subsequent steps of analysis. The list of monoderivatized peptides can also be downloaded to compare against cross-link assignments.
Fig. 2.
Side products of cross-linking. A, Pr Lys ɛ-amines preferentially react with the N-succinimide functional group of GMBS at near-neutral pH to form product 1. Hydrolysis of the maleimide group (product 2) competes either with cross-linking reactions by Pr nucleophiles (product 3) or with exogenous nucleophiles that are present as contaminants or quench reagents to halt cross-linking at specific times (product 4). Conversion of products 3 and 4 to their corresponding maleimic derivatives occurs after hydrolysis and subsequent ring opening of the S-substituted maleimide rings (products 5 and 6). B, targeting of the maleimido functional group is generally favored by Pr Cys thiols (product 7). Hydrolysis of the succinimide group competes with cross-linking reactions by Pr nucleophiles, resulting in monoderivatization of the thiol (product 8). A second mass addition to the thiol may be detected after conversion of the maleimide to the corresponding maleimic derivative (product 9).
The next step is to identify in the experimental mass list of the digested conjugate (E1) those masses corresponding to side products generated from both theoretical and experimental (Pr1 and Pr2 controls) sources. Digests of the Pr1 and Pr2 controls (remaining Pr1 and Pr2 monomers isolated from the quenched cross-linking reaction mixture by SDS-PAGE or other separation methods) contain monoderivatized, incompletely digested, and non-modified peptides as well as acrylamide contaminants, the last of which are not accounted for in the theoretical list. The masses from the protein controls are loaded into tables (E2 and E3) and together with a list of common contaminants (e.g. human keratins) are compared with E1 to yield the E4 table of masses. The masses of non-modified peptides (M1 and M2) derived from the theoretical cleavage of Pr1 and Pr2 are matched with those from E4 to generate E5 because the digestion products from the Pr1 and Pr2 controls may differ from those of their corresponding counterparts in the Pr1·Pr2 conjugate (e.g. constraints could be imposed on the protease by cross-linking and/or different extents of monoderivatization). Inasmuch as the extent of monoderivatization may differ for Pr1 and Pr2 in their respective experimental control (E2 and E3) and conjugate forms (E1), a comparison of E2 and E3 with E1 may not identify all masses corresponding to monoderivatized peptides. To account for such differences in the monoderivatized peptide pools, all possible monoderivatized peptides from Pr1 and Pr2 are generated theoretically (T7 and T8) by combining data tables T1, T2, and T6 with the combination subsequently identified in E5 thus annotated to yield E6. The masses in table E6 represent those masses that match either cross-linked peptides or cross-linked peptides with different extents of monoderivatization. To predict the possible composition of these products, the experimental masses from E6 are matched against all possible theoretical combinations of cross-linking (obtained in order of sequence from T1–T5, T9, and T10) and monoderivatization (T11).
Theoretical matches for each experimental mass are displayed in two different formats, the first of which is a statistical summary of the analysis showing the number of potential theoretical matches possible for seven different mass error ranges progressing in order of increasing mass accuracy from 0.1 to 0.00001-dalton error. In addition to the possible cross-linking combinations, potential theoretical matches corresponding to monoderivatized and/or non-modified peptides are also shown, providing the investigator the opportunity to either reject or select mass data for further analysis. In the second format, all possible combinations of cross-linking ± monoderivatization are listed in the final prediction table in order of increasing mass error, allowing for further analysis of all possible assignments by fragmentation, labeling, or other methods of choice. A Web-based version of the search engine using several commonly used cross-linkers is now available for beta testing.
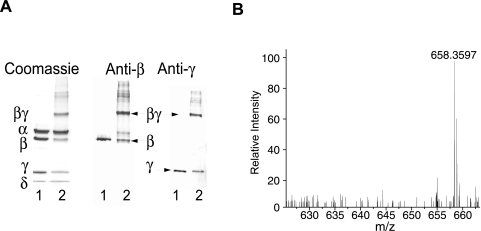
Previously we demonstrated that GMBS is an affinity cross-linker of the (αβγδ)4 PhK complex and that it selectively targets the β and γ subunits in activated forms of the kinase (23). In that same study, we isolated intermolecular peptide conjugates from a βγ dimer that corresponded to cross-linking between the C terminus of γ and the N terminus of β; however, other peptide m/z signals were also observed that could not be eliminated as contaminants, side products of cross-linking, or intermolecular conjugates of β and γ, suggesting that either one or both subunits might be intramolecularly cross-linked within the βγ dimer. To test this possibility, PhK was cross-linked with a 10-fold molar excess of GMBS over αβγδ protomer for 2.5 min at pH 8.2 (Fig. 3A, Lane 2). As described previously, cross-linking resulted in the formation of a major conjugate corresponding to a βγ dimer by apparent mass on SDS-polyacrylamide gels (massTHEOR = 170 kDa, 5.9% error) and cross-reactivity against PhK subunit-specific mAbs (Fig. 3A). To determine potential regions of intramolecular cross-linking of β and γ in the βγ conjugate, this band was excised from a preparative SDS-polyacrylamide gel and digested in gel with trypsin (23). The resulting tryptic peptides were extracted, resolved by reverse phase chromatography, and analyzed by FT-ICR MS.
Fig. 3.
Cross-linking of PhK with GMBS and detection of a potential cross-linked peptide by FT-ICR MS and search engine analyses of a tryptic digest of the major βγ conjugate. A, PhK (Lane 1) was cross-linked with GMBS (Lane 2) and resolved by SDS-PAGE. Parallel samples were transferred to PVDF membranes and probed with mAbs against all of the subunits. All major conjugates cross-reacted only with anti-β and anti-γ mAbs, not with anti-α or anti-δ (δ = integral calmodulin subunit) mAbs. Cross-linking of PhK by GMBS resulted primarily in the formation of a major conjugate corresponding to a βγ dimer by apparent mass (170 kDa) and cross-reactivity (23). B, the cross-linked βγ dimer was digested in gel with trypsin and analyzed by FT-ICR MS. The monoisotopic peak for a peptide with a signal at m/z 658.3597 (∼10× above background) was further subjected to a tandem MS step, and the resulting spectrum was compared against common contaminants as well as tryptic and non-enzymatic hydrolysis products of all the PhK subunits using the Sequest algorithm (37) included in Bioworks 3.2 (ThermoFinnigan). The absence of significant matches observed for the 658 m/z signal in either the Sequest data sets or mass lists obtained from digests of non-cross-linked β and γ subunit controls (A, Lane 2) together indicated the presence of a potential cross-linked peptide.
The mass list from the MS run was uploaded to the cross-link search engine and analyzed as described above for potential β intramolecular conjugates; a parallel screen unsuccessfully analyzed for intramolecular conjugates of γ. A small pool of potential intramolecular β candidates remained after the statistical elimination of experimental masses corresponding to side products of cross-linking, contaminants, and non-modified peptides. To verify and expand our statistical approach to eliminating side products, tandem mass spectra of all candidate ions were analyzed using the Sequest algorithm to search for contaminants and for non-modified peptides arising either from tryptic or non-enzymatic cleavage of the subunits (37). Of the remaining potential conjugates, only those signals with peptide-characteristic isotopic envelopes that were clearly above background (>6×) were further analyzed (Fig. 3B). A signal at m/z 658.3597 (intensity = 3.72e4) for a doubly charged ion best matched a mass (m/zTHEOR = 658.3524, 11.10-ppm error) predicted for a peptide corresponding to cross-linking between the N-terminal 21–23 (TKR) and C-terminal 1037–1042 (DESRLK) residues of β (Fig. 3B). Because the probability of hits increases with mass error, we used a set of internal standards corresponding to non-GMBS-modified tryptic peptides of γ (five total) and β (18 total) to assess the dynamic range in mass accuracy for the MS run and to estimate the potential error for peaks with similar intensities to the 658 signal. The average intensity (2.33e4 ± 1.33e4) and error (5.45 ± 4.80 ppm, ranging from 1.30 to 19.17 ppm) demonstrated that the error value calculated for the β intramolecular conjugate was well within the range observed for the standards and only 0.85 ppm outside the sum of their average errors and standard deviations (Eavg + S.D.). Given the number of possible modifications of the cross-linker (Fig. 2), the large combined mass of the βγ conjugate (170 kDa), and the large number of potential combinations of cross-linked and monoderivatized peptides that may account for a single ion (for an example see Table I), we analyzed all possible combinations of cross-linking (βγ intermolecular and β and γ intramolecular) ± monoderivatization, progressing to twice the value (20.50 ppm) calculated for Eavg + S.D. Using this extended error range, only one match (m/zTHEOR = 658.3726, 19.6-ppm error) was observed that corresponded to a monoderivatized γ peptide (γ267–275: FLVVQPQKR + a Lys-274 ɛ-amine-substituted form of product 9; Fig. 2). No further matches were observed until progressing to ∼30 ppm (3 × Eavg + S.D.); match predictions in this range were rejected as they were all far outside the error range measured for the internal standards.
Table I.
Possible conjugates (±monoderivatization) that may correspond to a single signal (e.g. m/z 2509.3101) identified by CrossSearch from a digest of the PhK βγ dimer cross-linked with GMBS
The two separate cross-linked peptides of each conjugate are shown separated by a colon (:).
| m/z calculated | Error | Sequence | Cross-link
|
Monoderivatization
|
||
|---|---|---|---|---|---|---|
| Number | Structurea | Number | Structurea,b | |||
| ppm | ||||||
| 2509.3140 | 1.6 | RVK:IYGHWVKKGQQQNR | 1 | 3 | 1 | 9 |
| 2509.3240 | 5.6 | DPYALR:EILGRGVSSVVRR | 1 | 3 | 1 | 2 or 8 |
| 2509.3279 | 7.1 | PVTREIVIR:IYYQYRR | 1 | 3 | 1 | 9 |
| 2509.3391 | 11.6 | HFSPRGK:KVSGHPNIIQLK | 1 | 3 | 1 | 2 or 8 |
| 2509.2705 | 15.8 | IYYQYR:DPYALRPLRR | 1 | 3 | 1 | 2 or 8 |
| 2509.2705 | 15.8 | IYYQYRR:DPYALRPLR | 1 | 3 | 1 | 2 or 8 |
| 2509.2412 | 27.4 | HFSPR:GQQQNRAALFENTPK | 1 | 3 | ||
| 2509.4153 | 41.9 | KIMR:PVTREIVIRDPYALR | 1 | 3 | ||
| 2509.2012 | 43.4 | RCIHK:HFSPRGKFK | 2 | 5 | 2 | 2 or 8, 9 |
The numbers indicated in the column refer to the corresponding structure shown in Fig. 2. Side-chain substitutions will vary based on the nucleophilic chains of Arg, Cys, Lys, Tyr, or His being present in the peptides shown for each conjugate.
Structures with the same mass are indicated as “a” or “b.”
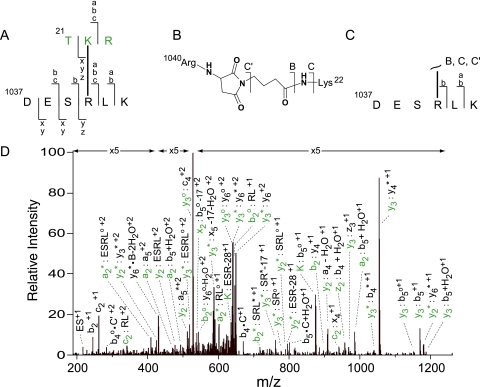
The tandem mass spectrum of the m/z signal at 658 was first analyzed using the best match ion as a template to identify fragment masses. All major peak assignments were made within the error limits of the collision cell (supplemental Table 1). Both the total ion mass and the observed fragmentation pattern confirmed the sequence assignments for each region (21–23 and 1037–1042) of the β subunit and showed the incorporation of 1 mol of the GMBS reagent, which covalently coupled Lys-21 and Arg-1040 (Fig. 4A). Modification of these residues is consistent with the inability of trypsin to hydrolyze the amide bonds immediately following each residue (1). The chemistry of cross-linking was deduced from cleavage products resulting from disruption of the amide bonds of the intervening cross-link (Fig. 4, B and C), demonstrating that Lys-22 and Arg-1040 add, respectively, to the succinimide ester and maleimide groups to form the expected amide linkage and Michael adduct (8). This same mode of addition to each reactive group of GMBS was observed in a previous study showing cross-linking between Lys and Arg side-chain donors from the γ subunit and a peptide mimetic of the β subunit in a PhK·β peptide complex (23). Targeting of the GMBS maleimide functional group by Arg has been attributed previously to the tight initial binding of GMBS by PhK, i.e. affinity cross-linking (23), and is consistent with several reports that suggest side-chain guanidinyl nitrogens may act as nucleophiles in the formation of Arg-containing conjugates (39–41), whereas the kinetically preferred addition of Lys ɛ-amines over that of other protein nucleophilic side chains to succinimide esters is well characterized (8).
Fig. 4.
MS/MS analysis of the signal at m/z 658.3597 identifying a conjugate comprising residues 21–23 and 1037–1042 of the regulatory β subunit of PhK. A, fragmentation pattern of the cross-linked peptide. Small letters denote ions arising from amide cleavages of the peptide backbone and are color-coded with the respective peptide in the conjugate pair (green and black for residues 21–23 and 1037–1042, respectively). B, structure of the GMBS cross-link between Lys-22 and Arg-1040. The capital letters indicate ions resulting from amide cleavage of the reagent and ɛ-amine of Lys-22. C, fragmentation pattern of ions corresponding to cleavage (∼) of the intervening cross-link. D, composition of ions identifying the cross-linked peptide. Intact covalent links formed between peptides are indicated by a colon (:), and covalent links between fragments of the cross-link and peptide are indicated by a centered dot (·). For singly charged ions, it should be noted that one of the two peptide ions covalently attached to either functional group of GMBS is a neutral product of the indicated backbone amide cleavage. * and o, loss of ammonia and water.
The second best match (∼2× the error of the first) corresponding to a monoderivatized γ267–275 peptide was also evaluated; however, only a small percentage (28%) of the fragments in the tandem mass spectrum shown in Fig. 4D could be assigned to potential fragments of this peptide, and of those few matched, almost all were assigned with overall greater mass error than fragments predicted for the β intramolecular cross-link (supplemental Table 1). The results strongly indicate that the β subunit undergoes intramolecular cross-linking by GMBS in the PhK complex and are consistent with a previous report demonstrating the same general type of cross-linking for this subunit in phosphorylated forms of the kinase (42). The evidence reported herein for cross-linking between the N and C termini of β represents the first report of a tertiary structural element for this subunit within the PhK complex (Fig. 5).
Fig. 5.
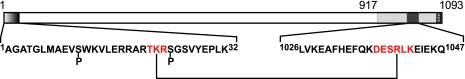
Domain map of the β subunit of PhK showing regions of intramolecular cross-linking by GMBS. Seryl residues within the N-terminal region that are phosphorylated by either cAMP-dependent protein kinase or PhK autophosphorylation are indicated by a P below them. The N-terminal 32 residues of β, indicated by gradation from white to black, represent a region of the subunit that has been shown to regulate homodimeric β interactions in two-hybrid screens (23). A region required for homodimeric interactions of β in two-hybrid screens corresponds to residues 917–1093 (light gray) and includes a stretch of residues (1026–1047) that are a predicted to have high propensity for forming a coiled-coil domain (23). GMBS intramolecular cross-linking of these two regions in the β subunit of the activated PhK complex is indicated by peptides (shown in red lettering) cross-linked through residues Lys-22 and Arg-1040.
DISCUSSION
The computational requirements for detecting cross-linked peptides from the vast array of products that arise from the cross-linking and subsequent digestion of proteins are considerable, especially when the proteins cross-linked are not small. Several approaches have been developed to predict the identity of potential conjugates from the mass output measured for digests of cross-linked proteins (29–33); however, they either require a general understanding of the chemistry involved or the use of specific labeling techniques (6, 14–19, 27, 28). Most search strategies reported to date have focused on the analysis of products generated from the digestion of conjugates of relatively low mass (<60 kDa); however, as the mass of a conjugate increases, so does the number of potential covalent linkages it may contain (e.g. intra- and intermolecular cross-linking, monoderivatizations, and combinations of the lot). In fact, a conservative estimate suggests that the number of possible combinations may increase exponentially to the third power of the number of tryptic peptides generated (6). We report herein the development of CrossSearch, a user-friendly data base search engine that provides the foundation of an overarching strategy to detect cross-linked peptides from either low or high mass (≥100 kDa) conjugates. Our strategy comprises three major steps: prediction, elimination of side products of cross-linking, and verification of cross-link assignments.
To fully test the capabilities of CrossSearch, cross-linking of the 1.3-MDa PhK complex was carried out with the heterobifunctional cross-linker GMBS followed by analysis of the resulting major conjugate, a βγ dimer. In the first step of the analysis, the extensive data base of potential linkage combinations generated by CrossSearch for the 170-kDa PhK βγ conjugate virtually assured multiple assignments for each experimental mass often with several assignments falling within an error range of 5–6 ppm (Table I). Similarly multiple matches for precursor ions within this error range were also predicted for conjugates of the 176-kDa NDC complex (6). Various labeling strategies have been used successfully to reduce the pool of potential cross-links for low mass conjugates (6, 14–19, 27, 28); however, analysis of conjugates with masses over 100 kDa formed by cross-linking of the aforementioned NDC80 complex with both non-labeled and deuterated forms of a cross-linker suggests that such strategies do not adequately reduce the amount of experimental data to a level that readily facilitates manual inspection and verification of all possible cross-link candidates (6). No matter the reagent used, the study herein and those of others (6, 27) demonstrate that a rigorous assessment of the mass error for candidate m/z signals should be carried out to reduce the multiplicity of assignments generated by large conjugate data bases. Such an assessment is particularly pertinent for low intensity signals corresponding to cross-linked peptides. For large protein complexes like PhK, it is difficult to achieve high yields of cross-linking between specific sites on adjacent protein pairs (dimeric conjugates being the easiest to analyze) because dimers are generally consumed in the formation of multiple, larger low yield multimeric conjugates in the ongoing process of cross-linking (1). Our approach using non-modified peptide m/z signals with intensities bracketing those of candidate ions compensates for the dynamic range of error in mass measurements (43, 44), and it allows for removal with high confidence of those assignments with errors 2× the average mass error.
The size of the predicted data base can also be reduced significantly by eliminating the many possible side products of cross-linking, which in sum provide a significant source of potential false positives. CrossSearch provides a statistical basis for eliminating such products by mass matching using an adjustable mass error window, allowing values to be entered corresponding to the accuracy estimated for the mass spectrometer used. Choice of the correct error value is an important parameter: values that are either too small or large result correspondingly in the loss of correct assignments or the incorporation of false positive assignments. To alleviate this complication, a pool of non-modified and monoderivatized peptides is generated independently from the experimental data with the Sequest algorithm using low confidence correlation scores (see “Experimental Procedures”). The use of low correlation scores simultaneously allows for maximum capture of side products (potential false positives) and a means of comparing cross-link assignments from CrossSearch against potential side products generated from Sequest. In the event that hits from either source are observed for a single experimental m/z signal, fragmentation analyses are then carried out to verify the assignment as being correct. Although the methods described above may reduce the number of potential candidates by as much as a factor of 10, the number of remaining candidates may still be very large, depending on the mass of the conjugate and the mole ratio of cross-linker to proteins used in the cross-linking experiment. An additional strategy used herein to limit the number of potential side reactions is to limit the mole ratio of cross-linker to protein. In screens of the PhK complex against a library of cross-linkers, GMBS was determined to be an affinity cross-linker that bound to PhK and formed significant amounts of a single conjugate at only a 10-fold excess of cross-linker over the αβγδ tetramer (23). This MSEC method used herein is an alternative approach to the use of labeled cross-linkers and has multiple advantages over labeling strategies. (i) Cross-linkers with varying chemistries and cross-linking spans may used. (ii) It accommodates the use of zero-length cross-linkers, which in the absence of high resolution reference structures are the best reagents for determining actual contact sites between interacting proteins. (iii) Conventional cross-linkers are more economical than their labeled counterparts. The major disadvantage of MSEC is that shared by all cross-linkers, namely the ability to selectively form a conjugate of interest.
The final step in the analysis, verification of assignments, is carried out using a combination of programs. The fragmentation patterns of candidates are first evaluated using MS2Links (31), a program that allows for rapid analysis of cross-linked peptides, ranking possible assignments for each ion fragment m/z signal in order of error. That program automatically calculates peptide backbone cleavages for the candidate peptide sequences entered; however, the user must provide mass information relating to the cross-linker, its potential cleavage products, and the amino acid side chains that are likely targeted by the reagent. In our analysis of GMBS, we loaded all possible fragments arising from cleavage of the amide bonds of the reagent (23) as well as those reported for structural analogs of GMBS (45). All assignments in this phase of the analysis are checked manually in what is the most time-consuming part of the process. Recently an algorithm has been developed that may speed up this process by directly scoring the fragmentation patterns of cross-linked peptides and by providing estimates for the probability of false positive predictions; manual inspection of the data is still required albeit on a more limited basis (6).
After eliminating probable false positives and side products from the βγ conjugate data base, previously reported conjugates corresponding to intermolecular cross-linking between the C terminus of γ and the N terminus of β were observed (23). In addition to these intersubunit cross-links, we also observed one intramolecular β conjugate, corresponding to cross-linking between the N and C termini of this subunit.
The low number of cross-links detected is consistent with the stoichiometry of cross-linking imposed using the MSEC approach; however, because the total sequence coverage estimated for the conjugate was ∼50% (using both CrossSearch and Sequest for verification of assignments), the presence of additional cross-links in the conjugate is certainly possible.
The cross-link reported herein between Lys-22 and Arg-1040 of the β subunit provides the first glimpse of the tertiary structure of either of the two large, homologous, regulatory α and β subunits of PhK (46). The N-terminal region containing Lys-22 is unique to the β subunit, contains the major activating phosphorylation site of PhK (Ser-26) and a second phosphorylation site (Ser-11), and is relatively near (≤12 Å) the active site of the catalytic γ subunit as well as its C-terminal regulatory domain (23). We previously reported that its first 31 residues mediate self-association of the β subunit in that either their deletion or the mutation of the phosphorylatable Ser-11 and Ser-26 allows self-association to be observed in yeast two-hybrid screens (23). In those screens deletion of the C-terminal 177 residues of the 1093-residue β subunit, which contains Arg-1040, totally eliminated self-association. The most straightforward explanation for those two-hybrid results would be that the full-length wild type β subunit is unable to self-associate because its N terminus interacts with its C terminus, thus blocking self-association of the latter. Such a conclusion could not be reached from two-hybrid data, however, because observed homodimeric interactions could represent either intrasubunit or intersubunit interactions, which would be indistinguishable unless a single small region self-associated (47). The possibility of intersubunit interactions is of special concern with PhK because ββ dimers can be readily formed in activated PhK by a short cross-linker (42). Our current data corroborate and extend the previous two-hybrid results by supporting the concept of an intrasubunit interaction between the N and C termini of the β subunit. Further the results herein considered together with those two-hybrid data suggest a structural model for PhK in which the regulatory N terminus of the β subunit may participate in opposing interactions with the catalytic γ subunit and with its own C terminus, depending upon its phosphorylation state. The detection and identification of a cross-linked peptide from the βγ digest demonstrates the utility of the CrossSearch strategy in the analysis of large proteins by cross-linking.
Supplementary Material
Footnotes
Published, MCP Papers in Press, February 16, 2008, DOI 10.1074/mcp.M800020-MCP200
The abbreviations used are: PhK, phosphorylase kinase; GMBS, N-[γ-maleimidobutyryloxy]succinimide ester; MSEC, minimal stoichiometric excess of cross-linker; mAb, monoclonal antibody; NDC, nuclear division cycle; Pr, protein; LC ESI, liquid chromatography electrospray ionization; PHP, hypertext preprocessor; theor., theoretical; XHTML, extensible hypertext markup language.
This work was supported by National Institutes of Health Grant DK32953 (to G. M. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Nadeau, O. W., and Carlson, G. M. (2005) Protein interactions captured by chemical cross-linking, in Protein-Protein Interactions, A Molecular Cloning Manual (Golemis, E., and Adams, P. D., eds) 2nd Ed., pp. 105–127, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 2.Sinz, A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25 663–682 [DOI] [PubMed] [Google Scholar]
- 3.Nadeau, O. W. (2006) Protein interaction analysis: chemical cross-linking, in Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine (Ganten, D., and Ruckpaul, K., eds) pp. 1506–1509, Springer, Berlin
- 4.Soderblom, E. J., and Goshe, M. B. (2006) Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78 8059–8068 [DOI] [PubMed] [Google Scholar]
- 5.Nadeau, O. W., and Carlson, G. M. (eds) (2002) Protein-protein interactions, in Protein-Protein Interactions, A Molecular cloning Manual (Golemis, E., ed) 1st Ed., pp. 75–91, Cold Spring Harbor Press, Cold Spring Harbor, NY
- 6.Maiolica, A., Cittaro, D., Borsotti, D., Sennels, L., Ciferri, C., Tarricone, C., Musacchio, A., and Rappsilber, J. (2007) Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell. Proteomics 6 2200–2211 [DOI] [PubMed] [Google Scholar]
- 7.Wong, S. S. (1993) Chemistry of Protein Conjugation and Cross-linking, pp. 214–215, CRC Press, Boca Raton, FL
- 8.Hermanson, G. T. (1996) Bioconjugate Techniques, p. 148, Academic Press, San Diego, CA
- 9.Swaim, C. L., Smith, J. B., and Smith, D. L. (2004) Unexpected products from the reaction of the synthetic cross-linker 3,3′-dithiobis(sulfosuccinimidyl propionate), DTSSP with peptides. J. Am. Soc. Mass Spectrom. 15 736–749 [DOI] [PubMed] [Google Scholar]
- 10.Reynolds, K. J., Yao, X., and Fenselau, C. (2002) Proteolytic 18O labeling for comparative proteomics: evaluation of endoprotease Glu-C as the catalytic agent. J. Proteome Res. 1 27–33 [DOI] [PubMed] [Google Scholar]
- 11.Staes, A., Demol, H., Van Damme, J., Martens, L., Vandekerckhove, J., and Gevaert, K. (2004) Global differential non-gel proteomics by quantitative and stable labeling of tryptic peptides with oxygen-18. J. Proteome Res. 3 786–791 [DOI] [PubMed] [Google Scholar]
- 12.Hurst, G. B., Lankford, T. K., and Kennel, S. J. (2004) Mass spectrometric detection of affinity purified crosslinked peptides. J. Am. Soc. Mass Spectrom. 15 832–839 [DOI] [PubMed] [Google Scholar]
- 13.Sinz, A., Kalkhof, S., and Ihling, C. (2005) Mapping protein interfaces by a trifunctional cross-linker combined with MALDI-TOF and ESI-FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 16 1921–1931 [DOI] [PubMed] [Google Scholar]
- 14.Chu, F., Mahrus, S., Craik, C. S., and Burlingame, A. L. (2006) Isotope-coded and affinity-tagged cross-linking (ICATXL): an efficient strategy to probe protein interaction surfaces. J. Am. Chem. Soc. 128 10362–10363 [DOI] [PubMed] [Google Scholar]
- 15.Taverner, T., Hall, N. E., O'Hair, R. A., and Simpson, R. J. (2002) Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking, and mass spectrometry. J. Biol. Chem. 277 46487–46492 [DOI] [PubMed] [Google Scholar]
- 16.Collins, C. J., Schilling, B., Young, M., Dollinger, G., and Guy, R. K. (2003) Isotopically labeled crosslinking reagents: resolution of mass degeneracy in the identification of crosslinked peptides. Bioorg. Med. Chem. Lett. 13 4023–4026 [DOI] [PubMed] [Google Scholar]
- 17.Petrotchenko, E. V., Olkhovik, V. K., and Borchers, C. H. (2005) Isotopically coded cleavable cross-linker for studying protein-protein interaction and protein complexes. Mol. Cell. Proteomics 4 1167–1179 [DOI] [PubMed] [Google Scholar]
- 18.Seebacher, J., Mallick, P., Zhang, N., Eddes, J. S., Aebersold, R., and Gelb, M. H. (2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5 2270–2282 [DOI] [PubMed] [Google Scholar]
- 19.Soderblom, E. J., Bobay, B. G., Cavanagh, J., and Goshe, M. B. (2007) Tandem mass spectrometry acquisition approaches to enhance identification of protein-protein interactions using low-energy collision-induced dissociative chemical crosslinking reagents. Rapid Commun. Mass Spectrom. 21 3395–3408 [DOI] [PubMed] [Google Scholar]
- 20.Back, J. W., Hartog, A. F., Dekker, H. L., Muijsers, A. O., de Koning, L. J., and de Jong, L. (2001) A new crosslinker for mass spectrometric analysis of the quaternary structure of protein complexes. J. Am. Soc. Mass Spectrom. 12 222–227 [DOI] [PubMed] [Google Scholar]
- 21.Chowdhury, S. M., Munske, G. R., Tang, X., and Bruce, J. E. (2006) Collisionally activated dissociation and electron capture dissociation of several mass spectrometry-identifiable chemical cross-linkers. Anal. Chem. 78 8183–8193 [DOI] [PubMed] [Google Scholar]
- 22.Tang, X., Munske, G. R., Siems, W. F., and Bruce, J. E. (2005) Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal. Chem. 77 311–318 [DOI] [PubMed] [Google Scholar]
- 23.Nadeau, O. W., Anderson, D. W., Yang, Q., Artigues, A., Paschall, J. E., Wyckoff, G. J., McClintock, J. L., and Carlson, G. M. (2007) Evidence for the location of the allosteric activation switch in the multisubunit phosphorylase kinase complex from mass spectrometric identification of chemically crosslinked peptides. J. Mol. Biol. 365 1429–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadeau, O. W., Sacks, D. B., and Carlson, G. M. (1997) The structural effects of endogenous and exogenous Ca2+/calmodulin on phosphorylase kinase. J. Biol. Chem. 272 26202–26209 [DOI] [PubMed] [Google Scholar]
- 25.Nadeau, O. W., Sacks, D. B., and Carlson, G. M. (1997) Differential affinity cross-linking of phosphorylase kinase conformers by the geometric isomers of phenylenedimaleimide. J. Biol. Chem. 272 26196–26201 [DOI] [PubMed] [Google Scholar]
- 26.Carlson, G. M. (1984) Precautions when determining kinetically the order of inactivation of enzymes by functionally irreversible inhibitors. Biochim. Biophys. Acta 789 347–350 [DOI] [PubMed] [Google Scholar]
- 27.Anderson, G. A., Tolic, N., Tang, X., Zheng, C., and Bruce, J. E. (2007) Informatics strategies for large-scale novel cross-linking analysis. J. Proteome Res. 6 3412–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao, Q., Xue, S., Doneanu, C. E., Shaffer, S. A., Goodlett, D. R., and Nelson, S. D. (2006) Pro-CrossLink. Software tool for protein cross-linking and mass spectrometry. Anal. Chem. 78 2145–2149 [DOI] [PubMed] [Google Scholar]
- 29.de Koning, L. J., Kasper, P. T., Back, J. W., Nessen, M. A., Vanrobaeys, F., Van Beeumen, J., Gherardi, E., de Koster, C. G., and de Jong, L. (2006) Computer-assisted mass spectrometric analysis of naturally occurring and artificially introduced cross-links in proteins and protein complexes. FEBS J. 273 281–291 [DOI] [PubMed] [Google Scholar]
- 30.Lee, Y. J., Lackner, L. L., Nunnari, J. M., and Phinney, B. S. (2007) Shotgun cross-linking analysis for studying quaternary and tertiary protein structures. J. Proteome Res. 6 3908–3917 [DOI] [PubMed] [Google Scholar]
- 31.Schilling, B., Row, R. H., Gibson, B. W., Guo, X., and Young, M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14 834–850 [DOI] [PubMed] [Google Scholar]
- 32.Wefing, S., Schnaible, V., and Hoffmann, D. (2006) SearchXLinks. A program for the identification of disulfide bonds in proteins from mass spectra. Anal. Chem. 78 1235–1241 [DOI] [PubMed] [Google Scholar]
- 33.Young, M. M., Tang, N., Hempel, J. C., Oshiro, C. M., Taylor, E. W., Kuntz, I. D., Gibson, B. W., and Dollinger, G. (2000) High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 97 5802–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King, M. M., and Carlson, G. M. (1981) Synergistic activation by Ca2+ and Mg2+ as the primary cause for hysteresis in the phosphorylase kinase reactions. J. Biol. Chem. 256 11058–11064 [PubMed] [Google Scholar]
- 35.Wilkinson, D. A., Marion, T. N., Tillman, D. M., Norcum, M. T., Hainfeld, J. F., Seyer, J. M., and Carlson, G. M. (1994) An epitope proximal to the carboxyl terminus of the α-subunit is located near the lobe tips of the phosphorylase kinase hexadecamer. J. Mol. Biol. 235 974–982 [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson, D. A., Norcum, M. T., Fizgerald, T. J., Marion, T. N., Tillman, D. M., and Carlson, G. M. (1997) Proximal regions of the catalytic γ and regulatory β subunits on the interior lobe face of phosphorylase kinase are structurally coupled to each other and with enzyme activation. J. Mol. Biol. 265 319–329 [DOI] [PubMed] [Google Scholar]
- 37.Eng, K. J., McCormack, A. L., and Yates, J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5 976–989 [DOI] [PubMed] [Google Scholar]
- 38.Chalkley, R. J., Baker, P. R., Huang, L., Hansen, K. C., Allen, N. P., Rexach, M., and Burlingame, A. L. (2005) Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting, quadrupole collision cell, time-of-flight mass spectrometer. II. New developments in Protein Prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol. Cell. Proteomics 4 1194–1204 [DOI] [PubMed] [Google Scholar]
- 39.Beavis, R. C., Kneirman, M. D., Sharknas, D., Heady, M. A., Frank, B. H., and DeFelippis, M. R. (1999) A novel protein cross-linking reaction in stressed Neutral Protamine Hagedorn formulations of insulin. J. Pharm. Sci. 88 331–336 [DOI] [PubMed] [Google Scholar]
- 40.Hoffman, T. (1998) 4-Alkylidene-2-imino-5-[4-alkylidene-5-oxo-1,3-imidazol-2-inyl]aza-methylidene-1,3-imidazolidine—a novel colored substructure in melanoidins formed by Maillard reactions of bound arginine with glyoxal and furan-2-carboxaldehyde. Biochem. J. 46 3896–3901 [Google Scholar]
- 41.Tessier, F. J., Monnier, V. M., Sayre, L. M., and Kornfield, J. A. (2003) Triosidines: novel Maillard reaction products and cross-links from the reaction of triose sugars with lysine and arginine residues. Biochem. J. 369 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzgerald, T. J., and Carlson, G. M. (1984) Activated states of phosphorylase kinase as detected by the chemical cross-linker 1,5-difluoro-2,4-dinitrobenzene. J. Biol. Chem. 259 3266–3274 [PubMed] [Google Scholar]
- 43.Makarov, A., Denisov, E., Lange, O., and Horning, S. (2006) Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom. 17 977–982 [DOI] [PubMed] [Google Scholar]
- 44.Peterman, S. M., Dufresne, C. P., and Horning, S. (2005) The use of a hybrid linear trap/FT-ICR mass spectrometer for on-line high resolution/high mass accuracy bottom-up sequencing. J. Biomol. Tech. 16 112–124 [PMC free article] [PubMed] [Google Scholar]
- 45.Novak, P., Haskins, W. E., Ayson, M. J., Jacobsen, R. B., Schoeniger, J. S., Leavell, M. D., Young, M. M., and Kruppa, G. H. (2005) Unambiguous assignment of intramolecular chemical cross-links in modified mammalian membrane proteins by Fourier transform-tandem mass spectrometry. Anal. Chem. 77 5101–5106 [DOI] [PubMed] [Google Scholar]
- 46.Kilimann, M. W., Zander, N. F., Kuhn, C. C., Crabb, J. W., Meyer, H. E., and Heilmeyer, L. M., Jr. (1988) The α and β subunits of phosphorylase kinase are homologous: cDNA cloning and primary structure of the β subunit. Proc. Natl. Acad. Sci. U. S. A. 85 9381–9385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayers, N. A., Wilkinson, D. A., Fitzgerald, T. J., and Carlson, G. M. (1999) Self-association of the α subunit of phosphorylase kinase as determined by two-hybrid screening. J. Biol. Chem. 274 35583–35590 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.