Abstract
Absolute quantification of peptides by mass spectrometry requires a reference, frequently using heavy isotope-coded peptides as internal standards. These peptides have traditionally been generated by chemical stepwise synthesis. Recently a new way to supply such peptides was described in which nucleotide sequences coding for the respective peptides are concatenated into a synthetic gene (QconCAT). These QconCATs are then expressed to produce a polypeptide consisting of concatenated peptides, purified, quantified by various methods, and then digested to yield the final internal standard peptides. Although both of these methods for peptide production are routinely used for absolute quantifications, there is currently no information regarding the accuracy of the quantifications made in each case. In this study, we used sets of synthetic and biological peptides in parallel to evaluate the accuracy of either method. We also addressed some technical issues regarding the preparation and proper utilization of such standard peptides. Twenty-five peptides derived from the Caenorhabditis elegans proteome were selected for this study. Twenty-four were successfully chemically synthesized. Five QconCAT genes were designed, each a concatenation of the same 25 peptides but each in separate, different randomized order, and expressed via in vitro translation reactions that contained heavy isotope-labeled lysine and arginine. Three of the five QconCATs were successfully produced. Different digestion conditions, including various detergents and incubation conditions, were tested to find those optimal for the generation of a reproducible and accurate reference sample mixture. All three QconCAT polypeptides were then digested using the optimized conditions and then mixed in a 1:1 ratio with their synthetic counterparts. Multireaction monitoring mass spectrometry was then used for quantification. Results showed that the digestion protocol had a significant impact on equimolarity of final peptides, confirming the need for optimization. Under optimal conditions, however, most QconCAT peptides were produced at an equimolar ratio. A few QconCAT-derived peptides were largely overestimated due to problems with solubilization or stability of the synthetic peptides. Although the order in which the peptide sequences appeared in the QconCAT sequence proved to affect the success rate of in vitro translation, it did not significantly affect the final peptide yields. Overall neither the chemical synthesis nor the recombinant genetic approach proved to be superior as a method for the production of reference peptides for absolute quantification.
Systems biology depends intimately on the ability to make quantitative measurements of the transcriptome, proteome, and metabolome. Quantitative proteomics has grown in the last 6–7 years to become a major source of information for systems biology studies where iterative measurements of reproducible sets of proteins from differentially perturbed biological states are essential.
Quantification in proteomics is typically performed by a means of relative measurement (1). Mass spectrometry-based methods use light and heavy isotope-coded reagents, such as ICAT, global internal standard technology, or isobaric tags for relative and absolute quantitation, to tag the peptides for mass spectrometric analysis (2–4). Label-free quantification has also been established as a reliable method for relative quantification in recent years (5–8). Despite many attractive features of these technologies, relative quantification methods suffer from serious limitations that constitute a major bottleneck in large scale proteomics (9). For example, for all relative quantification methods, the -fold changes of proteins in the sample of interest (disease, treated, etc.) are measured versus a normal reference sample. Any error in determining the normal state or preparing the normal sample will lead to serious errors in quantitative measurements made. Also for isotope tagging experiments, only a small number of samples can be compared with the reference in a single experiment. Although the reference sample could be prepared in bulk and used as a universal reference to compare many samples, its storage and preservation over time becomes a challenge. In label-free quantification methods, the experimental conditions need to be kept constant at all times to assure pattern reproducibility. Considering the complicated work flow and the diverse instrument platforms used for label-free proteomics experiments, pattern reproducibility within and across experiments in a single group and between groups becomes a major challenge. Absolute quantification, on the other hand, does not have any of the above limitations (10) because it allows comparison of data from any experiment (no reference/control needed), provides boundary conditions for different experiments, and makes data comparable between laboratories. In addition, absolute quantification is especially important in the biomarker discovery work flow because it provides necessary information for assay development.
The gold standard for absolute quantification of proteins and peptides is amino acid analysis. Unfortunately this method is slow and requires considerable amounts of starting material that is destroyed in the analysis and knowledge of the protein or peptide sequence (11). In addition, the quantified analyte needs to be purified to homogeneity. With the emergence of mass spectrometry-based proteomics, new possibilities for large scale absolute quantification of proteins have emerged. These methods depend on synthetic, isotopically labeled peptides as internal standards. Target peptides of interest are synthesized in a heavy labeled form and added to the sample of interest in known quantities prior to mass spectral analysis (12). Ion intensities for the light and heavy peptides are measured, and absolute quantity of the light peptides, representing the biological sample, can be calculated from the ratio of the respective signal intensities. Currently heavy isotope-coded peptides can be provided either by chemical synthesis or biological expression. Each method has limitations worth noting. Success of chemical synthesis is sequence-dependent. Chemical synthesis is also expensive and time-consuming. Synthesized peptides need to be quantitatively resolubilized before being used. In addition, some peptides are labile or may change due to artifactual modifications over time. These and other issues could potentially introduce errors in absolute measurements made.
In 2006, Pratt and co-workers (13–16) introduced a new technology to produce isotopically labeled reference peptides by genetically engineering them. In this approach, a synthetic gene is produced and expressed in Escherichia coli in a medium containing heavy lysine and arginine, resulting in the expression of a midsize protein in which peptides of interest are concatenated (13). After purification and concentration determination, the artificial protein, termed QconCAT, is digested to yield synthetic equivalent peptides that can be added to the mixture of interest as the internal standard (15). This method also has limitations. Not all QconCATs are successfully expressed even when DNA sequences are optimized for expression. Purification and handling can also be complicated when the expressed QconCATs are poorly soluble. Generating an equimolar mixture of peptides from an intact QconCAT is the next bottleneck. Complete digestion is required for generation of an equimolar peptide mixture. Another source of variation for tryptic digests are post-translational modifications (PTMs)1 that may occur during digestion. All these sources of variations (miscleavages and PTMs) can affect the equimolarity of the final peptide mixture and result in serious discrepancies between the real peptide concentration and the calculated value. Considering the cost and potential sources of error in both methods, it seemed prudent to evaluate these methods for their reliability and accuracy.
In an attempt to evaluate both peptide generation platforms, 25 tryptic peptides selected from the Caenorhabditis elegans proteome were selected for both chemical and biological production. Chemical synthesis yielded 24 peptides in their isotopically light form (synthesis of one peptide failed). Five QconCAT sequences were assembled by concatenating the same peptides in different random orders of which three were successfully translated in vitro. Purified QconCATs were then digested under conditions that had been optimized for each protein and mixed with a purportedly equimolar mixture of the 24 synthetic peptides. MRM mass spectrometry was then used to calculate the ratios between the light and heavy versions of the same peptides to evaluate the assumed equimolarity of peptides mixtures. Results showed that the equimolarity of the QconCAT digest was most significantly affected by digestion conditions, whereas resolubilization and peptide modifications were the most critical factors affecting equimolarity of synthetic peptides.
EXPERIMENTAL PROCEDURES
Materials
Synthetic peptides in their isotopically light form (see Table I for a complete list) and l-[13C6]lysine hydrochloride and l-[13C6]arginine hydrochloride were purchased from Sigma. TFA, SilverSNAP stain for mass spectrometry, SDS, urea, guanidinium HCl, and Coomassie Blue (Bradford) protein assay kits were purchased from Pierce. NuPAGE 4–12% Bis-Tris gel, 20× NuPAGE MOPS buffer, NuPAGE lithium dodecyl sulfate sample buffer, NuPAGE sample reducing agent, SeeBlue Plus2 prestained standard, and the gel running apparatus were purchased from Invitrogen. Rapigest SF and mixed cation exchange (MCX) trap cartridges were acquired from Waters Corp. (Milford, MA). HPLC grade ACN and HPLC grade water with 0.1% formic acid were purchased from Mallinckrodt Chemical Works. The PepMap100 reversed-phase C18 column (0.075 × 150 mm) was purchased from Dionex, Inc. (Sunnyvale, CA). The CAPTRAP C18 trap cartridge was purchased from Michrom Bioresources, Inc. (Auburn, CA). Mass spectrometry grade Trypsin Gold was purchased from Promega (Madison, WI). Mass spectra were acquired using a Tempo 2D nano-LC/4000QTrap system (Applied Biosystems, Foster City, CA). All spectra were obtained in the positive ion mode.
Table I.
List of selected peptides used to generate five QconCAT constructs
Only three QconCATs were expressed successfully. Peptide 25 failed in chemical synthesis. The method of Hopp and Woods (24) was used to calculate hydrophobicity. Numbers in columns Q1–Q5 are the order of each peptide in that construct.
| No. | Synthetic peptides | Molecular weight | Hydrophobicity | Q1 | Q2 | Q3 | Q4 | Q5 |
|---|---|---|---|---|---|---|---|---|
| 1 | APMPQHMQEMDEHQR | 1863.7814 | 0.5 | 10 | 6 | 20 | 9 | 25 |
| 2 | ATIAGGGVIPHIHR | 1397.7891 | −0.5 | 13 | 20 | 12 | 7 | 19 |
| 3 | AYQEALDIAK | 1120.5764 | 0.2 | 3 | 2 | 3 | 13 | 1 |
| 4 | DSSFLFTNPVPTVETAPPLR | 2187.1211 | −0.2 | 23 | 21 | 1 | 22 | 10 |
| 5 | DVASVFNNLLR | 1246.6669 | −0.3 | 2 | 11 | 5 | 21 | 21 |
| 6 | GYGFVIDDITGEDLFVHQSNLNMQGFR | 3071.4446 | −0.2 | 1 | 17 | 14 | 19 | 9 |
| 7 | HRPEQVAAEEAEAAR | 1662.8073 | 0.9 | 24 | 22 | 23 | 6 | 2 |
| 8 | IANEELANDVR | 1242.6204 | 0.6 | 7 | 8 | 21 | 5 | 6 |
| 9 | IPVDEETEIQILTDEIDSAPEAR | 2582.2598 | 0.7 | 18 | 14 | 19 | 23 | 24 |
| 10 | LILTSAPDTGLVPLSK | 1623.9446 | −0.3 | 11 | 12 | 13 | 20 | 14 |
| 11 | LTSLGVIGALVK | 1169.7383 | −0.6 | 4 | 1 | 24 | 15 | 18 |
| 12 | MANYIATFK | 1057.5266 | −0.7 | 19 | 19 | 6 | 4 | 11 |
| 13 | MGVDEAFYTLVR | 1399.6805 | −0.2 | 12 | 3 | 7 | 8 | 22 |
| 14 | NNDIGNLIEQFR | 1431.7106 | 0.2 | 20 | 5 | 10 | 2 | 20 |
| 15 | NSVGLGGGLAER | 1128.5887 | 0.1 | 8 | 4 | 4 | 17 | 16 |
| 16 | SQAMEELAHR | 1170.5451 | 0.5 | 22 | 18 | 2 | 10 | 8 |
| 17 | TLVQLDEILSDMK | 1503.7854 | 0.2 | 16 | 9 | 16 | 24 | 15 |
| 18 | TYWQFPLDAFAVGTYSETK | 2223.0524 | −0.5 | 5 | 7 | 25 | 12 | 4 |
| 19 | VETVSPEANILNSDQIK | 1855.9526 | 0.2 | 25 | 15 | 11 | 1 | 12 |
| 20 | VTEQGQELSNEER | 1517.6957 | 0.9 | 17 | 10 | 15 | 16 | 13 |
| 21 | VVEQGTSSEVDFEWIPR | 1976.9479 | 0.2 | 15 | 13 | 18 | 18 | 3 |
| 22 | VYVGGLPSDATSQELEEIFDR | 2324.1171 | 0.2 | 14 | 24 | 17 | 14 | 5 |
| 23 | YAASVAEVAHGGGGPK | 1469.7262 | −0.1 | 9 | 25 | 9 | 11 | 23 |
| 24 | YDDMAAAMK | 1014.4150 | 0.3 | 21 | 23 | 22 | 25 | 17 |
| 25 | TASVLISGLGSVGVEIAK (chemical synthesis failed) | 1699.9719 | −0.3 | 6 | 16 | 8 | 3 | 7 |
Methods
Resolubilization of Synthetic Peptides—
Solubility of synthetic peptides was tested using aqueous and organic buffers. The aqueous buffer recommended by the manufacturer was 0.1 m ammonium bicarbonate. We also used an organic buffer containing 50% ACN, 1% formic acid for comparison.
Selection and Construction of QconCATs—
Five QconCATs were constructed by random concatenation of the 25 selected peptide sequences (see Table I for the list of peptides and order by which they appeared in each construct). The subsequent amino acid sequences were then flanked with a leader N-terminal sequence (MSVTK-) and a C-terminal sequence (-CHHHHHH). The corresponding DNA sequence for each construct was designed by CODA Genomics who optimized for codon pairs (17, 18). The assembled genes were subcloned from PCR-Blunt II-TOPO (Invitrogen) via PCR into PEXP5-CT-TOPO (Invitrogen) and confirmed by sequencing.
Protein expression was performed via in vitro translation using the Roche Applied Science RTS 500 ProteoMaster E. coli high yield kit and 15 μg of plasmid using l-[13C6]lysine and l-[13C6]arginine as the heavy isotope label source. The in vitro translation reaction was incubated for 18 h at 30 °C while shaking at 1000 rpm. The insoluble protein product was isolated via centrifugation for 10 min at 16,000 × g of the in vitro translation reaction volume. The pellet was resuspended in 1× PBS, pH 8.0, containing 8 m urea and 0.1% Triton X-100 and bound to His6 IMAC resin (nickel-nitrilotriacetic acid, Qiagen). The resin was washed with buffer containing PBS, 8 m urea, and 40 mm imidazole, and then the protein was eluted with PBS, 8 m urea, and 250 mm imidazole.
Precipitation of QconCATs for Buffer Exchange—
20 μl of 20% trichloroacetic acid was added to 20 μl of purified QconCAT and incubated on ice for 30 min. The acidified solution was then centrifuged at 8000 rpm at 4 °C for 15 min, and the supernatant was carefully removed. 300 μl of cold acetone was added to the tube. The sample was then vortexed and again centrifuged for 5 min at 4 °C. Acetone was decanted, and the pellet was dried in a speed vacuum concentrator. Protein concentration in a representative vial was measured using the BCA protein assay.
Conditional Testing of Trypsin Digestion—
Several digestion conditions were tested to establish the optimum digestion protocol. Digestion conditions were diversified by using five different denaturing agents (urea, guanidinium HCl, ACN, Rapigest, and SDS) and three different incubation conditions (overnight at 37 °C, sonication, and microwave radiation). Three aliquots were used for each denaturing agent so each could be tested under different incubation conditions. The contents of each sample (∼800 fmol) were resuspended in 20 μl of buffer containing one of the denaturing agents (6 m urea, 6 m guanidinium HCl, 80% ACN, 0.1% Rapigest, and 0.05% SDS) in 50 mm ammonium bicarbonate and 5 mm DTT. The contents of each vial were then sonicated for 5 min and incubated at 37 °C for 2 h. After the solutions cooled to room temperature, 100 μl of 50 mm ammonium bicarbonate was added to vials containing urea or guanidinium HCl. To be consistent, 100 μl of solubilization solution without DTT were added to the other vials as well (see Table II). 0.3 μg of trypsin was then added to each vial. One vial from each denaturing agent set was incubated at 37 °C overnight. The second vial from each set was digested using a CEM Discover® System (CEM Biosciences Division, Matthews, NC) using a program that applied 50 °C via microwave radiation for 15 min, and the third vial was sonicated at 50% amplitude for 15 min in a Branson Sonifier 250 bath sonicator (Krackeler Scientific, Inc, Albany, NY). Trypsin activity was inhibited by lowering the pH below 2 by addition of 0.5% TFA. In the case of Rapigest digestion, solutions were incubated at 37 °C for 45 min. The samples were then centrifuged at 4 °C, and the supernatant was removed for further analysis. In the case of SDS digestion, the trypsin digestion pH was lowered by addition of 1% TFA, and digestion solutions was cleaned by a mixed cation exchange trap cartridge.
Table II.
List of denaturing agents, incubation conditions, and -fold dilution used in each digestion protocol
| Denaturing agent | Incubation conditions: 37 °C overnight, sonication for 15 min, or microwave radiation for 15 min |
|---|---|
| Urea (6 m) | Diluted 6× with 50 mm ammonium bicarbonate before digestion |
| Guanidinium HCl (6 m) | Diluted 6× with 50 mm ammonium bicarbonate before digestion |
| ACN (80%) | Diluted 6× with 80% ACN, 50 mm ammonium bicarbonate before digestion |
| Rapigest (0.1%) | Diluted 6× with 50 mm ammonium bicarbonate, 0.1% Rapigest before digestion |
| SDS (0.05%) | Diluted 6× with 50 mm ammonium bicarbonate, 0.05% SDS before digestion |
SDS-PAGE of Intact and Digested QconCATs—
5 μl of digested and intact QconCATs were added to separate Eppendorf tubes each containing 2.5 μl of NuPAGE (Invitrogen) SDS sample buffer and 1 μl of NuPAGE sample reducing agent. Sample tubes were incubated at 75 °C for 10 min and then separated on a NuPAGE 4–12% Bis-Tris precast gel in a gel box filled with 1× NuPAGE MOPS running buffer. Separation was accomplished by applying 200 V at constant current for 50 min. Protein and peptide spots were stained using SilverSNAP stain for mass spectrometry from Pierce following the manufacturer's protocol.
LC/MS/MS Analysis—
Tryptic peptides generated as a result of QconCAT digestion were separated on an Agilent Zorbax SB-C18 column (0.075 × 150 mm) using a Proxeon easy-nLC system (Odense, Denmark) at 200 nl/min. Solvent A was 0.1% formic acid in deionized H2O, and solvent B was 98% ACN, 0.1% formic acid in deionized H2O. Flow from the column was directed to a Q-STAR pulsar i work station (Applied Biosystems, Framingham, MA) equipped with a nano-ESI source. Peptides were separated in a 60-min linear gradient (from 10 to 35% solvent B), and MS/MS spectra were acquired in positive ion mode at 2700 V of ionization voltage and 20 units of curtain gas at a sampling rate of one spectrum per second. The top three peptides with charges ranging from 2+ to 4+ were selected for CID. Precursor ions once selected for CID were excluded from reselection for 60 s. The centroid MS/MS peak list was generated using Analyst QS 1.1 software (Applied Biosystems, Foster City, CA) using the default parameters. MASCOT 1.9 was used for all database searches.
The following search parameters were used for peptide identification: the database included the QconCAT sequence and trypsin, proteolysis was achieved with trypsin, up to two miscleavages were allowed, heavy lysine and arginine (6 Da heavier than light) were considered in all cases as fixed modifications, no variable modification was included, the peptide mass tolerance was set at ±1.2 Da, mass tolerance for fragment ions was set at ±0.6 Da, the charge was specified for each peptide, and monoisotopic peaks were used for identification. The MASCOT scoring system was used as a measure of identification certainty. The cutoff score was set to allow 5% false positive because the purpose of the MS/MS search was not to identify the peptides but to confirm their presence and to evaluate the digestion efficiency.
LC/MRM Analysis—
A list of 10 or more MRM transitions for each peptide was generated using only y ions. Synthetic peptides (provided by the manufacturer in vials, each containing 1 nmol of pure peptide) were resuspended in 50 μl of 50% ACN, 1% formic acid and mixed in a 1:1 ratio. The 24 synthetic peptides were then separated on an LC Packings PepMap100 reversed-phase C18 column (0.075 × 150 mm) using a Tempo 2D nano-LC system from Eksigent Technologies, LLC (Dublin, CA) at 200 nl/min, and flow from the column was directed to a 4000QTrap work station (Applied Biosystems, Framingham, MA) equipped with a nano-ESI source. The three most intense transitions were selected from the MRM spectra for each peptide to generate the final list of MRM transitions. A list of MRM transitions for heavy peptides was generated from the list of MRM transitions for light peptides by adding 6 mass units to peptide and transition ions. The lists of light and heavy transitions were then combined to generate the final MRM transition list (For a complete list of heavy and light transitions see supplemental Table 1.). QconCAT digests were mixed in 1:1 ratio with an equimolar mixture of the synthetic peptides. Original QconCAT:synthetic mixing ratios were calculated based on estimation of intact QconCAT concentration but were later normalized using average MRM ratios. The final mixture, containing heavy and light peptides, was subjected to MRM analysis using the complete heavy and light MRM transition list. All MRM experiments were done in triplicate.
Quantification—
The quantification function of the Applied Biosystems (Foster City, CA) Analyst software (version 1.4.2) was used to calculate the area under the peak for light and heavy peptides as well as their ratio. Peaks were manually checked, and peak integrations were adjusted accordingly. The light to heavy peak area ratios were calculated for each transition and exported to Excel where the average of peak ratios for three transitions was calculated to yield the heavy to light peptide ratios. These calculations were done for triplicate MRM experiments to calculate the error for each ratio measurement and evaluate the reproducibility.
RESULTS
Analytical Strategy
The goal of this study was to measure the deviation from equimolarity of peptide mixtures generated by either resolubilization/mixing of synthetic peptides or digestion of QconCATs. Twenty-five peptides from C. elegans were selected to cover a wide range of mass and hydrophobicity (Table I). Although peptides containing labile amino acids such as methionine are not ideal targets for quantification, every work flow has its own goals and limitations, and the luxury of choice is not guaranteed. Hence we included several methionine-containing peptides as part of our platform evaluation. The peptides were submitted to Sigma-Genosys for chemical synthesis in their unlabeled, natural isotope forms, and each peptide sample was quantified by amino acid analysis. Synthesized peptides were resolubilized and mixed in equimolar fashion. A list of MRM transitions was generated for each peptide, and the three most intense transitions were identified for each peptide.
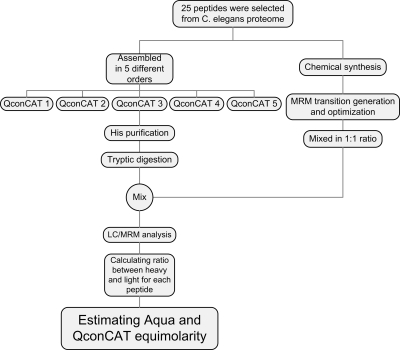
To investigate the effect of peptide order on the digestion efficiency of QconCATs the same 25 peptides were assembled in five different orders to produce five QconCATs of the same amino acid and peptide content but different sequence (Table I). The sequences of the respective synthetic genes were then optimized, synthesized, and translated in vitro in a medium containing heavy arginine and lysine. Resultant heavy isotope-coded QconCATs were then purified from the medium using the polyhistidine tag that was added to the end of each sequence. Purified QconCATs were then digested with trypsin. The equimolar mixture of heavy peptides derived from each QconCAT and the light peptides from the synthetic peptide batch were then analyzed by LC/MRM, and ratios between light and heavy peptides were calculated to evaluate the equimolarity of both mixtures (heavy and light). A schematic of the analytical strategy used is shown in Fig. 1.
Fig. 1.
Analytical strategy devised to compare the accuracy of both chemical and biological peptide generation platforms by measuring equimolarity of peptides generated in each method.
Preparation and MRM Analysis of Synthetic Peptides
Preparing an Equimolar Mixture of Synthetic Peptides—
Synthetic peptides were provided by the manufacturer in vials each containing 1 nmol of lyophilized peptide. The manufacturer's instructions for resolubilization of peptides suggested 0.1 m ammonium bicarbonate as a suitable solubilizing agent for all peptides in the set. Equal volumes from each peptide were mixed to prepare a supposedly equimolar mixture. Alternatively peptides were resolubilized in 50% ACN, 1% formic acid, combined to prepare an equimolar mixture, and concentrated under vacuum to remove the acetonitrile. Both mixtures were analyzed by MRM under the same conditions. When MRM signal intensities for a mixture prepared by following the manufacturer's solubilization instructions (mainly ammonium bicarbonate) were compared with the intensities for the same fragment ions from a mixture of peptides prepared using 50% ACN, 1% formic acid, a significant difference was observed. Results showed that the buffer containing organic solvent was a better solubilizing agent. Thus for all following experiments, peptides were resolubilized in 50% ACN, 0.1% formic acid.
Optimization of MRM Transitions—
To generate a list of MRM transitions for each peptide, the fragment ion spectrum was used as starting information. All singly charged y ions were listed in an optimization MRM method. Synthetic peptides were resuspended in 50 μl of 50% ACN, 1% formic acid and mixed in a 1:1 ratio. The equimolar mixture of synthetic peptides was then separated on an LC Packings PepMap100 reversed-phase C18 column (0.075 × 150 mm) using a Tempo 2D nano-LC system from Eksigent (Eksigent Technologies, LLC) at 200 nl/min. Flow from the column was directed to a 4000QTrap work station (Applied Biosystems, Framingham, MA) equipped with a nano-ESI source. The area under the peak for each MRM transition was calculated using Analyst (1.4) software, and the three most intense transitions for each peptide were selected for the QconCAT peptide quantification. Equivalent transitions for heavy isotope-coded peptides from the QconCAT digests were generated and added to the list of light transitions to create the final MRM transition list. (A complete list of transitions for the heavy and light peptides is compiled in supplemental Table 1.)
Expression and Purification of QconCATs
To investigate the effect of peptide order in the digestion efficiency of QconCATs, the order of the 25 selected peptides was scrambled and assembled in five different proteins of the same amino acid composition but varying sequence (Table I). DNA sequences encoding the designed QconCAT sequences were optimized by removal of translational “pause” sites, thus improving translation efficiencies (17, 18). QconCATs in heavy isotope-labeled form were produced via in vitro translation, incorporating heavy lysine and arginine, such that each resultant tryptic peptide would be obtained in a heavy labeled form. One of the major advantages of in vitro translation is the exclusive incorporation of heavy lysine and arginine due to the absence of other sources of lysine and arginine in the medium. Hence the isotope purity of QconCAT peptides is determined by the purity of heavy lysine and arginine, which in this case was 98% according to the manufacturer. The His6 tag at the C terminus of each construct allowed for confirmation of complete protein generation and for rapid purification using IMAC. The resultant eluates were tested by dot-blotting using a polyhistidine-specific antibody to ensure the presence of the target protein (data not shown). QconCAT purity was then assessed by SDS-PAGE (Fig. 2a). QconCATs appeared to be pure enough to proceed, so we did not consider further purification necessary for the following reasons. 1) The impurities present constituted only a small portion of the proteins present in mixture (samples were relatively pure). 2) Any impurity that had a different sequence from QconCATs would not interfere with the equimolarity of the QconCAT peptides. 3) Truncations and incompletely translated QconCATs that could potentially throw off the equimolarity of the digest lack the C terminus necessary for IMAC purification. 4) Other impurities that could potentially interfere with our study were pieces of incomplete QconCATs from only the C-terminal portion of the construct, which could only be generated post-translationally under degrading conditions, which we had avoided.
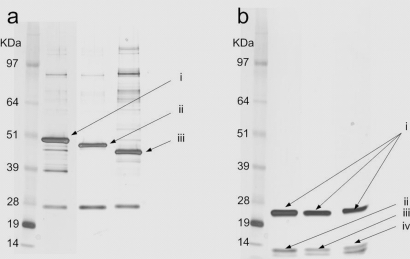
Fig. 2.
a, separation of intact QconCATs by SDS-PAGE to evaluate mass accuracy and purity. i, QconCAT 1; ii, QconCAT 2; iii, QconCAT 3. b, separation of digested QconCATs to test the digestion completeness. i, trypsin; ii, QconCAT 1 digest; iii, QconCAT 2 digest; iv, QconCAT 3 digest.
Only three of the five QconCATs were successfully translated. All three QconCATs appeared to have a molecular mass of around 50 kDa as expected (Fig. 2a). There was, however, a slight difference in electrophoretic mobility probably due to variation in folding (19). The translation failure of two of the five could be due to several factors. Peptide order may play a role in translation success rate. Alternatively the codon pair optimization may also play a role. However, the in vitro translation strategy proved effective in simplifying the purification process due to the fact that only essential components for protein translation are present versus an in vivo translation system.
Digestion and Characterization of QconCATs
Sequence Confirmation—
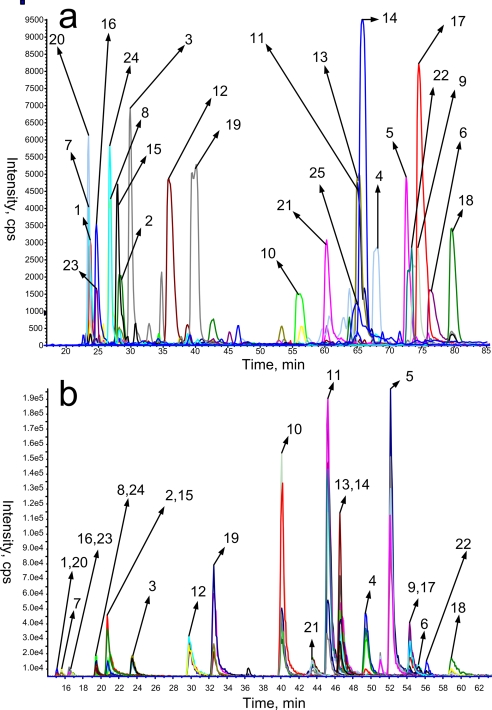
Although the presence of three QconCAT proteins was confirmed by immunoblotting, the accuracy of their sequence was still unknown. We thus examined the QconCATs by tandem mass spectrometry. Following elution from the IMAC column and reduction of cysteines, trypsinization was performed (at 1:20 enzyme to substrate ratio) using a combination of sonication and overnight incubation at 37 °C. Digestion was then terminated by addition of TFA. The degree of digestion was assessed by SDS-PAGE (Fig. 2b). Two bands are visible in each lane, one for trypsin and the other at the bottom of the gel, indicating the presence of small peptides. Notably the bands representing the staring material were no longer detectable. All three QconCAT digests were then analyzed by LC/MS/MS using an Applied Biosystems (Foster City, CA) Q-Star Pulsar i. Each sample was separated on a 60-min gradient, and the three most intense precursor ions were selected for MS/MS. The MS/MS data were searched using MASCOT. In all three cases, all 25 peptides were identified (For a complete list of identified peptides and their MASCOT score, see supplemental Table 2.). As an example the extracted ion chromatogram of all 25 peptides from QconCAT 2 is shown in Fig. 3a. Notably the data confirmed the presence of peptide TASVLISGLGSVGVEIAK (Fig. 3a, marked as 25), which could not be generated by chemical synthesis. These results indicate that from the three QconCATs that could be expressed all the expected peptides could be detected. This is in contrast to the set of chemically synthesized peptides where one peptide synthesis failed.
Fig. 3.
a, extracted ion chromatogram of the 25 QconCAT peptides identified by database search of MS/MS spectra from QconCAT 2. b, MRM mass spectra acquired for a 1:1 mixture of 24 synthetic peptides and QconCAT 2 digest. Ratios between area under the peak for the light (synthetic) and heavy (QconCAT) peptides were used to calculate the ratio between the two. cps, counts per second.
Optimization of Digestion—
Although QconCAT digestion yielded all the expected peptides, their presence alone does not indicate equimolarity, a requirement for the QconCAT approach if it is to be useful for absolute quantification. Protocol dependence has been well documented for tryptic digestion (20). Denaturing agents, as well as trypsin quality and incubation conditions, are all known to affect the quality (fewer peptides with miscleavages) and quantity (equimolarity of resultant peptides) of digestion (21–23). It is also known that not all proteins behave the same under the same trypsinization conditions. Because our goal was to evaluate the QconCATs as a tool for absolute quantification, it was important to optimize for any factors that might affect the equimolarity of the resultant QconCAT peptides. Although we assume that digestions will never be truly 100% efficient and accurate, conditions can be tailored to best fit the construct of interest. Because these QconCATs only varied in the order of peptides, we assumed that if we optimized digestion conditions for one of the QconCATs, it will be the same for the others. QconCAT 2 was used for digestion optimization.
We thus evaluated a wide range of digestion conditions, utilizing five common denaturing agents: urea, guanidinium HCl, ACN, Rapigest, and SDS. Each denaturing condition was also evaluated under three common incubation conditions: microwave radiation, sonication, and 37 °C incubation. Thus in total 15 different digestion conditions were tested. Digestion volumes were kept constant for all conditions tested, and all digestions were terminated by acidification. Because SDS is not compatible with mass spectrometry, it had to be removed from the digestion solution via a mixed cation exchange trap cartridge. A molar equivalent of the 24 synthetic peptide mixture was also added to the SDS-based digests prior to the cleaning step to ensure that any effect from differential peptide recoveries on the final yield would not complicate data interpretation.
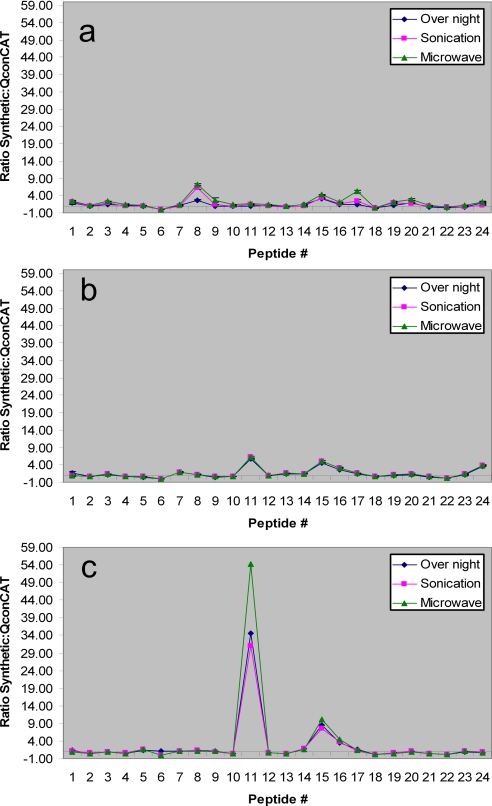
Each mixture was then analyzed by LC/MRM using the method generated previously with the list of optimized transitions. An LC/MRM ion chromatogram for one of the 15 QconCAT digests (overnight digestion with urea as denaturing agent) mixed with 24 synthetic peptides mixtures is shown as an example in Fig. 3b. The injection volumes were adjusted so that the transition with lowest intensity would be detectable, and each sample was analyzed three times. The area under each transition was measured using the IntelliQuan algorithm in the Analyst (1.4) software. Each integration was manually reviewed and adjusted to best fit the peak, and the ratio between each light and heavy transition could thus be calculated. Because we measured three transitions per peptide, we averaged the ratios for the three transitions selected for each peptide, and again for the three replicate runs, to generate the final ratio for each peptide. Standard deviations were calculated for the three replicate runs. These results for digestions using urea, ACN, and Rapigest as denaturing agents are shown in Fig. 4. (The final ratios for the other six digestion protocols are provided as supplemental materials.)
Fig. 4.
Synthetic:QconCAT ratio for nine of 15 digestion protocols. a, digestion using urea as denaturing agent. b, digestion using ACN as denaturing agent. c, digestion using Rapigest as denaturing agent. Overnight digestion with urea resulted in ratios closest to equimolarity. The presented data represent mean results for triplicate analyses.
When QconCAT peptides were not generated equimolarly, light to heavy peptide ratios were found to be higher than 1. There were also peptides that appeared at ratios of less than 1. The only explanation for such cases is deviation from equimolarity among synthetic peptides. Peptides 6 and 22 are clear examples of such cases. Obviously this issue (whether synthetic peptides can be used to generate equimolar peptide mixtures) is an equally important concern for absolute quantification using a synthetic heavy isotope-coded peptide platform. A lower presence of some synthetic peptides in the mixture could be explained in several ways, including problems with solubilization of the peptides, peptide degradation, and peptide modification. Several solubilization strategies including use of high organic solvent-containing buffers and extensive sonication steps were tested with no improvement in the observed molar ratio of lower abundance peptides (data not shown). Following the detailed instructions provided by the manufacturer of the synthetic peptides for their solubilization also did not improve their recovery into solution. We thus concluded that such peptides are either bound tightly to the tube walls or are somehow degraded or modified. Peptide 6 for example is an exceptionally long peptide (GYGFVIDDITGEDLFVHQSNLNMQGFR) with a single methionine in its sequence. To investigate whether this peptide was degraded or modified, this peptide was analyzed separately by LC/MS/MS. The MS/MS data showed that both peptide fragmentation and methionine oxidation had occurred at considerable levels prior to MS analysis (data not shown). However, the relative level of degradation and modification could not be determined exactly because the side products have different and unknown ionization efficiencies.
On the other hand, the QconCAT data showed that urea as denaturing agent, coupled with overnight incubation at 37 °C, resulted in peptide ratios closest to the desired equimolarity (Fig. 4a). The average ratio for all peptides in this case was calculated to be 1.23 with a standard deviation equal to 0.68.
We observed that urea-containing digests utilizing microwave radiation or sonication were not as complete. Digestions using guanidinium HCl as denaturing agent showed a similar trend, but overall the results were not quite as good. ACN was also found to be an effective denaturing agent for tryptic digestion. A notable observation, however, was the similarity of the peptide ratios seen for the different incubation methods when ACN was used, suggesting an improved rate of digestion in the presence of ACN (Fig. 4b). QconCAT digests using Rapigest and SDS were not quite as complete. Because the Rapigest removal step requires heating for 30–45 min at a pH lower than 2, this step may account for some peptide loss. As a result, in some cases, up to a 50-fold difference between heavy and light peptides was observed as a function of the digestion conditions (Fig. 4c).
Sequence Dependence and QconCAT Digestion
As mentioned above, peptide order in the QconCATs impacted the in vitro translation success. Similarly changing the peptide order could potentially affect the digestion efficiency. It was thus important to investigate any peptide sequence order dependence for digestion because, without complete digestion, QconCATs are not useful for absolute quantification. Because all three QconCATs expressed in this experiment had the same peptide content and digestion conditions were already optimized for one of the constructs (QconCAT 2), peptide sequence order dependence could be tested.
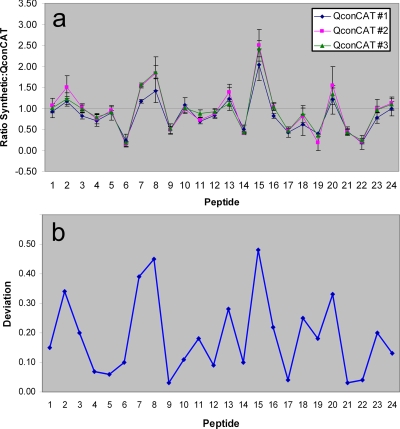
All three QconCATs were digested using the optimal conditions. Quantification of heavy peptides from the QconCAT digests was accomplished as before using the unlabeled synthetic peptides and MRM, and ratios measured as light (synthetic) to heavy (QconCAT) (Fig. 5a) were calculated. All ratios were expected to be 1:1 as an equimolar mixture of synthetic peptides was mixed with an equimolar QconCAT digest. Any ratio with a value sufficiently different from 1:1 would indicate that one of the peptides of interest was not present in an equimolar amount (Fig. 5b). Six of the 24 peptides were found to be such cases. The highest ratio observed was 2.5 for peptide 15. It is not clear why this peptide (NSVGLGGGLAER) was specifically generated in lower amounts during QconCAT digestions. The fact that this peptide was observed at similar ratios for all three QconCATs rules out the possibility of a digestion artifact. On the other hand, the next three QconCAT peptides that appeared in higher ratios show a common property. Peptides 7 (ratio, 1.5), 8 (ratio, 1.8), and 20 (ratio, 1.5) are all very hydrophilic peptides. But again the relationship between peptide hydrophilicity and digestion efficiency is not clear.
Fig. 5.
a, synthetic (light):QconCAT (heavy) peptide ratio evaluated using MRM. Each peptide was monitored using three transitions, and each run was repeated in triplicates. b, differences in peptide ratios observed between the three QconCATs.
On the other hand, although we did find some evidence that there are variations in QconCAT folding (slight differences in mass of intact QconCAT determined by gel electrophoresis; see Fig. 2) it did not seem to have a significant impact on QconCAT digestion efficiency. All three QconCATs showed a very similar ratio pattern (Fig. 5a). In a few cases (peptides 7, 8, 18, and 20) some variation was observed. However, in the least reproducible cases, the variation in ratio did not exceed 25% of the average ratio (Fig. 5b). These results suggest that peptide order may not affect the digestion efficiency significantly and further support the indication that QconCATs may be a more generally applicable technique than conventional peptide synthesis.
DISCUSSION
The purpose of this study was to evaluate the current internal standard peptide production platforms for mass spectrometry-based absolute quantification. Mass spectral absolute quantification requires the use of heavy isotope-labeled peptides at known concentrations. Chemical and biological methods can both be used to produce these heavy peptides. Each method has strengths and weaknesses, which were evaluated in this study and which are discussed below.
Peptide Generation—
The most common way of supplying heavy labeled peptides is chemical synthesis. Chemical synthesis has a number of limitations, and as a result many peptides cannot be synthesized. Similarly some peptides can be synthesized but not purified sufficiently well for their use in a quantitative experiment. In addition, chemical synthesis is expensive and time-consuming and requires special facilities. Biological expression of concatenated peptides is an alternate method of peptide production. In this approach, sets of peptides can be expressed in groups, and hence the method is not as expensive and is much faster. Although there are fewer limitations for biological generation of certain peptides, limitations exist for expression of certain QconCATs. Our results showed that altering the order of peptides may result in successful expression in some such cases but requires reconstruction of the DNA sequence. Thus it might be more practical to use several QconCATs for the same peptide content to increase the probability of expression success. Overall in our opinion, the QconCAT approach appears to be a more versatile method for generation of peptides for quantitative proteomics but is limited by predetermining the mixture for an experiment.
Concentration of Standard Peptides—
Amino acid analysis is commonly used to determine synthetic peptide concentrations; again this is expensive and time-consuming for large scale experiments. Synthetic peptides are usually provided lyophilized. Resolubilization can be complicated and can also affect the experimental results dramatically because there is no easy way to determine whether the entire peptide content of a vial was resolubilized. PTMs and degradations are also factors that can affect peptide concentrations and are again difficult to detect because each peptide is different in nature. On the other hand, the concentration of QconCAT-derived peptides can be calculated from the concentration of intact QconCAT itself. Thus the only requirement for deriving the equimolar peptide mixture at the same concentration as the QconCAT is complete tryptic digestion. Tryptic digestion is known to be protocol- and protein-dependent. Thus, ideally a range of conditions should be tested for each new QconCAT. Unfortunately this may often be impractical. However, the data presented here suggest that digestion was fairly comparable for QconCATs that are expressed, and because, by design, tryptic cleavage sites will be evenly distributed throughout a QconCAT gene, it is quite possible that digestion might be similar for most QconCATs once initially optimized.
Preparation of Standard Peptide Mixtures—
A significant advantage of synthetic peptides is having control over the concentration of each peptide when making a mixture. The concentration of each peptide needs to be adjusted individually for following reasons. 1) Different peptides have different ionization efficiencies. 2) Proteins in biological samples, most extremely in body fluids, exist across a wide range of concentrations. Thus, for absolute quantification, standard peptides have to be added in quantities that fall within the linear dynamic range of mass spectrometry methods. Because QconCAT-generated peptides, by definition, exist at an equimolar ratio, individual peptide concentrations cannot be adjusted. This is one advantage individual peptide chemical synthesis has over the QconCAT approach. Thus to generate peptide mixtures from QconCATs that could effectively be used for quantification of several proteins, the target peptides need to be carefully selected sometimes, based on criteria that may not be known. Practical considerations for each peptide production platform are presented in Table III.
Table III.
Practical considerations when choosing peptide production platforms
| No. | Considerations |
|---|---|
| Chemical synthesis | |
| 1 | Synthetic peptides are expensive |
| 2 | Synthesis usually takes longer time |
| 3 | Peptide synthesis occasionally fails |
| 4 | Resolubilization is not always 100% |
| 5 | Degradation and PTMs occur as a result of lyophilization/storage and are not easy to detect |
| QconCAT platform | |
| 1 | Several QconCAT constructs may be needed to guarantee success |
| 2 | Generation of equimolar peptide mixture requires complete tryptic digestion, which requires optimization of digestion conditions |
| 3 | QconCAT peptides cannot be used when significant -fold changes in peptide concentrations are required for quantification |
| 4 | It is not practical to generate QconCATs comprised of more than 30–40 peptides |
Conclusion—
Twenty-five peptides were concatenated in different orders to generate five different QconCATs, three of which were successfully translated into intact proteins. This result indicated that in a case of expression failure peptides might be rearranged to achieve successful expression. One of the peptides included in the QconCAT peptide list had failed in chemical synthesis but was successfully generated by the QconCAT method showing that the QconCAT approach can generate peptides that are not amenable for chemical synthesis. When QconCATs were subjected to tryptic digestions using different denaturing agents and incubation protocols, equimolarity of the resultant peptide mixture was quite variable, indicating the importance of determining optimal digestion conditions prior to peptide use in a quantitative experiment. If QconCAT digestions are not tested each time, this could lead to inaccuracies in subsequent quantifications made. Synthetic peptides were also found to have serious limitations when used for accurate quantifications. These limitations mostly arise from issues with solubility, degradation, peptide PTMs, and the fact that some peptide sequences are not amenable to chemical synthesis. When optimal QconCAT digestion conditions were used, most peptides generated from the QconCAT appeared to exist at equimolar amounts. Even peptides that existed in lesser amounts were well within accepted ranges for effective quantification (2.5-fold variations). Finally we found that the order of the peptides in the QconCAT sequence did not have a significant effect on digestion efficiency. Careful examination of both methods of quantification proves that there is no gold standard, and the method of quantification needs to be selected carefully on a case by case basis.
Supplementary Material
Footnotes
Published, MCP Papers in Press, December 17, 2007, DOI 10.1074/mcp.M700495-MCP200
The abbreviations used are: PTM, post-translational modification; MRM, multireaction monitoring; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; 2D, two-dimensional.
This work was funded in part with federal funds from the NHLB, National Institutes of Health, under Contract N01-HV-28179 and by NIAID, National Institutes of Health, Grant R01-AI51344 (to J. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Aebersold, R., and Mann, M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2.Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H., and Aebersold, R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty, A., and Regnier, F. E. (2002) Global internal standard technology for comparative proteomics. J. Chromatogr. A 949, 173–184 [DOI] [PubMed] [Google Scholar]
- 4.Ross, P. L., Huang, Y. L. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., Khainovski, N., Pillai, S., Dey, S., Daniels, S., Purkayastha, S., Juhasz, P., Martin, S., Bartlet-Jones, M., He, F., Jacobson, A., and Pappin, D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 5.Rinner, O., Mueller, L. N., Hubalek, M., Mueller, M., Gstaiger, M., and Aebersold, R. (2007) An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat. Biotechnol. 25, 345–352 [DOI] [PubMed] [Google Scholar]
- 6.Wang, G., Wu, W. W., Zeng, W., Chou, C., and Shen, R. (2006) Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J. Proteome Res. 5, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 7.Askenazi, M. (July 12, 2006) Iterative Base Peak Framing of Mass Spectrometry Data. World Patent WO/2006/130368
- 8.Kipping, M., Pollack, L., and Langridge, J. (2005) Label free quantitative proteomics: high resolution electrospray LC-MS for functional proteome analysis. BIOspektrum 11, 780–781 [Google Scholar]
- 9.Puetz, S., Reinders, J., Reinders, Y., and Sickmann, A. (2005) Mass spectrometry-based peptide quantification: applications and limitations. Expert Rev. Proteomics 2, 381–392 [DOI] [PubMed] [Google Scholar]
- 10.Wienkoop, S., and Weckwerth, W. (2006) Relative and absolute quantitative shotgun proteomics: targeting low-abundance proteins in Arabidopsis thaliana. J. Exp. Bot. 57, 1529–1535 [DOI] [PubMed] [Google Scholar]
- 11.Fountoulakis, M., and Lahm, H. (1998) Hydrolysis and amino acid composition analysis of proteins. J. Chromatogr. A 826, 109–134 [DOI] [PubMed] [Google Scholar]
- 12.Barnidge, D. R., Hall, G. D., Stocker, J. L., and Muddiman, D. C. (2004) Evaluation of a cleavable stable isotope labeled synthetic peptide for absolute protein quantification using LC-MS/MS. J. Proteome Res. 3, 658–661 [DOI] [PubMed] [Google Scholar]
- 13.Pratt, J. M., Simpson, D. M., Doherty, M. K., Rivers, J., Gaskell, S. J., and Beynon, R. J. (2006) Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 1, 1029–1043 [DOI] [PubMed] [Google Scholar]
- 14.Pratt, J. M., Beynon, R. J., and Gaskell, S. J. (July 12, 2006) Artificial Protein, Method for Absolute Quantification of Proteins and Uses Thereof. World Patent WO/2006/128492
- 15.Beynon, R. J., Doherty, M. K., Pratt, J. M., and Gaskell, S. J. (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 [DOI] [PubMed] [Google Scholar]
- 16.Rivers, J., Simpson, D. M., Robertson, D. H., Gaskell, S. J., and Beynon, R. J. (2007) Absolute multiplexed quantitative analysis of protein expression during muscle development using QconCAT. Mol. Cell. Proteomics 6, 1416–1427 [DOI] [PubMed] [Google Scholar]
- 17.Gutman, G. A., and Hatfield, G. W. (1989) Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86, 3699–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irwin, B., Heck, J. D., and Hatfield, G. W. (1995) Codon pair utilization biases influence translational elongation step times. J. Biol. Chem. 270, 22801–22806 [DOI] [PubMed] [Google Scholar]
- 19.Goldenberg, D. P., and Creighton, T. E. (1984) Gel electrophoresis in studies of protein conformation and folding. Anal. Biochem. 138, 1–18 [DOI] [PubMed] [Google Scholar]
- 20.Strader, M. B., Tabb, D. L., Hervey, W. J., Pan, C., and Hurst, G. B. (2006) Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal. Chem. 78, 125–134 [DOI] [PubMed] [Google Scholar]
- 21.Bermejo, P., Capelo, J. L., Mota, A., Madrid, Y., and Camara, C. (2004) Enzymatic digestion and ultrasonication: a powerful combination in analytical chemistry. Trends Anal. Chem. 23, 654–663 [Google Scholar]
- 22.Vesper, H. W., Mi, L., Enada, A., and Myers, G. L. (2005) Assessment of microwave-assisted enzymatic digestion by measuring glycated hemoglobin A1c by mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2865–2870 [DOI] [PubMed] [Google Scholar]
- 23.Yu, Y., Gilar, M., Lee, P. J., Bouvier, E. S. P., and Gebler, J. C. (2003) Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal. Chem. 75, 6023–6028 [DOI] [PubMed] [Google Scholar]
- 24.Hopp, T. P., and Woods, K. R. (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. U. S. A. 78, 3824–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.