Abstract
Signal transduction involves posttranslational modifications and protein-protein interactions, which can be studied by proteomics. In Arabidopsis, the steroid hormone (brassinosteroid (BR)) binds to the extracellular domain of a receptor kinase (BRI1) to initiate a phosphorylation/dephosphorylation cascade that controls gene expression and plant growth. Here we detected early BR signaling events and identified early response proteins using prefractionation and two-dimensional (2-D) DIGE. Proteomic changes induced rapidly by BR treatments were detected in phosphoprotein and plasma membrane (PM) fractions by 2-D DIGE but not in total protein extracts. LC-MS/MS analysis of gel spots identified 19 BR-regulated PM proteins and six proteins from phosphoprotein fractions. These include the BAK1 receptor kinase and BZR1 transcription factor of the BR signaling pathway. Both proteins showed spot shifts consistent with BR-regulated phosphorylation. In addition, in vivo phosphorylation sites were identified for BZR1, two tetratricopeptide repeat proteins, and a phosphoenolpyruvate carboxykinase (PCK1). Overexpression of a novel BR-induced PM protein (DREPP) partially suppressed the phenotypes of a BR-deficient mutant, demonstrating its important function in BR responses. Our study demonstrates that prefractionation coupled with 2-D DIGE is a powerful approach for studying signal transduction.
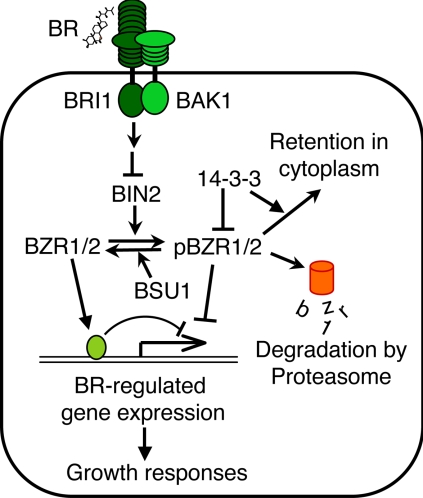
Perception and response to extracellular signals are crucial for growth and survival of all organisms. Signals are transduced intracellularly through mechanisms that include posttranslational protein modification and protein-protein interaction. Detection of such signaling events using proteomics methods can identify proteins involved in signal transduction but is technically challenging because of the low abundance of signaling proteins. Brassinosteroids (BRs)1 are plant hormones that play important roles in multiple plant developmental processes. Mutant plants with a defect in BR synthesis or signal transduction show a wide range of phenotypes including dwarfism, reduced fertility, light-grown morphology in the dark, and delayed senescence (1). BRs structurally resemble animal steroid hormones but act through a distinct signaling mechanism (2). Although animal steroid hormones are perceived by nuclear receptors, BRs are perceived by a cell surface receptor-like kinase named BRI1 (2, 3). BRs bind to the extracellular domain of BRI1 to activate its kinase and downstream BR signal transduction (4), which involves another receptor-like kinase named BAK1 (5, 6); a GSK3-like kinase, BIN2 (7); a phosphatase, BSU1 (8); and two transcription factors, BZR1 and BZR2 (also known as BES1) (9, 10). In the absence of BR, the inhibitory BIN2 kinase phosphorylates BZR1 and BZR2, and phosphorylation inhibits the function of BZR1 and BZR2 through multiple mechanisms, which include inhibition of DNA binding activity, degradation by the proteasome, and cytoplasmic retention by the 14-3-3 proteins that bind to the phosphorylated BZR1 and BZR2 (11–13). BR binding causes dimerization and activation of BRI1 and BAK1 (4, 14), leading to dephosphorylation of BZR1 and BZR2 (10, 15) presumably by inhibition of BIN2 or activation of BSU1 through unknown mechanisms. Dephosphorylated BZR1 and BZR2 accumulate in the nucleus, bind to DNA, and regulate gene expression (Fig. 1) (16, 17). There are still gaps in our understanding of the BR signaling pathway, and in particular, how activated receptor kinases regulate the cytoplasmic BIN2 kinase and BSU1 phosphatase remains unclear (13). A proteomics study of BR-regulated proteins can potentially identify additional components of the BR signal transduction pathway.
Fig. 1.
A model for the BR signaling pathway in Arabidopsis. Arrows and bars represent actions of promotion and inhibition, respectively. BR binds to the extracellular domain of the BRI1 receptor kinase to increase its kinase activity and induce dimerization with and phosphorylation of the BAK1 receptor kinase. The signal is transduced through an unknown mechanism to inhibit BIN2 kinase or activate BSU1 phosphatase, leading to dephosphorylation of BZR1 and BZR2/BES1 (denoted together as BZR1/2). BIN2 phosphorylation of BZR1/2 (pBZR1/2, phosphorylated BZR1 or BZR2/BES1) inhibits its DNA binding, targets it for proteasomal degradation, and increases its cytoplasmic retention by promoting binding to the 14-3-3 proteins. BR-induced dephosphorylation increases nuclear accumulation and DNA binding activity of BZR1 and BZR2/BES1, leading to BR-responsive gene expression and growth responses (13).
Two-dimensional gel electrophoresis (2-DE) is a powerful proteomics technique to differentially display protein expression and posttranslational modifications. 2-DE can separate thousands of proteins based on their differences in charge and size (18, 19). A recent improvement of the technique is called two-dimensional (2-D) DIGE (20). In 2-D DIGE, protein samples to be compared are covalently labeled with different fluorescence dyes (Cy2, Cy3, and Cy5), mixed together, and separated in a single gel of 2-DE. Gel images with identical protein spot pattern are acquired by scanning the gel at a specific wavelength for each dye, and spot intensities can be compared directly between samples and quantified using image analysis softwares (20). The accuracy of 2-D DIGE for quantitative proteomics has been confirmed by analyzing the same samples using immunoblotting (21, 22) or metabolic stable isotope labeling (23). One of the advantages of 2-D DIGE is that it can detect not only changes of protein quantity but also posttranslational modifications that change the charge or size of the protein (23, 24). 2-D DIGE has been widely used for identifying disease markers and proteins involved in biological responses (25, 26). 2-DE has been used to monitor molecular responses induced by the activation or inhibition of specific signaling pathways (27). However, because 2-DE has a bias toward abundant proteins, signaling proteins of low abundance, such as kinases and transcription factors, often cannot be detected in 2-DE analysis of total cellular proteins (26, 28). A study of BR-regulated proteins using 2-D DIGE analysis of total proteins failed to detect any of the known components of the BR signaling pathway or early BR-responsive proteins (22). Therefore, enrichment of low abundance proteins using prefractionation methods seems to be required for identifying components of signal transduction pathways in Arabidopsis.
In this study, we analyzed BR-regulated proteins in the plasma membrane (PM) and phosphoprotein fractions using 2-D DIGE and identified BR-regulated phosphorylation changes of components of the BR signaling pathway. Our study demonstrates that signal transduction components can be identified using 2-D DIGE after prefractionation. We identified a number of novel BR early response proteins and showed a functional role for one of the BR-regulated PM proteins in BR promotion of plant growth. Our study demonstrates that prefractionation followed by 2-D DIGE is a powerful approach for studying signal transduction.
EXPERIMENTAL PROCEDURES
Materials—
Polyethylene glycol 3350, Murashige-Skoog salt, and acetonitrile were from Sigma. Dextran T-500 was from Fisher Scientific. CyDye DIGE fluors, dimethylformamide, DTT, iodoacetamide, IPG strips, IPG buffers, and Deep Purple fluorescence dye were purchased from GE Healthcare. Sequencing grade trypsin was from Roche Applied Science.
Plant Materials and Growth Conditions—
The BR-deficient mutant det2-1 (29), transgenic plants expressing the mutant bzr1-1D gene fused to cyan fluorescent protein (mBZR1-CFP) under the native BZR1 promoter (15), and BAK1-GFP transgenic plants are in the Arabidopsis thaliana Columbia-0 ecotype background. The bak1-1 mutant is in the WS-2 background (6). Seeds were sterilized in 100% bleach for 10 min. After extensive washing with sterilized water, seeds were added to growth medium (1/2 Murashige-Skoog salt, 1.5% sucrose, pH 5.7) at a ratio of 50 mg of seeds/250 ml of medium. Seeds were cold-treated for 2 days to synchronize germination and grown in the dark for 4 days or under continuous light for 7 days on a shaker at a shaking speed of 90 rpm. BL (100 nm) or mock solvent (0.01% ethanol) was added to the culture to start the treatment.
Immobilized Metal Affinity Chromatography—
Phosphorylated proteins were enriched by IMAC (30) with a few modifications. Total proteins extracted with the phenol-methanol method (22) were dissolved in DIGE buffer (6 m urea, 2 m thiourea, 2% CHAPS) at about 6 mg/ml, diluted with wash buffer (6 m urea, 50 mm sodium acetate, pH 4.0, 0.25% CHAPS) to 1 mg/ml, and then incubated with gallium-charged chelating Sepharose fast flow beads (∼10 mg of protein/ml) (GE Healthcare) for 1 h at room temperature. The beads were washed on a column by gravity flow with 20 column volumes of wash buffer. Phosphoproteins were eluted with 2 column volumes of elution buffer (6 m urea, 50 mm Tris acetate, pH 7.4, 0.1 m EDTA, 0.1 m EGTA, 0.25% CHAPS) and precipitated with 5 volumes of methanol.
Plasma Membrane Protein Isolation—
A BR-treated sample and untreated control sample were processed in parallel for plasma membrane isolation according to Larsson et al. (31) with some modification. One-week-old liquid-grown seedlings were collected by filtering through a mesh. Seedlings were surface-dried with tissue paper, weighed, and added to grinding buffer (25 mm HEPES, 0.33 m sucrose, 10% glycerol, 0.6% polyvinylpyrrolidone, 5 mm ascorbic acid, 5 mm EDTA, 1 mm NaF, 1 mm sodium molybdate, 2 mm imidazole, 1 mm activated sodium vanadate, 5 mm DTT, 1 μm E-64, 1 μm bestatin, 1 μm pepstatin, 2 μm leupeptin, 1 mm PMSF, pH 7.5) at a ratio of 2 ml of grinding buffer/g of seedlings. Plant tissue was homogenized (PowerGen 700D, Fisher Scientific) at 20,000 rpm for 10 min on ice. The homogenate was filtered through one layer of Miracloth and then centrifuged at 10,000 × g for 10 min to remove tissue debris. Microsomal membranes were pelleted by centrifugation at 60,000 × g for 45 min. The pellet was resuspended in buffer R (5 mm potassium phosphate, pH 7.8, 0.25 m sucrose, 3 mm KCl, 0.1 mm DTT, 1 μm E-64, 1 μm bestatin, 1 μm pepstatin, 2 μm leupeptin, 1 mm PMSF) by pipetting up and down. Two-phase partitioning was performed by using a solution containing 6% polyethylene glycol 3350, 6% Dextran T-500, and 8 mm KCl. After partitioning, the upper U3 phase (31) solution was diluted with 10 volumes of buffer R and centrifuged at 100,000 × g for 1 h to collect the plasma membrane. The pellet was resuspended in 50–60 μl of buffer R without sucrose by slowly pipetting up and down and then mixed with 5 volumes of 100 mm ammonium acetate in 100% methanol to precipitate the protein. All the procedures were performed in the cold room.
Labeling of Proteins with CyDye—
Precipitated proteins were pelleted by centrifuging at 20,000 × g for 15 min. Supernatant was discarded, and 1 ml of ice-cold methanol was slowly added to the pellet to remove salt. After 30 min on ice, methanol was removed, and the pellet was air-dried for 1 min before adding DIGE buffer (6 m urea, 2 m thiourea, 4% CHAPS, 25 mm Tris-HCl, pH 8.8) to dissolve the protein. Protein concentration was measured using the Bradford method (Bio-Rad) and adjusted to 3–5 μg/μl. For labeling of the proteins, 50 pmol of CyDye was mixed with 10 μl of protein sample and incubated on ice for at least 2 h in the dark. The labeling reaction was terminated by adding 1 μl of 10 mm lysine.
Two-dimensional Electrophoresis and Image Scanning—
For analytical 2-D DIGE analysis, 20 μl of combined Cy3- or Cy5-labeled protein was mixed with 2-D DIGE buffer (6 m urea, 2 m thiourea, 4% CHAPS, 20 mm DTT, 0.5% IPG buffer) and used for IEF. IEF was performed on 24-cm IPG strips, pH 4–7. The running conditions were: rehydration for 2 h, 50 V for 10 h, step and hold at 500 V, 1000 V for 1 h each, and then at 8000 V until reaching a total of 80,000 V-h. Second dimension electrophoresis was performed using 10% SDS-polyacrylamide gels. The electrophoresis was performed at 50 V for 2 h and then at 110 V until the bromphenol blue front reached the end of the gel. Cy3- and Cy5-labeled images were acquired using a Typhoon 8600 scanner (GE Healthcare).
DIGE Image Analysis—
DIGE images were analyzed using DeCyder 6.5 software (GE Healthcare). Spot detection was performed using the DIA (differential in-gel analysis) module with an estimated number of spots set to 4500. After removing the artifact spots by manual editing, DIGE images were further analyzed using the DeCyder BVA (biological variation analysis) module. For each treatment, images from at least three biological repeats were used for statistical analysis of protein abundance.
Spot Picking and LC-MS—
Preparative IEF was performed with increased protein amount loaded into IPG strips for spot picking. Ten microliters of protein (5 μg/μl) were labeled with Cy5, mixed with ∼700 μg of unlabeled protein, and used for 2D electrophoresis as described above. After SDS-PAGE, the gel was stained with Deep Purple dye according to the manufacturer's instructions. The gel was then scanned with the Typhoon scanner using Cy3 and Cy5 channels. The Cy5 image was matched to the images of analytical gels to identify spots of interest. The Cy3 image (Deep Purple) was then aligned with the Cy5 image to mark the spots of interest for spot picking. The spots were excised using an Ettan Spot Picker (GE Healthcare) with a 1.4-mm picker head
Reversed-phase LC-MS/MS Analysis—
Protein spots were excised from gels and digested in-gel with trypsin as described previously (32). Briefly specific protein spots were washed twice with 50% acetonitrile in 25 mm ammonium bicarbonate (NH4HCO3) and vacuum-dried. The gel samples were next reduced with DTT (10 mm in 25 mm NH4HCO3 at 56 °C for 1 h) and alkylated with iodoacetamide (55 mm in 25 mm NH4HCO3 at room temperature for 45 min). Then the gel pieces were vacuum-dried, rehydrated in 8 μl of digestion buffer (10 ng/μl trypsin in 25 mm NH4HCO3), and covered with a minimum volume of NH4HCO3. After overnight digestion at 37 °C, peptides were extracted twice with a solution containing 50% acetonitrile and 5% formic acid. The extracted digests were vacuum-dried and resuspended in 5 μl of 0.1% formic acid in water. The digests were separated by nanoflow liquid chromatography using a 75-μm × 150-mm reversed-phase C18 PepMap column (Dionex-LC Packings, San Francisco, CA) at a flow rate of 300 nl/min in a NanoLC-1D Proteomics high performance liquid chromatography system (Eksigent Technologies, Livermore, CA) equipped with a FAMOS autosampler (Dionex-LC Packings). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. Following equilibration of the column in 5% solvent B, approximately one-fifth of each digest (1 μl) was injected, and then the organic content of the mobile phase was increased linearly to 40% over 30 min and then to 50% in 1 min. The column effluent was directed to a microionspray source attached either to a QSTAR Pulsar or to a QSTAR-XL mass spectrometer (Applied Biosystems/MDS Sciex, South San Francisco, CA). Peptides were analyzed in positive ion mode. MS spectra were acquired for 1 s in the m/z range between 310 and 1400. MS acquisitions were followed by 3-s CID experiments in information-dependent acquisition mode. For each MS spectrum, the most intense multiple charged peaks over a threshold of 30 counts were selected for generation of CID mass spectra. The CID collision energy was automatically set according to m/z ratio and charge state of the precursor ion. A dynamic exclusion window was applied that prevented the same m/z from being selected for 1 min after its acquisition. In some cases, the LC/MS/MS analyses were performed on an LTQ/FT instrument (Thermo Finnigan, Bremen, Germany) where the instrument alternated between acquiring a survey full MS scan in FT mode and three sets (on the three most intense multiple charged ions detected on the survey scan) of zoom scans (in FT mode) plus subsequent scans of the CID products of these same ions (using the linear trap).
QSTAR Pulsar data were analyzed with Analyst QS 1.1 software (Applied Biosystems/MDS Sciex), and peak lists were generated using the mascot.dll script (Mascot.dll 1.6b18, Applied Biosystems). Precursor mass tolerance for grouping was set to 0.2 amu. MS centroiding parameters were 50% peak height and 0.02 amu merge distance. MS/MS centroiding parameters were 50% peak height and 0.05 amu merge distance. LTQ/FT raw data were converted to ASCII peak lists by Mascot Distiller software version 2.1.0.0 (Matrix Science, Boston, MA). Parameters for MS processing were set as follows: peak half-width, 0.02; data points per Da, 100. For MS/MS data parameters were: peak half-width, 0.02; data points per Da, 100.
The peak lists were searched in in-house Protein Prospector version 4.25.4 (a public version is available). Enzyme specificity was set to trypsin, and the maximum number of missed enzyme cleavages per peptide was set at 2. Carbamidomethylation and formation of acrylamide adducts of cysteine, N-acetylation of the N terminus of the protein, oxidation of methionine, and phosphorylation of serine, threonine, or tyrosine were all allowed as variable modifications. Mass tolerance for QSTAR data was 150 ppm for precursor and 0.2 Da for fragment ions. For LTQ/FT data these parameters were set at 10 ppm and 0.8 Da, respectively.
The peak lists were searched against the Arabidopsis subset of the UniProt database as of April 19, 2007 (50,054 entries searched). Control searches of some of the files against the whole database (4,534,260 entries searched) confirmed the absence of contaminations from human keratins or other non-Arabidopsis proteins, so searches were routinely performed on the Arabidopsis subset.
In all protein identifications, a minimal protein score of 22, a peptide score of 15, and a minimal discriminant score threshold of 0.0 were used for initial identification criteria. Maximum expectation values (number of different peptides with scores equivalent to or better than the result reported that are expected to occur in the database search by chance) for accepting individual spectra were set to 0.01. When several accession numbers in the database matched the same set of peptides identified, the entries with the most descriptive name were reported. Individual isoforms of proteins were reported according to the detection of peptides unique to their sequences. If several isoforms shared the same set of peptides identified, they were all reported. For identifications based on one or two peptide sequences with high scores, the MS/MS spectrum was reinterpreted manually by matching all the observed fragment ions to a theoretical fragmentation obtained using MS Product (Protein Prospector) (33). The CID spectra of the peptides identified in Protein Prospector to contain posttranslational modifications were manually inspected for site assignment.
Overexpression of DREPP—
A full-length DREPP (At4g20260) cDNA coding sequence without stop codon was amplified using gene-specific primers and cloned into pENTR/SD/D-TOPO vectors. A DREPP-GFP fusion construct was generated by subcloning the cDNA fragment into the destination plasmid pMDC83 (34) through Gateway cloning system (Invitrogen). Agrobacterium strain GV3101 carrying the pMDC83-DREPP construct was transformed into Arabidopsis plants by floral dipping. The transgenic plants were selected by hygromycin resistance and verified by PCR genotyping and immunoblotting using GFP antibody.
Immunoblotting—
For immunoblotting, the SDS-PAGE gels were transferred to a nitrocellulose membrane and probed with different antibodies followed by detection with the SuperSignal West Dura Chemiluminescence System (Pierce). The polyclonal anti-GFP antisera were produced in rabbits using GFP protein expressed in Escherichia coli, and the polyclonal anti-phosphothreonine antibody was from New England Biolabs
RESULTS
Identification of BR-regulated Phosphoproteins Using Protein IMAC Followed by 2-D DIGE and LC-MS/MS—
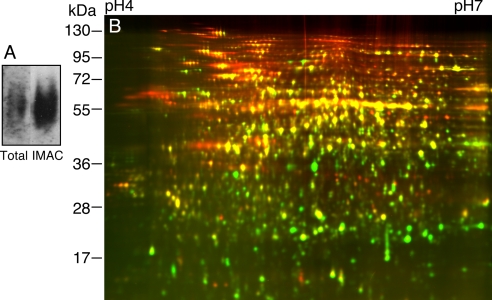
Protein phosphorylation has been shown to play an important role in BR signal transduction. We thus performed 2-D DIGE analysis of phosphoproteins purified using the IMAC method described previously (30). Seedlings of transgenic plants expressing the mBZR1-CFP protein under the control of BZR1 native promoter were used. Previous studies have shown that the dominant bzr1-1D mutation stabilizes the mBZR1 protein and that mBZR1 is dephosphorylated upon BR treatment (14). Furthermore our previous data showed that the presence of the CFP in the BZR1-CFP fusion protein does not impede phosphorylation/dephosphorylation behavior of the BZR1 protein (15). The seedlings were grown in the dark for 4 days, and total cellular protein was extracted using SDS buffer and precipitated by phenol and methanol. The protein pellet was resuspended in 6 m urea, 2 m thiourea, plus 2% CHAPS and subjected to IMAC purification of phosphoproteins as described under “Experimental Procedures.” Immunoblotting using anti-phosphothreonine antibodies indicated that the phosphorylated proteins were significantly enriched by IMAC (Fig. 2A). A comparison of the input with elution of the IMAC column by 2-D DIGE showed dramatically different profiles (Fig. 2B). Overall more proteins with high molecular weight were enriched than those with low molecular weight, likely due to higher phosphate content (per protein) of larger proteins as suggested by the prevalent patterns of rows of spots in the upper part of the gel. Several rows of spots showed more effective enrichment of spots at the acidic side than the basic side, whereas the whole gel image showed no overall bias for acidic proteins in the IMAC fraction. Such an observation suggests that under our experimental conditions the IMAC column selectively enriches phosphoproteins rather than acidic proteins.
Fig. 2.
Evaluation of IMAC enrichment of phosphoproteins. A, immunoblot of total protein and IMAC fraction using anti-phosphothreonine antibodies. B, 2-D DIGE comparison of total protein (Cy3, green) with IMAC fraction (Cy5, red). Proteins that were enriched in the phosphoprotein fraction appear red.
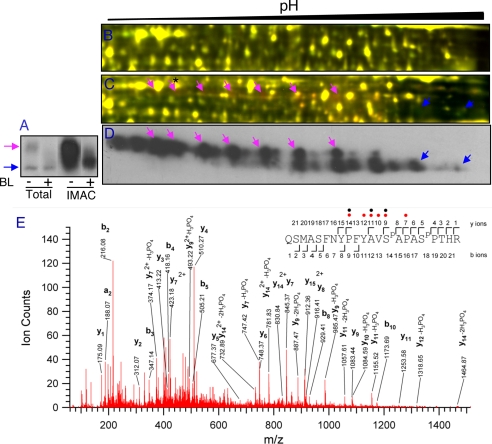
BR treatment rapidly induces dephosphorylation of BZR1 protein, which can be detected as a mobility shift in an immunoblot of SDS-PAGE gels (15) (Fig. 3A). However, when total protein samples were analyzed by 2-D DIGE, changes in mBZR1-CFP phosphorylation and accumulation could not be detected in the fluorescence images (Fig. 3B). IMAC significantly enriched the phosphorylated mBZR1-CFP proteins (Fig. 3A). 2-D DIGE analysis of the IMAC fractions of BR-treated and untreated samples detected a decrease and increase of rows of spots that match the pattern of mBZR1-CFP detected by immunoblotting (Fig. 3, C and D). DeCyder software analysis of the image indicated that BZR1-CFP showed the most dramatic changes among all detected protein spots (1.77-fold decrease of a phosphorylated mBZR1-CFP spot and 1.40-fold increase of an unphosphorylated mBZR1-CFP spot). Furthermore LC-MS/MS analysis of a spot at the acidic side confirmed its identify as BZR1 and identified two in vivo phosphorylation sites at Ser-220 and Ser-224 (Fig. 3E), which are among the 25 putative BIN2 phosphorylation sites of BZR1 (9). Consistent with multiple phosphorylation events in BZR1, we observed about 24 spots in the immunoblot of 2-DE (Fig. 3D). These results demonstrate that IMAC enrichment of phosphoproteins improves detection of low abundance proteins such as transcription factors in 2-D DIGE. The endogenous BZR1 protein was not detected likely because of its basic pI or low abundance.
Fig. 3.
BZR1 protein is detected by 2-D DIGE following IMAC enrichment of phosphoproteins. A, immunoblot of mBZR1-CFP in total protein and IMAC fractions of plants treated with mock solution (−) or 100 nm BL (+) for 2 h using anti-GFP antibodies. B and C, total protein (B) or phosphoprotein fractions (C) were analyzed by 2-D DIGE after labeling BL-treated and untreated samples with Cy3 and Cy5, respectively. D, gel of C was blotted and probed with anti-GFP antibodies. Cyan arrows show the hyperphosphorylated and blue arrows show the hypophosphorylated or unphosphorylated mBZR1-CFP proteins. Matching areas of 2-DE are shown in B–D. E, a spectrum of a BZR1 spot (marked by a star and arrow in C) obtained from a precursor ion with m/z value 863.72033+, corresponding to a double phosphorylated peptide spanning the residues Gln-207 to Arg-228 of BZR1. Phosphorylated residues (Ser-220 and Ser-224) are indicated in the peptide sequence as SP. Phosphate losses are marked with red circles (one loss) or black circles (two losses).
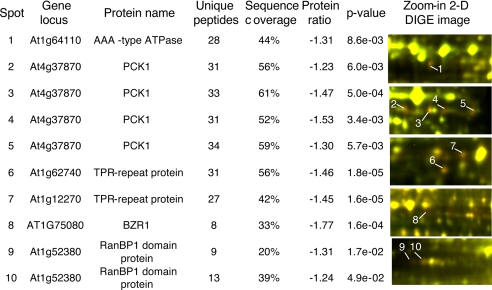
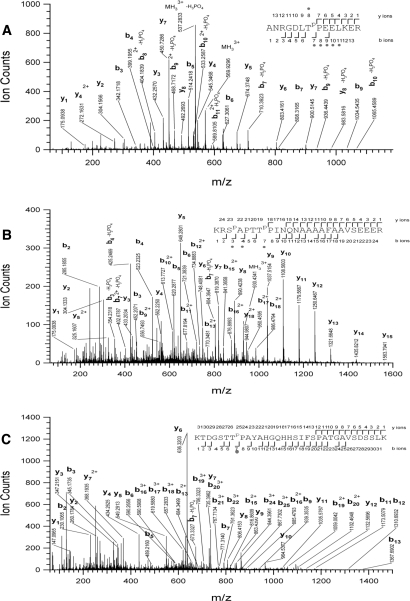
In addition to BZR1, five early BR-response proteins (within 2 h) were identified in the IMAC fractions (Fig. 4). These include phosphoenolpyruvate carboxykinase 1 (PCK1) (35), an AAA-type ATPase family protein, two putative tetratricopeptide repeat (TPR) proteins that are closest homologs of each other, and a putative RAN-binding protein (RanBP). Except the putative AAA-type ATPase, the other four genes were not BR-responsive at the RNA level based on microarray studies (36–39), suggesting posttranscriptional regulation. None of these early BR-response proteins were identified by 2-D DIGE analyses of total proteins (22). In addition to the BZR1 phosphorylation sites, three in vivo phosphorylation sites (Ser-62, Thr-66, and Thr-122) were found in PCK1, and one phosphorylation site (Thr-507 in At1g12270 and Thr-506 in At1g62740) was found in both TPR proteins (Table I and Fig. 5). These results demonstrate that prefractionation of phosphoproteins followed by 2-D DIGE can detect early response proteins of signal transduction pathways.
Fig. 4.
BR-regulated phosphoproteins identified in IMAC fractions by 2-D DIGE. Average protein abundance ratios of BR-treated (2 h) to mock-treated and p values of Student's t test were calculated from four biological repeat experiments. In the 2-D DIGE images, proteins down-regulated by BL appear red.
Table I.
Posttranslationally modified peptides observed
Listed are protein identification number (ID), precursor m/z and z observed, error, sequence identified, and E value for the peptide. Phosphorylated residues are labeled in the peptide sequence with a "P" in superscript to their right side.
| Protein ID | m/z | z | Error | Peptide sequence | Expectation (E value) |
|---|---|---|---|---|---|
| ppm | |||||
| At1g75080 | 863.7203 | 3 | 31 | QSMASFNYPFYAVSPAPASPPTHR | 7.0e−7 |
| At1g12270 | 569.9506 | 3 | 18 | ANRGDLTPPEELKER | 3.3e−4 |
| At1g62740 | 569.9493 | 3 | 16 | ANRGDLTPPEELKER | 7.4e−5 |
| At4g37870 | 930.4570 | 3 | 33 | KRSPAPTTPPINQNAAAAFAAVSEEER | 4.1e−11 |
| At4g37870 | 667.9269 | 5 | 22 | KTDGSTTPPAYAHGQHHSIFSPATGAVSDSSLK | 1.6e−5 |
Fig. 5.
Phosphorylation sites were identified by LC-MS/MS for BR-regulated proteins. Shown are tandem mass spectra obtained from precursor ions with m/z value 569.95063+, 930.45703+, and 667.92695+ corresponding, respectively, to a peptide spanning residues Ala-501 to Arg-514 of TPR protein (At1g62740) phosphorylated at Thr507 (A), a double phosphorylated peptide (at Ser-62 and Thr-66) spanning the residues Lys-60 to Arg-84 of PCK1 (B), and a peptide (phosphorylated in Thr-122) spanning residues Lys-116 to Lys-147 of PCK1 (C). Phosphorylated residues are labeled in the peptide sequence with a “P” in superscript to their right side. The observed sequence ions are displayed. Phosphate losses are marked with circles in the sequence.
2-D DIGE Analysis of BR-regulated Plasma Membrane Proteins—
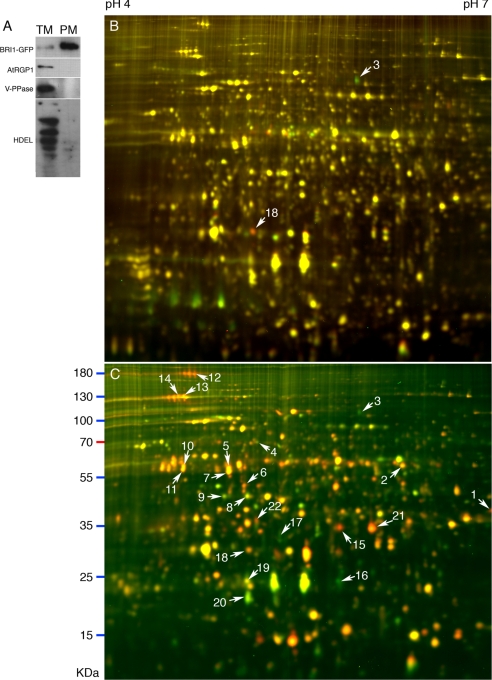
The two-phase partition method (31) was used to purify the plasma membrane. Transgenic plants expressing the BRI1-GFP fusion protein were used to monitor the purity of isolated plasma membrane. When equal amounts of microsomal proteins and plasma membrane proteins were separated by SDS-PAGE and probed with anti-GFP antibody, the signal of the PM sample was at least 10-fold higher than that of the microsomal sample (Fig. 6A). Further immunoblot analysis showed that marker proteins for Golgi (40), tonoplast (41), and endoplasmic reticulum (42) were excluded from the PM sample compared with the microsomal fraction (Fig. 6A). These results indicate that the PM fractions were highly purified.
Fig. 6.
2-D DIGE analyses of BR-regulated plasma membrane proteins. A, immunoblots of microsomal (TM) and plasma membrane (PM) proteins of the det2 mutant expressing BRI1-GFP fusion protein. Three micrograms of microsomal or plasma membrane proteins were separated by SDS-PAGE. Western blots were probed using antibodies against different membrane marker proteins (BRI1-GFP, PM; AtRGP1, Golgi; vacuolar H+-translocating pyrophosphatase (V-PPase), tonoplast; HDEL, ER). B and C, 2-D DIGE image of plasma membrane proteins from 7-day-old det2 seedlings treated with BL for 2 (B) or 24 h (C). BL-treated PM proteins were labeled by Cy5, and mock-treated PM proteins were labeled by Cy3. Spots identified by LC-MS/MS are marked by arrows and numbers (listed in Table II).
To identify BR-regulated plasma membrane proteins, we purified PM fractions from seedlings of the BR-deficient det2 mutant treated with BR or mock solution for 2 or 24 h. At least three sets of biological repeat samples were analyzed for each time point, and the DIGE images were analyzed using the BVA (biological variation analysis) module of the DeCyder 6.5 software. About 1900 spots were detected in each 2-D DIGE image, and 18 spots were up- or down-regulated by at least 1.2-fold (p < 0.05) after 2-h BR treatment (Fig. 6B). These spots were considered BR early response proteins. After 24 h of BL treatment, more than 240 spots were up- or down-regulated by at least 1.5-fold (p < 0.05) (Fig. 6C). By spot in-gel digestion and LC-MS/MS analysis, we obtained protein identities of 22 spots, which contained 19 distinct BR-responsive proteins (Table II). Seven of these proteins showed response to 2-h BR treatment (Table II), and one of them is the BR co-receptor BAK1 (Fig. 6B, spot 3). However, BRI1 was not identified possibly due to its low protein abundance or high molecular weight that results in poor separation in IEF or SDS-PAGE. Six of the 19 proteins from the PM fraction, including PATL1, PATL2, tubulin β-4 chain, tubulin α-6 chain, actin 2, and a kelch repeat-containing protein, were also identified in the analysis of total proteins (Table II and Ref. 22). The extents of protein changes in Table II are in agreement with the fact that BRs have been shown to induce mild transcriptional changes (36).
Table II.
BR-regulated proteins identified in plasma membrane fractions.
Statistically significant differences (ratio) are in bold.
| Spot no. | Gene locus | Protein name | E valuea | No. of peptidesb | Sequence coverage | 2 h | 24 h | ||
|---|---|---|---|---|---|---|---|---|---|
| Ratio | p value | Ratio | p value | ||||||
| % | |||||||||
| 1 | At1g09070 | Src2-like protein | 2.2e−5 | 4 | 15 | 1.15 | 2.4e−1 | 2.16 | 1.5e−2 |
| 2 | At1g63940 | Monodehydroascorbate reductase | 8.0e−9 | 20 | 38 | 1.13 | 2.1e−1 | 1.64 | 1.7e−4 |
| 3 | At4g33430 | BAK1 | 3.7e−5 | 15 | 18 | −2.16 | 1.1e−2 | −1.81 | 7.2e−3 |
| 4 | At3g13470 | GroEL protein | 3.5e−7 | 15 | 30 | 1.14 | 9.9e−3 | −1.83 | 1.1e−3 |
| 5 | At5g19770c | Tubulin α-3chain | 1.8e−6 | 10 | 23 | 1.28 | 1.9e−3 | 1.92 | 2.2e−3 |
| 6 | At3g16470 | Myrosinase-binding protein-like | 4.4e−7 | 5 | 14 | 1.10 | 6.3e−2 | 1.72 | 1.9e−2 |
| 7 | At4g14960 or At1g50010c,d | Tubulin α-6 or α-2 chain | 1.4e−8 | 18 | 33 | 1.19 | 8.0e−3 | 1.95 | 1.6e−4 |
| 8 | At5g09810 | Actin 7 | 6.6e−8 | 21 | 46 | 1.15 | 2.2e−2 | −1.72 | 1.9e−2 |
| 9 | At3g12780 | Phosphoglycerate kinase | 1.1e−6 | 15 | 35 | 1.09 | 1.6e−10 | −1.70 | 1.1e−4 |
| 10 | At5g44340c,d | Tubulin β-4 chain | 5.1e−7 | 24 | 36 | 1.29 | 1.6e−3 | 1.69 | 3.2e−3 |
| 11 | At4g20260e | DREPP family protein | 1.3e−7 | 3 | 11 | 1.25 | 1.1e−3 | 1.82 | 4.9e−4 |
| 12 | At1g22530d,e | Patellin-2 | 4.0e−7 | 32 | 49 | 1.25 | 2.4e−2 | 2.68 | 5.4e−3 |
| 13 | At1g72150d | Patellin-1 | 5.2e−6 | 14 | 26 | 1.30 | 1.9e−4 | 1.90 | 3.9e−3 |
| 14 | At1g72150d | Patellin-1 | 4.6e−7 | 17 | 28 | 1.32 | 2.8e−5 | 1.87 | 1.2e−2 |
| 15 | At1g54040d | Kelch repeat-containing protein | 3.2e−6 | 14 | 43 | 1.08 | 4.2e−2 | 3.38 | 5.8e−4 |
| 16 | At3g01290 | Band 7 family protein | 5.8e−8 | 13 | 42 | 1.11 | 1.8e−1 | −2.76 | 2.7e−3 |
| 17 | At3g16420 | Jacalin lectin family protein | 4.7e−7 | 5 | 20 | 1.14 | 6.9e−3 | −1.85 | 6.2e−5 |
| 18 | At3g16420 | Jacalin lectin family protein | 2.6e−6 | 5 | 20 | 2.26 | 3.7e−3 | 1.59 | 1.2e−1 |
| 19 | At5g14740 | Carbonic anhydrase 2 | 9.7e−10 | 19 | 59 | 1.19 | 8.2e−2 | −2.06 | 7.9e−5 |
| 20 | At3g56190 | α-SNAP 2 | 2.2e−7 | 15 | 53 | 1.11 | 8.1e−2 | −1.75 | 4.3e−2 |
| 21 | At1g54040d | Kelch repeat-containing protein | 7.6e−6 | 21 | 62 | 1.05 | 3.0e−1 | 3.49 | 3.5e−4 |
| 22 | At3g18780 or At1g49240d | Actin 2 or actin 8 | 7.0e−8 | 11 | 32 | 1.02 | 3.80e−1 | 1.99 | 6.6e−6 |
BR-induced Posttranslational Modification of BAK1—
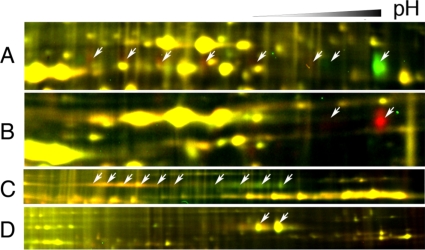
It has been shown that BR induces BAK1 phosphorylation but does not affect BAK1 protein level (14). It has also been proposed that BAK1 can undergo endocytosis (43). By increasing the image contrast, we observed at least six evenly spaced BR-induced (red) spots that have similar molecular weight but lower pI compared with spot 3 (Fig. 7A). These BR-induced spots are likely phosphorylated forms of BAK1. When plasma membrane protein of the T-DNA knock-out mutant bak1-1 (5) was compared with that of wild type using 2-D DIGE, spot 3 and another weaker spot with similar molecular weight but lower pI were absent in the bak1-1 plants (Fig. 7B), confirming their identity as the BAK1 protein. When BAK1-GFP transgenic plants were analyzed by 2-D DIGE, a row of smeary BAK1-GFP spots shifted to the more acidic side in the BR-treated sample (Fig. 7C). Immunoblotting of SDS-PAGE and 2-DE gels using anti-GFP antibodies confirmed that the charge state but not the level of BAK1-GFP protein in PM was affected by BR treatment (data not shown). These results indicate that BAK1 is posttranslationally modified upon BR treatment. Furthermore in bri1-5 mutant plants expressing BAK1-GFP (6), only the two most basic spots were detected, and they were not affected by BR treatment (Fig. 7D), indicating that functional BRI1 is required for BR-induced BAK1 modification. The data indicate that without BR activation BAK1 exists in two forms with different charges. Upon BR activation of BRI1, BAK1 changes to at least six modified (likely phosphorylated) forms.
Fig. 7.
BAK1 is posttranslationally modified after BR treatment. A, zoomed-in view of the region of Fig. 6B containing BAK1 (spot 3). Arrows show potential shifted BAK1 spots that disappear (green spots) or appear (red spots) after BL treatment. B, BAK1 spots disappear in bak1-1 mutant. PM proteins were isolated from wild type and T-DNA knock-out mutant bak1-1 and labeled with Cy5 and Cy3, respectively. Arrows show the BAK1 spots. C and D, plasma membrane proteins isolated from transgenic plant expressing the BAK1-GFP fusion protein in Columbia (C) or bri1-5 (D) were labeled with Cy5 (BL-treated) or Cy3 (mock-treated) and analyzed by 2-D DIGE. Arrows show the BAK1-GFP protein spots.
Functional Study of BR-regulated Proteins—
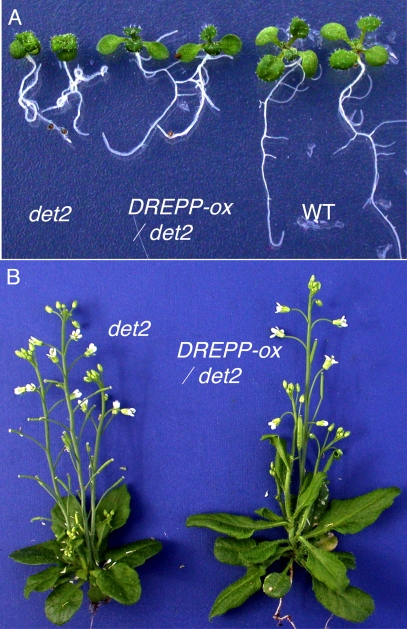
To study the potential role of BR-regulated proteins found in this study, we overexpressed a number of genes selected from Table II in Arabidopsis. From these transgenic plants, we observed that overexpression of a DREPP-GFP fusion protein in det2 plants partially rescued the dwarf phenotype of det2. Three of 11 transgenic lines showed elongated petioles and leaves and larger plant size compared with the untransformed det2 plants (Fig. 8). The increase of plant size correlated with the expression levels of the DREPP-GFP fusion protein in the transgenic lines and in a segregating T2 population (data not shown). Taken together, these results indicate that the BR-induced DREPP protein mediates BR promotion of plant growth.
Fig. 8.
Overexpression of DREPP suppresses the det2 mutant. A, 10-day-old det2-1 mutant, det2-1-overexpressing DREPP-GFP (DREPP-ox/det2), and wild type (WT) plants. B, 5-week-old det2-1- and det2-1-overexpressing DREPP-GFP (DREPP-ox/det2).
DISCUSSION
Identification of signal-induced changes in protein modification or subcellular localization is critical for understanding signal transduction pathways that regulate various biological processes. Proteomics, by surveying large number of proteins, offers great potential for studying signal transduction. However, detecting changes in signaling proteins, which are present at low abundance in cells, is challenging and requires efficient protein fractionation/enrichment as well as sensitive and quantitative proteomics detection methods. 2-D DIGE analysis of total protein samples failed to identify proteins that respond rapidly to BR treatment (22). Apparently the relative abundance of BR signaling proteins is too low in total protein samples to be detected by 2-D DIGE. Using 2-D DIGE analysis of phosphoproteins purified by IMAC and PM proteins purified by two-phase partitioning, we identified BZR1, BAK1, and novel BR-responsive proteins that play a role in BR promotion of plant growth. Our study demonstrates that prefractionation of PM or phosphoproteins followed by 2-D DIGE is a powerful approach for identifying signaling components and signal-transducing biochemical events in Arabidopsis.
IMAC has been widely used for proteomics study of phosphoproteins. In most cases, proteins are first digested by trypsin, and then phosphopeptides are enriched by IMAC before analysis by mass spectrometry. Enrichment of undigested phosphoprotein by IMAC for 2-DE analysis has not been widely used (26). Recently Collins et al. (30) used a protein IMAC method to enrich phosphoproteins, which were then digested by trypsin and analyzed either directly by LC-MS/MS or by a second peptide IMAC enrichment before LC-MS/MS analysis. Using a similar protein IMAC protocol, we successfully enriched Arabidopsis phosphoproteins as shown by the enrichment of phosphorylated BZR1 and proteins detected by the anti-phosphothreonine antibody. Interestingly IMAC enrichment is more biased for proteins of high molecular weight. This is likely due to the higher probability of having phosphorylated residues in a large protein than in a small protein. Large proteins tend to exist in rows of evenly spaced spots in 2-DE presumably due to multiple phosphorylation residues.
One advantage of 2-D DIGE is its ability to detect and quantify change in protein phosphorylation. Each phosphorylation event reduces the pI of protein and causes a shift of the protein spot toward the acidic side along the IEF dimension in a 2-DE gel. In 2-D DIGE, stoichiometry of phosphorylation can be determined by quantifying the spot intensity of each of the phosphorylation forms. Both BAK1 and BZR1 were detected as such a row of green and red shifted spots on 2-D DIGE images consistent with their BR-regulated phosphorylation changes. 2-D DIGE and immunoblotting detected 24 mBZR1-CFP spots shifted along IEF dimension (Fig. 3D). This result agrees with the previous prediction of 25 putative phosphorylation sites in BZR1 for GSK3 kinases (15). Of these 25 predicted GSK3 kinase phosphorylation sites, two phosphorylated residues were identified by LC-MS/MS analysis of a BZR1 spot at the acidic side.
Our 2-D DIGE analyses of the PM proteins suggest that BAK1 can be phosphorylated at least at six phosphorylation sites after BL treatment, whereas previous studies have identified four in vitro autophosphorylation sites in BAK1 (14). Most BAK1 proteins in the cell are phosphorylated after 2-h BR treatment as shown by the 2.16-fold reduction of the unphosphorylated BAK1 (Table II), suggesting that BAK1 plays a specific role in BR signaling. However, recent studies demonstrated dual roles of BAK1 in BR and disease signaling. The bak1 mutant was shown to have increased susceptibility to bacterial and fungal pathogens and reduced response to bacterial flagellin (44, 45). Furthermore BAK1 was shown to dimerize with FLS2, the receptor kinase-perceiving flagellin, upon flagellin treatment (45). Whether the flagellin-induced FLS2-BAK1 dimerization increases BAK1 phosphorylation remains unknown. A recent quantitative phosphoproteomics study by Benschop et al. (46) using metabolic labeling identified 472 phosphoproteins in the PM. Of these, 76 phosphoproteins showed flagellin-induced changes in phosphorylation. In contrast to our study, Benschop et al. (46) identified a very high percentage (16%) of phosphoproteins that respond rapidly to flagellin treatment (10 min) but found no phosphorylation of known signaling proteins of the pathway such as FLS2, BAK1, or mitogen-activated protein kinases (46). It is unclear whether such differences in the results are due to differences of the signaling pathways or reflect differences in the sensitivity and specificity of the methods used in each study.
There has been a trend that 2-D gel-based approaches are replaced by LC-MS/MS-based profiling methods. It is widely believed that LC-MS/MS has better coverage of the proteome than 2-DE because in theory some proteins such as hydrophobic membrane proteins cannot be separated in 2-DE, whereas MS can detect any protein. However, neither approach can cover the whole proteome of a eukaryotic organism. Typically about 1000 proteins are covered using either method. Our study supports the notion that 2-D DIGE and LC-MS/MS are highly complementary proteomics methods, each with advantages and shortcomings.
Most of the BL-regulated PM proteins identified in this study showed no BR response at the RNA level in previous studies (Table II). Only two of the 19 BL-regulated PM proteins have been reported previously to be BR-responsive at the RNA levels (Table II). A search of the microarray database (Genevestigator) showed that transcript levels of four genes (At3g01290, At1g22530, At1g72150, and At4g20260) are regulated by 3-h BL treatment. For those proteins that show no BR response at the RNA level, BR is likely to regulate their protein accumulation or localization to the PM.
Six of the 19 BL-regulated PM proteins, including PATL1 (At1g72150), PATL2 (At1g22530), tubulin β-4 chain (At5g44340), tubulin α-6 chain (At4g14960), actin 2 (At3g18780), and a kelch repeat-containing protein (At1g54040), were also identified in the total protein samples but only after long time BR treatments (22). Among them, PATL1, PATL2, and tubulin β-4 showed responses to 2-h BR treatment in PM fractions but not to 3-h BR treatment in total proteins (22). It is possible that low complexity of the PM proteome reduces background and increases accuracy of quantitation. Alternatively these proteins might change more dramatically in PM than in the cytoplasm.
Besides the known BR signaling proteins BAK1 and BZR1, several novel early BR-responsive proteins were identified in this study, including PCK1, an AAA-type ATPase family protein, two putative TPR proteins, a RanBP1 domain-containing protein, and jacalin, although the exact role of these proteins in BR-regulated early cell response is still unclear. PCK1 converts oxaloacetate to pyruvate and plays a role in fueling early seedling growth via gluconeogenesis in Arabidopsis (35, 47). PCK is regulated by reversible phosphorylation, and dephosphorylation increases its decarboxylase activity in C4 plant Panicum maximum (48). In this study, the BR-induced spot shift of PCK1 is consistent with dephosphorylation of PCK1. Thus, it is likely that BR induces PCK1 dephosphorylation and activates its decarboxylase activity, which fuels BR-promoted seedling growth via gluconeogenesis. RAN is an important Ras-like GTPase whose activity is modulated by its interacting proteins such as RCC1 and RanBP (49). RAN and its interacting proteins are involved in diverse processes, including nucleocytoplasmic transport, spindle assembly, and postmitotic nuclear envelope assembly (49). It would be interesting to find out whether BL regulates cytosol-nuclear trafficking of its signal component (such as BZR1) by regulating the activity of RAN. The two TPR proteins, each containing three TPR motifs, are the closest homologs of each other. TPR motif is a highly degenerate 34-amino acid peptide with amphipathic α-helices involved in protein-protein interactions and can serve as a scaffold for the assembly of multiprotein complexes (50). TPR proteins are involved in biosynthesis and signal transduction of different plant hormones, such as ETO1 in ethylene production (51), SPINDLY in gibberellin response (52), and TWD1 in auxin signaling (53). One of the jacalin lectin-related proteins identified in PM showed very rapid response to BR treatment. Jacalins are known to bind glycoproteins, but their functions in plants remain unknown. Because SPINDLY encodes an O-linked N-acetylglucosamine transferase, it is tempting to speculate that BR-regulated jacalin might be involved in glycosylation-mediated signaling processes. The functions of these early BR-response proteins merit further study.
DREPP was originally identified as a developmentally regulated plasma membrane polypeptide in tobacco (54). In Arabidopsis, DREPP protein (At4g20260) has been identified in several proteomics studies of plasma membrane proteins (55–57), and its RNA was shown to be induced by BR (36). However, the function of DREPP has remained unknown. In this study, we identified DREPP as a BR-inducible plasma membrane protein. The suppression of det2 mutant phenotypes by DREPP overexpression demonstrates that DREPP plays an important role in BR promotion of plant growth.
In conclusion, using prefractionation of plasma membrane and phosphoproteins followed by the 2-D DIGE analysis, we identified signal-induced phosphorylation and dephosphorylation of two known BR signaling proteins, BAK1 and BZR1. We also identified a number of novel early BR-response proteins and demonstrated function for one of the BR-regulated PM proteins, DREPP. This study demonstrates that 2-D DIGE coupled with prefractionation is a very powerful approach for studying signal transduction pathways, particularly for identifying signal-induced changes in protein modification.
Acknowledgments
We thank Dr. Philip A. Rea (University of Pennsylvania) for providing polyclonal anti-vacuolar H+-translocating pyrophosphatase antibodies, Dr. Richard Napier (Crop Improvement and Biotechnology, Warwick, UK) for providing monoclonal anti-HDEL antibody, Dr. Natasha V. Raikhel (University of California, Riverside) for providing AtRGP1 antibody, Dr. Jia Li (University of Oklahoma) for providing bak1-1 seeds, Jianming Li (University of Michigan) for providing BAK1::BAK1-GFP seeds, and Dr. Zuxiong Chen (Stanford University) for assistance with image analysis. The University of California San Francisco Mass Spectrometry Facility (A. L. Burlingame, Director) is supported by the Biomedical Research Technology Program of the National Center for Research Resources, National Institutes of Health (Grants NCRR RR01614, RR012961, and RR019934).
Footnotes
Published, MCP Papers in Press, January 8, 2008, DOI 10.1074/mcp.M700358-MCP200
The abbreviations used are: BR, brassinosteroid; BL, brassinolide; GSK3, glycogen synthase kinase 3; PM, plasma membrane; TPR, tetratricopeptide repeat; 2-D, two-dimensional; 2-DE, two-dimensional gel electrophoresis; CFP, cyan fluorescent protein; GFP, green fluorescent protein; PCK, phosphoenolpyruvate carboxykinase; RanBP, RAN-binding protein; DREPP, developmentally regulated plasma membrane polypeptide; AAA, ATPases associated with diverse cellular activities; LTQ, linear quadropole ion trap; RAN, Ras-related nuclear protein.
This work was supported by United States Department of Energy Grant DE-FG02-04ER15525 and National Science Foundation Grant NSF 0724688 (to Z.-Y. W. and A. L. B.), by National Institutes of Health Grant 2R01 GM066258 (to Z.-Y. W.), and by the National Natural Science Foundation of China (to S. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Clouse, S. D., and Sasse, J. M. (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451 [DOI] [PubMed] [Google Scholar]
- 2.Thummel, C. S., and Chory, J. (2002) Steroid signaling in plants and insects—common themes, different pathways. Genes Dev. 16, 3113–3129 [DOI] [PubMed] [Google Scholar]
- 3.Wang, Z. Y., Wang, Q., Chong, K., Wang, F., Wang, L., Bai, M., and Jia, C. (2006) The brassinosteroid signal transduction pathway. Cell Res. 16, 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinoshita, T., Cano-Delgado, A., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171 [DOI] [PubMed] [Google Scholar]
- 5.Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 [DOI] [PubMed] [Google Scholar]
- 6.Nam, K. H., and Li, J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 [DOI] [PubMed] [Google Scholar]
- 7.Li, J., and Nam, K. H. (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301 [DOI] [PubMed] [Google Scholar]
- 8.Mora-Garcia, S., Vert, G., Yin, Y., Cano-Delgado, A., Cheong, H., and Chory, J. (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, Z. Y., Nakano, T., Gendron, J., He, J. X., Chen, M., Vafeados, D., Yang, Y. L., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513 [DOI] [PubMed] [Google Scholar]
- 10.Yin, Y., Wang, Z. Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., and Chory, J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191 [DOI] [PubMed] [Google Scholar]
- 11.Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., Guan, S., Lalonde, S., Sun, Y., Gendron, J. M., Chen, H., Shibagaki, N., Ferl, R. J., Ehrhardt, D., Chong, K., Burlingame, A. L., and Wang, Z. Y. (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., and Wang, Z. Y. (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U. S. A. 104, 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gendron, J. M., and Wang, Z. Y. (2007) Multiple mechanisms modulate brassinosteroid signaling. Curr. Opin. Plant Biol. 10, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, X., Goshe, M. B., Soderblom, E. J., Phinney, B. S., Kuchar, J. A., Li, J., Asami, T., Yoshida, S., Huber, S. C., and Clouse, S. D. (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17, 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, J. X., Gendron, J. M., Yang, Y., Li, J., and Wang, Z. Y. (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 99, 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S., Gendron, N., Sun, C. Q., and Wang, Z. Y. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259 [DOI] [PubMed] [Google Scholar]
- 18.O'Farrell, P. H. (1975) High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250, 4007–4021 [PMC free article] [PubMed] [Google Scholar]
- 19.Unlu, M., Morgan, M. E., and Minden, J. S. (1997) Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 [DOI] [PubMed] [Google Scholar]
- 20.Tonge, R., Shaw, J., Middleton, B., Rowlinson, R., Rayner, S., Young, J., Pognan, F., Hawkins, E., Currie, I., and Davison, M. (2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1, 377–396 [DOI] [PubMed] [Google Scholar]
- 21.Alfonso, P., Dolado, I., Swat, A., Nunez, A., Cuadrado, A., Nebreda, A. R., and Casal, J. I. (2006) Proteomic analysis of p38α mitogen-activated protein kinase-regulated changes in membrane fractions of RAS-transformed fibroblasts. Proteomics 6, Suppl. 1, S262–S271 [DOI] [PubMed] [Google Scholar]
- 22.Deng, Z., Zhang, X., Tang, W., Oses-Prieto, J. A., Suzuki, N., Gendron, J. M., Chen, H., Guan, S., Chalkley, R. J., Peterman, T. K., Burlingame, A. L., and Wang, Z. Y. (2007) A proteomic study of brassinosteroid response in Arabidopsis. Mol. Cell. Proteomics 6, 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolkman, A., Dirksen, E. H., Slijper, M., and Heck, A. J. (2005) Double standards in quantitative proteomics: direct comparative assessment of difference in gel electrophoresis and metabolic stable isotope labeling. Mol. Cell. Proteomics 4, 255–266 [DOI] [PubMed] [Google Scholar]
- 24.Casati, P., Zhang, X., Burlingame, A. L., and Walbot, V. (2005) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol. Cell. Proteomics 4, 1673–1685 [DOI] [PubMed] [Google Scholar]
- 25.Tannu, N. S., and Hemby, S. E. (2006) Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat. Protoc. 1, 1732–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersten, B., Agrawal, G. K., Iwahashi, H., and Rakwal, R. (2006) Plant phosphoproteomics: a long road ahead. Proteomics 6, 5517–5528 [DOI] [PubMed] [Google Scholar]
- 27.Lewis, T. S., Hunt, J. B., Aveline, L. D., Jonscher, K. R., Louie, D. F., Yeh, J. M., Nahreini, T. S., Resing, K. A., and Ahn, N. G. (2000) Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6, 1343–1354 [DOI] [PubMed] [Google Scholar]
- 28.Rossignol, M. (2006) Proteomic analysis of phosphorylated proteins. Curr. Opin. Plant Biol. 9, 538–543 [DOI] [PubMed] [Google Scholar]
- 29.Chory, J., Nagpal, P., and Peto, C. A. (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins, M. O., Yu, L., Husi, H., Blackstock, W. P., Choudhary, J. S., and Grant, S. G. (2005) Robust enrichment of phosphorylated species in complex mixtures by sequential protein and peptide metal-affinity chromatography and analysis by tandem mass spectrometry. Sci. STKE 2005, pl6. [DOI] [PubMed] [Google Scholar]
- 31.Larsson, C., Sommarin, M., and Widell, S. (1994) Isolation of highly purified plant plasma membranes and the separation of inside-out and right-side-out vesicles. Methods Enzymol. 228, 451–469 [Google Scholar]
- 32.Rosenfeld, J., Capdevielle, J., Guillemot, J. C., and Ferrara, P. (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203, 173–179 [DOI] [PubMed] [Google Scholar]
- 33.Clauser, K. R., Baker, P., and Burlingame, A. L. (1999) Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 34.Curtis, M. D., and Grossniklaus, U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penfield, S., Rylott, E. L., Gilday, A. D., Graham, S., Larson, T. R., and Graham, I. A. (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16, 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., and Yoshida, S. (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mussig, C., Fischer, S., and Altmann, T. (2002) Brassinosteroid-regulated gene expression. Plant Physiol. 129, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nemhauser, J. L., Mockler, T. C., and Chory, J. (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2, E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vert, G., Nemhauser, J. L., Geldner, N., Hong, F. X., and Chory, J. (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201 [DOI] [PubMed] [Google Scholar]
- 40.Delgado, I. J., Wang, Z., de Rocher, A., Keegstra, K., and Raikhel, N. V. (1998) Cloning and characterization of AtRGP1. A reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 116, 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rea, P. A., Britten, C. J., and Sarafian, V. (1992) Common identity of substrate binding subunit of vacuolar H-translocating inorganic pyrophosphatase of higher plant cells. Plant Physiol. 100, 723–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napier, R. M., Fowke, L. C., Hawes, C., Lewis, M., and Pelham, H. R. (1992) Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J. Cell Sci. 102, 261–271 [DOI] [PubMed] [Google Scholar]
- 43.Russinova, E., Borst, J. W., Kwaaitaal, M., Cano-Delgado, A., Yin, Y., Chory, J., and de Vries, S. C. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16, 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kemmerling, B., Schwedt, A., Rodriguez, P., Mazzotta, S., Frank, M., Qamar, S. A., Mengiste, T., Betsuyaku, S., Parker, J. E., Mussig, C., Thomma, B. P., Albrecht, C., de Vries, S. C., Hirt, H., and Nürnberger, T. (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 45.Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D., Felix, G., and Boller, T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500 [DOI] [PubMed] [Google Scholar]
- 46.Benschop, J. J., Mohammed, S., O'Flaherty, M., Heck, A. J., Slijper, M., and Menke, F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 47.Rylott, E. L., Gilday, A. D., and Graham, I. A. (2003) The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol. 131, 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker, R. P., Chen, Z. H., Acheson, R. M., and Leegood, R. C. (2002) Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant Guinea grass. Plant Physiol. 128, 165–172 [PMC free article] [PubMed] [Google Scholar]
- 49.Sazer, S., and Dasso, M. (2000) The ran decathlon: multiple roles of Ran. J. Cell Sci. 113, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 50.D'Andrea, L. D., and Regan, L. (2003) TPR proteins: the versatile helix. Trends Biochem. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 51.Wang, K. L., Yoshida, H., Lurin, C., and Ecker, J. R. (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950 [DOI] [PubMed] [Google Scholar]
- 52.Jacobsen, S. E., Binkowski, K. A., and Olszewski, N. E. (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 93, 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Geisler, M., Kolukisaoglu, H. U., Bouchard, R., Billion, K., Berger, J., Saal, B., Frangne, N., Koncz-Kalman, Z., Koncz, C., Dudler, R., Blakeslee, J. J., Murphy, A. S., Martinoia, E., and Schulz, B. (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14, 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Logan, D. C., Domergue, O., Teyssendier de la Serve, B., and Rossignol, M. (1997) A new family of plasma membrane polypeptides differentially regulated during plant development. Biochem. Mol. Biol. Int. 43, 1051–1062 [DOI] [PubMed] [Google Scholar]
- 55.Marmagne, A., Rouet, M. A., Ferro, M., Rolland, N., Alcon, C., Joyard, J., Garin, J., Barbier-Brygoo, H., and Ephritikhine, G. (2004) Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3, 675–691 [DOI] [PubMed] [Google Scholar]
- 56.Carter, C., Pan, S., Zouhar, J., Avila, E. L., Girke, T., and Raikhel, N. V. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16, 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson, C. J., Hegeman, A. D., Harms, A. C., and Sussman, M. R. (2006) A quantitative analysis of Arabidopsis plasma membrane using trypsin-catalyzed 18O labeling. Mol. Cell. Proteomics, 5, 1382–1395 [DOI] [PubMed] [Google Scholar]
- 58.Konishi, H., and Komatsu, S. (2003) A proteomics approach to investigating promotive effects of brassinolide on lamina inclination and root growth in rice seedlings. Biol. Pharm. Bull. 26, 401–408 [DOI] [PubMed] [Google Scholar]
- 59.Huang, B., Chu, C. H., Chen, S. L., Juan, H. F., and Chen, Y. M. (2006) A proteomics study of the mung bean epicotyl regulated by brassinosteroids under conditions of chilling stress. Cell. Mol. Biol. Lett. 11, 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]