Abstract
The model organism zebrafish (Danio rerio) is particularly amenable to studies deciphering regulatory genetic networks in vertebrate development, biology, and pharmacology. Unraveling the functional dynamics of such networks requires precise quantitation of protein expression during organismal growth, which is incrementally challenging with progressive complexity of the systems. In an approach toward such quantitative studies of dynamic network behavior, we applied mass spectrometric methodology and rigorous statistical analysis to create comprehensive, high quality profiles of proteins expressed at two stages of zebrafish development. Proteins of embryos 72 and 120 h postfertilization (hpf) were isolated and analyzed both by two-dimensional (2D) LC followed by ESI-MS/MS and by 2D PAGE followed by MALDI-TOF/TOF protein identification. We detected 1384 proteins from 327,906 peptide sequence identifications at 72 and 120 hpf with false identification rates of less than 1% using 2D LC-ESI-MS/MS. These included only ∼30% of proteins that were identified by 2D PAGE-MALDI-TOF/TOF. Roughly 10% of all detected proteins were derived from hypothetical or predicted gene models or were entirely unannotated. Comparison of proteins expression by 2D DIGE revealed that proteins involved in energy production and transcription/translation were relatively more abundant at 72 hpf consistent with faster synthesis of cellular proteins during organismal growth at this time compared with 120 hpf. The data are accessible in a database that links protein identifications to existing resources including the Zebrafish Information Network database. This new resource should facilitate the selection of candidate proteins for targeted quantitation and refine systematic genetic network analysis in vertebrate development and biology.
Embryonic development is governed by highly coordinated changes in the expression of large protein sets. Resolving the programs controlling these changes at the molecular level can provide important insights into the principles of human biology and disease. The zebrafish (Danio rerio) is an attractive vertebrate model organism for studies into the molecular mechanisms of development (2), pathology (3), and pharmacology (4, 5). Its high fecundity and short generation times render it particularly amenable to large scale forward genetics approaches (6), which study gene-function relationships free of a priori assumptions using genome-wide random chemical (7) or retroviral (8) mutagenesis. Subsequent mapping and positional cloning of mutated genes can link them to observed phenotypes. Definitive proof for such gene-function relationships is typically afforded by reverse genetics experiments. These depress gene expression either transiently using antisense morpholinos or using stable mutants systematically identified in a gene of interest by a method referred to as “targeting induced local lesions in genomes” (TILLING) (9).
Zebrafish genetics has been facilitated substantially by the increasing availability of genome sequence information (Sanger Institute's D. rerio sequencing project) as assembly of the 1.7-Gb genome sequence nears completion. The utility of the genome sequence increases with the quality of annotation of protein-encoding genes. Genome annotations are supported by alignments of experimentally documented transcript or protein sequences specific for the zebrafish genome, by alignments of homologous transcript or protein sequences, and ab initio by computational gene prediction (10). While computational predictions tend to be rather imprecise, many putative zebrafish genes have human orthologs, and large regions of synteny exist between human and zebrafish chromosomes underlining the relevance of the zebrafish to the analysis of higher vertebrates (11). However, conservation across species is not limited to protein-coding regions (12); thus, these annotations are not compelling. Currently ∼17,300 genes have been annotated based on the highest quality of evidence (species-specific transcript data such as expressed sequence tags), ∼2500 are unknown or predicted on the basis of evidence from closely related species, and ∼1500 genes are computationally predicted (Ensembl Assembly Zv7, April 2007 (13)). Direct detection of translated peptides by tandem mass spectrometry methods has proved valuable in the annotation of genomes as they allow prediction or refinement of gene structures and can resolve difficulties in annotating from cDNA alternative splice forms and overlapping gene sequences (14–18).
Genetic screens for perturbed phenotypes have generated numerous mutants with defects in pathways affecting development, physiology, and behavior (6). Many mutant phenotypes are reminiscent of human disease and have proved invaluable in deciphering signaling cascades implicated in cardiovascular (3, 19), renal (20), and gastrointestinal biology (21) and in cancer (22). Initial forward genetic screens have phenotyped embryos based on morphology as optical clarity during external zebrafish embryogenesis facilitates visual analysis during their rapid development (6). More recently, assays visualizing biological functions in vivo such as the metabolic processing of fluorescent marker lipids (23) or the regenerative response to injury (24) and assays detecting dysregulated gene expression (25) have discovered mutant phenotypes not primarily amenable to visual inspection. Metabolic screens based on the accurate quantitation of biomarkers (e.g. using highly sensitive MS) that have been performed in chemically mutated mice (26, 27) have yet to be applied to zebrafish (28). Studies into the genetics of protein expression are largely constrained by the availability of specific antibodies. Mass spectrometry-based proteomics methods have the potential to overcome these hurdles; however, this requires first an accurate characterization of proteins accessible to targeted quantitative analysis (29).
We applied mass spectrometric proteomics methodology and statistical analysis (30) to create profiles of proteins expressed during zebrafish embryonic development. A major problem in two-dimensional chromatography-tandem mass spectrometry (“shotgun”) proteomics approaches is that many protein identifications have low reproducibility if the sensitivity of detection is not carefully balanced against rates of false identification error by using multiple replicates and rigorous statistics (31). Thus, we applied a novel empirical Bayesian algorithm to integrate data sets from multiple search programs and multiple biological replicates (30). The data are accessible in a fully searchable database (see supplemental data for the URL database link under Instructions for Downloading) that links protein identifications to existing resources including the Zebrafish Information Network (ZFIN)1 database (32).
EXPERIMENTAL PROCEDURES
Sample Preparation—
Methods for breeding and raising zebrafish embryos were followed as described previously (33). Embryos were obtained from natural matings of wild-type tail long fin fish. Embryos from four independent matings were collected 72 and 120 h postfertilization (hpf), snap frozen in liquid nitrogen, and stored at −80 °C. Sample preparation, protein digestion, and chromatographic separation were performed as described previously (30). Briefly, five tubes of 20–30 embryos were lysed (7 m urea, 2 m thiourea, 4% CHAPS (all Amersham Biosciences), 100 mm DTT (Bio-Rad), Phosphatase Inhibitor Mixture 11 (Sigma), Complete protease inhibitor (Roche Applied Science)) and homogenized (TissueLyser, Qiagen; 2 × 3 min, 30 Hz), and proteins were precipitated and resuspended in 0.2% (w/v) RapiGest™ SF (Waters Co., Milford, MA) in 50 mm ammonium bicarbonate (Sigma). Samples containing 2.0 mg of protein were reduced (5 mm DTT, Bio-Rad), alkylated (5 mm iodoacetamide; Bio-Rad), and trypsin-proteolyzed using a 1:20 (w/w) enzyme-to-protein ratio (Promega, Madison, WI) at 37 °C for 16 h. Prior to strong cation exchange chromatography the pH was lowered to 3.
Two-dimensional Chromatography and Mass Spectrometry—
Off-line fractionation and LC-MS/MS were performed as described previously (30). The sample was loaded onto a PolySulfoethyl A column (100 mm × 4.6 mm, 5 μm, 300 Å; PolyLC, Columbia, MD) at a flow rate of 0.2 ml/min with mobile phase A (10 mm ammonium formate, 25% acetonitrile, pH 3). A linear gradient for 80 min was run to 100% mobile phase B (500 mm ammonium formate, 25% acetonitrile, pH 6.8). Sixty 1.6-min fractions were collected with a Foxy Jr. (Dionex, Sunnyvale, CA) automated fraction collector. Fractions with low peptide concentration were combined to yield a total of 40 fractions, which were lyophilized and stored at −80 °C. Lyophilized peptides were reconstituted with 0.1% formic acid, 5% acetonitrile for reversed phase liquid chromatography onto a Thermo Finnigan liquid ion trap mass spectrometer (Thermo Finnigan, San Jose, CA) using electron spray ionization. Each fraction was injected onto a Vydac C18 column (Everest 150 mm × 1 mm inner diameter, 300 Å, 5 μm; Bodmann, Aston, PA) with mobile phase A (0.1% formic acid, 0.01% trichloroacetic acid in water). A gradient (30 μl/min) was run over 180 min from 3 to 70% mobile phase B (0.1% formic acid, 0.01% trichloroacetic acid in acetonitrile). Nitrogen was used as the sheath (75 p.s.i.) and auxiliary (10 units) gas with the heated capillary at 180 °C. The mass spectrometer was operated in a data-dependent MS/MS mode (m/z 300–2000) in which the top seven ions were subjected to fragmentation at 27% normalized CID energy. Dynamic mass exclusion was enabled with a repeat count of 2 every 45 s for a list size of 250.
Analysis of LC-MS/MS Data—
The data sets of each sample were searched against the International Protein Index (IPI) D. rerio protein sequence database (version 3.07; number of protein sequences, 45,388; number of amino acid residues, 23,104,717) for peptide sequences using two independent algorithms, SEQUEST 3.1 (ThermoFinnigan, San Jose, CA) and MASCOT 2.1.04 (Matrix Sciences, Boston, MA). Raw mass spectra were converted to DTA peak lists using BioWorks Browser 3.2 (ThermoFinnigan) with the following parameter settings: peptide mass range, 300–5000 Da; threshold, 10; precursor mass, ±1.4 Da; group scan, 1; minimum group count, 1; minimum ion count, 15. Searches specified that peptides should have a maximum of two internal tryptic cleavage sites with methionine oxidation and cysteine carbamidomethylation as possible modifications. SEQUEST searches specified that peptides should possess at least one tryptic terminus and used a peptide mass tolerance of ±1.4 Da and a fragment ion tolerance of 0. MASCOT searches specified tryptic digestion and used a peptide mass tolerance of ±1.5 Da and a fragment ion tolerance of ±0.1 Da. The search results were converted into pepXML format. Peptide identification probabilities for both SEQUEST and MASCOT searches were calculated by executing PeptideProphet as implemented in the Trans-Proteomic Pipeline version 2.8 (Institute for Systems Biology, Seattle, WA) (34). SEQUEST results were processed using the “-Ol” tag, which uses ΔCn* values unchanged.
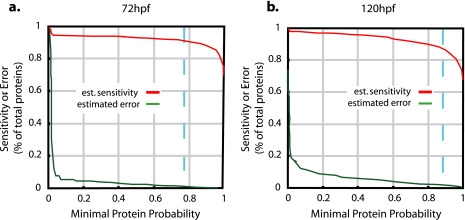
Results from both searches (SEQUEST and MASCOT) and all biological replicates were combined in a single statistical analysis of protein expression per developmental stage using Empirical Bayes Protein Identifier (EBP) 1.0 as described previously (30). Briefly EBP estimates both sensitivity and false identification rate and has been validated empirically for analysis of zebrafish liquid ion trap mass spectrometry data using a reversed/forward sequence database search approach (30). EBP combines the probabilities of correct peptide identification across multiple peptide searches using a function that returns the maximum probability from consensus identifications and penalizes non-consensual identifications. Different charge states of the same peptide are treated as a single identification. The statistical model parameters include protein length, estimated protein abundance, the size of the search database, and the number of peptide sequence identifications in the data set. For each protein in the database, an expression probability is estimated using an “expectation-maximization” algorithm. Replicates are integrated by simultaneously estimating multiple sets of model parameters. Peptides whose sequence matches multiple proteins are integrated in the analysis using “Occam's razor,” a principle by which the smallest set of probable proteins is chosen that is sufficient to explain the peptide sequence identifications. When proteins cannot be reliably distinguished by unique peptides, they are reported as a protein group. EBP analyses were run on combined SEQUEST and MASCOT results and biological replicates for each developmental stage using its default settings except for the calculation of the number of trypsin digests per protein, which specified peptides with at least one tryptic terminus. The default settings specify that only peptide identifications with probabilities greater than 0.5 are used in the calculation of protein identification probabilities. This is equivalent to using only those spectra for which SEQUEST and MASCOT reached consensus for the most likely peptide identification. Only proteins with expression probabilities corresponding to a false identification rate of less than 0.01 (1%) were reported. This was equivalent to expression probabilities of p > 0.77 and p > 0.87 in the given data sets, 72 and 120 hpf (Fig. 1). Spectra of protein identifications that met these criteria but were based on a single unique peptide were manually inspected. Thirty-eight such protein identifications were excluded after inspection. All single unique peptide identifications that remained in the data set are summarized in supplemental Table 3 as specified by the MCP data submission guidelines. Graphical representations of the corresponding annotated MS/MS spectra were extracted from the Trans-Proteomic Pipeline (Institute for Systems Biology, Seattle, WA) and the EBP plug-in using the Ruby 1.8.5 scripting language. Thus, result files were parsed for the peptide sequence with the highest PeptideProphet probability score for each charge state. Up to three such spectra were extracted if multiple spectra shared the highest probability value. HTML pages were created to display the resulting spectra images and hyperlinks included in supplemental Table 3 that open these pages. These pages and the table are included in the compressed (zipped) file (Lucitt_Supplemental_Table_3.zip) that installs the table and correct subfolder structure for viewing the linked MS/MS spectra when unpacked.
Fig. 1.
Estimated sensitivity and error of protein identification by 2D LC-MS/MS. a, integrated analysis of peptide sequence information obtained by MASCOT and SEQUEST searches of three replicate 72-hpf samples resulted in the identification of 1112 unique proteins with an estimated (est.) rate of less than 1% incorrectly identified proteins at an expression probability of p > 0.77 (dashed blue line). b, the analysis of the 120-hpf samples resulted in 867 unique protein identifications with an estimated error rate of less than 1% at an expression probability of p > 0.87 (dashed blue line).
Two-dimensional Gel Electrophoresis—
Two biological replicate samples of each developmental stage were analyzed. Proteins were resuspended in 30 mm Tris, pH 8.5, 7 m urea, 2 m thiourea, 4% CHAPS (GE Healthcare) for differential display using the DIGE technology (GE Healthcare) and in 7 m urea, 2 m thiourea, 4% CHAPS, 40 mm DTT for preparative gels. Fifty micrograms of protein from 72- and 120-hpf embryos were labeled with 400 pmol of Cy3 and Cy5 DIGE fluorophores (GE Healthcare), respectively. Reverse labeling was used to normalize for label differences. An internal control containing 25 μg of protein from both 72- and 120-hpf embryos was labeled with 400 pmol of Cy2 and included in each analytical gel. Samples were rehydrated into 24-cm, pH range 3–10, non-linear IPG strips (GE Healthcare) for 14 h at 30 V. A step and hold isoelectric focusing was completed as follows: 500 V for 500 Vhrs, 1000 V for 1000 Vhrs, 3000 V for 6000 Vhrs, 5000 V for 10,000 Vhrs, and 8000 V for 70,000 Vhrs. The cysteine sulfhydryl groups were reduced and carbamidomethylated (50 mm DTT for 15 min, 240 mm iodoacetamide for 15 min room temperature). IPG strips were then placed on top of an 8–16% precast polyacrylamide gel (Jule, Inc., Milford, CT). SDS-PAGE (Ethan DALT Twelve, GE Healthcare) was carried out at 2 watts/gel for 30 min followed by 4 watts/gel until the bromphenol dye front ran off the gel. Preparative gels contained 500 μg of unlabeled protein for identification of differentially regulated protein spots. These samples were dialyzed against 7 m urea, 2 m thiourea for 2 h prior to tow-dimensional (2D) PAGE, and gels were stained with Novex Colloidal Blue stain (Invitrogen). DIGE gels were imaged (Typhoon 9410, GE Healthcare) using excitation/emission wavelengths of 488/520 nm for Cy2, 532/580 nm for Cy3, and 633/670 nm for Cy5. The Colloidal Blue-stained gels were imaged using a wavelength of 633 nm.
Image Analysis—
Images were analyzed with Progenesis PG200 software (Nonlinear Dynamics, Newcastle, UK) according to the manufacturer's instructions applying a “cross-stain analysis” on the DIGE gels. Thus, two multiplex groups (groups of images derived from the same gel) were defined as follows: multiplex group one: gel 1, 72-hpf Cy3, 120-hpf Cy5, and internal control Cy2; multiplex group two: gel 2, 72-hpf Cy5, 120-hpf Cy3, and internal control Cy2). Three replicate groups were defined as replicate 1 (72-hpf Cy3 from gel 1 and 72-hpf Cy5 from gel 2), replicate 2 (120-hpf Cy5 from gel 1 and 120-hpf Cy3 from gel 2), and replicate 3 (internal standard labeled with Cy2 from gels 1 and 2). A reference gel was automatically selected by the software using the default settings and is based on an internal standard image. The maximum number of gels in which a spot was allowed absent within the replicate group parameters was selected to be 0. Spot detection, background subtraction, warping, matching, and normalization were all set at the default settings of the software. Where possible, unmatched spots were edited on each multiplex group based on a three-dimensional view of the spot, afterward normalization was restored, and the reference gels were updated. Differences in average normalized volume between 72 and 120 hpf of 3-fold or more were considered for protein identification.
Protein Identification from 2D Gels—
A total of 164 spots considered differentially regulated based on the above criteria and 379 spots that were considered not to be differentially regulated were excised for identification by MALDI-TOF/TOF. Spots of interest were robotically excised into 96-well plates using an Ettan Spot Picker (GE Healthcare). Gel plugs were washed with 100 μl of Milli-Q water for 15 min and three times with 100 μl of 25 mm ammonium bicarbonate, 50% ACN for 30 min while Vortex mixing. Plugs were then dehydrated in 100% ACN for 10 min and allowed to air dry. This was followed by reduction with 10 mm DTT in 50 mm ammonium bicarbonate at 60 °C for 30 min followed by alkylation with 100 mm iodoacetamide in 50 mm ammonium bicarbonate for 45 min at room temperature in the dark. Wash steps as mentioned above were repeated, and gel plugs were dehydrated with 100% ACN. Twenty micrograms of sequencing grade modified trypsin (Promega) was solubilized in 40 mm ammonium bicarbonate, 5% ACN to a concentration of 20 ng/μl. Ten microliters of the trypsin solution was added to each plug and allowed to rehydrate the gel plugs on ice for 30 min and then incubated at 37 °C overnight. Digestion buffer was removed to a new 96-well plate, and 50 μl of 1% TFA in 50% ACN was added to the gel plugs and sonicated for 30 min. This extract was removed and combined with the digestion buffer and dried in a SpeedVac concentrator (Jouan, RC1022, Thermo Savant, Milford, MA) for 45 min. Peptides were then resuspended in 15 μl of 0.5% TFA in Milli-Q water. Peptides were solid phase-extracted (Millipore reverse phase ZipTipC18) according to the manufacturer's instructions. Samples were eluted into a 96-well plate with 4 μl of a 0.1% TFA, 50% ACN solution.
One microliter of the eluate was premixed with 2 μl of α-cyano-4-hydroxycinnamic acid matrix (3 mg/ml in 10 mm ammonium phosphate, 50% acetonitrile, 0.1% TFA) and spotted in duplicate on a MALDI target plate (Opti-TOF® 192-well insert, Applied Biosystems, Foster City, CA). MALDI-TOF MS and tandem TOF/TOF MS were performed on a Voyager 4700 instrument (Applied Biosystems). Thus, two peptide mass fingerprint (PMF) spectra per gel spot were generated from separate MALDI plate wells. The spectra were acquired in the reflector mode by averaging 3000 laser shots per spectrum (mass range, 800–4000 Da; focus mass, 2000 Da). Spectra were smoothed (Gaussian filter width, 9; target resolution at 1300 m/z, 20,000) for internal calibration to trypsin autolytic peptides (m/z 842.510, 1045.564, 1940.935, 2211.105, 2239.136, 2299.179, and 2807.300) and only peaks that exceeded a signal-to-noise ratio of 100 (local noise window, 200 m/z) and a half-maximal width of 2.9 bins were considered. A minimum of two monoisotopic trypsin peaks were required to calibrate each spectrum to a mass accuracy within 20 ppm. Failure to meet these criteria resulted in the application of the external plate calibration that was performed prior to each run and required matching of six standard peptide ion masses (m/z 904.468, 1296.685, 1570.677, 2093.087, 2465.199, and 3657.929) from six calibration spots (4700 Mass Standard kit, catalog number 4333604, Applied Biosystems). The laser power for PMF acquisition was adjusted to produce an average intensity of ∼7000 for the m/z 2093.087 standard ion (ACTH-(1–17)) across the six calibration spots prior to each run. Data-dependent MS/MS analyses, using PSD on one replicate PMF spectra set and CID on the other replicate, was performed on the 15 most abundant peptide ions (excluding trypsin autolysis ions) to generate amino acid sequence information. MS/MS spectra were integrated over 3000 laser shots in the 1-kV positive ion mode with the metastable suppressor turned on. Air at the medium gas pressure setting (1.25 × 10−6 torr) was used as the collision gas in the CID on mode. An internal calibration of MS/MS spectra was attempted on at least two ions of the immonium ion series and the y1 ions of arginine, lysine, and histidine (m/z: Arg immonium, 70.066, 87.081, 100.088, and 112.088; Arg y1, 175.119; Lys immonium, 84.081, 101.108, and 129.103; Lys y1, 147.113; His immonium, 110.072 and 138.067) or reverted to the external calibration, which was performed prior to each PSD or CID run on four fragmentation ions of Glu1-fibrinopeptide B (m/z: precursor, 1570.677; y1, 175.120; y4, 480.257; y6, 684.347; y9, 1056.475). The laser intensity for the MS/MS spectra acquisition was adjusted to an intensity of ∼4000 of the y9 ion (m/z 1056.475) prior to each run.
The Global Proteome Server (GPS) Explorer 3.5 build 321 software (Applied Biosystems) was used to extract peaks from raw spectra using the following settings: MS peak filtering: mass range, 800–4000 Da; minimum signal-to-noise ratio, 10; peak density filter, 50 peaks/200 Da; maximum number of peaks, 65; MS/MS peak filtering: mass range, 60–20 Da below precursor mass; minimum signal-to-noise ratio, 10; peak density filter, 50 peaks/200 Da; maximum number of peaks, 65. A combined MS peptide fingerprint and MS/MS peptide sequencing search was performed against the IPI D. rerio version 3.07 database (number of protein sequences, 45,388; number of amino acid residues, 23,104,717) using the MASCOT 2.1.04 search algorithm. These searches specified trypsin as the digestion enzyme and allowed for carbamidomethylation of cysteine, partial oxidation of methionine residues (all variable modifications), and one missed trypsin cleavage. The monoisotopic precursor ion tolerance was set to 50 ppm, and the MS/MS ion tolerance was set to 0.05 Da. The output was limited to the 10 best hits. MS/MS peptide spectra with a minimum ion score confidence interval ≥95% were accepted; this was equivalent to a median ion score cutoff of ∼27 in the data set. Protein identifications were accepted with a statistically significant MASCOT protein search score ≥65 that corresponded to an error probability of p < 0.01 in our data set. All possible protein identifications from replicate analyses that met the above criteria were reported for each gel spot. However, the protein identification with the highest score was selected in the case of redundant protein identifications.
The raw mass spectra were exported to mzXML using the PzMsXML script (Nathan Edwards, University of Maryland Center for Bioinformatics and Computational Biology, College Park, MD). Annotated PMF spectra were produced by combining the spectra file formats for raw and processed peaks, mzXML and Mascot generic format, respectively. Peak annotations and modification information for identified peptides was extracted from the result summary table (supplemental Table 4). The Ruby scripting language was used to parse these files, send the spectra and annotation information to the R statistical tool (The R Project for Statistical Computing) for plotting, and creation of the HTML result pages. Annotated spectra for the tandem mass spectrometry experiments were obtained by transforming the dynamic MASCOT Web pages into static content using Ruby and saved locally to the drive. Hyperlinks to both PMF and MS/MS pages are included in supplemental Table 4. These pages and the table are included in a compressed (zipped) file (Lucitt_Supplemental_Table_4.zip) that installs the table and correct subfolder structure for viewing the linked spectra when unpacked.
Protein Classification—
Proteins were classified using the Gene Ontology (GO) functional annotations for cellular component, molecular function, and biological process (35). Annotation categories were taken from level three in the GO trees. GO enrichment analysis was conducted by calculating for each category the probability that the number of annotations in the protein list could have arisen by chance, assuming an underlying hypergeometric distribution (36). Pathway analysis (Ingenuity Systems, Redwood City, CA) was used to search for enrichment of proteins in canonical and metabolic signaling pathways. IPI protein sequences were BLAST searched against the RefSeq human and mouse protein sequence databases, and BLAST results were used for mapping in the Ingenuity Systems pathway database.
Zebrafish Proteomics Database—
A database was constructed parsing IPI records, GenBank™ records, the NCBI taxonomy, and GO ontology into a BioSQL relational database schema. The schema was extended to include the experimental result data and key word searching capabilities as well as optimized for the Web application. The web site itself was constructed using the Ruby on Rails web application framework. The zebrafish proteomics database can be accessed on line (see supplemental data for the URL database link under Instructions for Downloading).
RESULTS
Zebrafish Embryonic Protein Expression Profiles—
We performed 2D LC-MS/MS experiments to generate an accurate repository of proteins reliably detectable in zebrafish embryos at 72 and 120 hpf. Replicate samples for each embryonic developmental stage were analyzed. The acquired raw mass spectra data sets for each replicate were matched to peptide sequences in the IPI D. rerio database using the search engines MASCOT and SEQUEST. This generated 327,906 possible peptide identifications. Peptide sequence identifications generated from both search algorithms for each replicate were combined in an integrated analysis using the EBP algorithm for protein assignment to augment sensitivity and error control (30).
This approach identified 1112 unique proteins at 72 hpf and 867 unique proteins at 120 hpf with false identification rates of less then 1% and sensitivities of 91.7 and 88.2% (Fig. 1 and supplemental Tables 1, 2, and 3). An additional 45 proteins at 72 hpf and 31 proteins at 120 hpf were as likely to be expressed but were indistinguishable from homologous proteins based on the peptide evidence. Eighty-six percent of the identified proteins at 72 hpf and 82% at 120 hpf were based on gene models derived from transcript or protein sequences specific for the zebrafish genome (Ensembl Assembly Zv7, April 2007 (13)). Hypothetical proteins or proteins predicted by comparison with other genomes constituted 13% of the detected proteins at 72 hpf and 17% at 120 hpf.
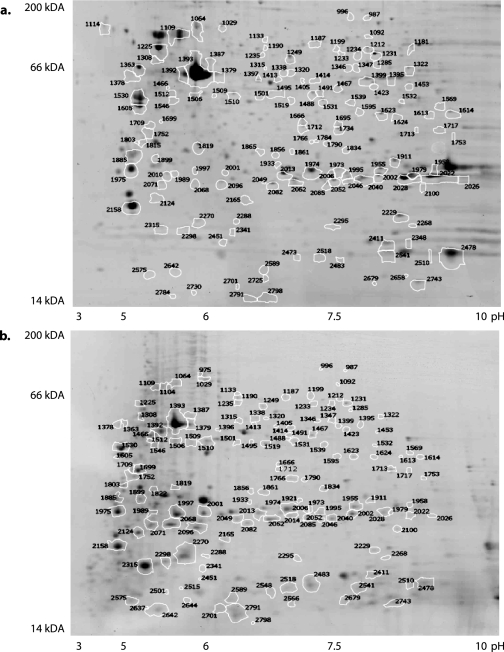
The separation of proteins at the peptide level by 2D LC-MS/MS may preclude discrimination of homologous proteins, such as distinct isoforms or modified forms of a protein. Gel-based proteomics techniques allow more readily the distinction of similar proteins based on their migration pattern in the electrical field. Thus, we ran protein samples from both developmental stages on 2D gels. In total, 348 unique proteins at 72 hpf and 317 unique proteins at 120 hpf were identified from 2D gels using MALDI-TOF/TOF tandem mass spectrometry with an error probability of less than 0.01 (Fig. 2 and supplemental Table 4). Approximately 85% of the detected proteins were annotated at the highest level of quality, and 15% were hypothetical or predicted proteins with similarity to sequences of other species (Ensembl Assembly Zv7, April 2007 (13)).
Fig. 2.
2D PAGE reference images of 72- and 120-hpf protein lysates. Annotated 2D gels for 72-hpf (a) and 120-hpf (b) zebrafish embryo proteins based on DIGE expression differences are shown.
Proteins with high quality annotations identified by either method included structural proteins (e.g. myosin, tubulin, actin, annexin, lamin B2, matrilin 4 precursor, septin 6, cofilin 2, and cytokeratin), heat shock proteins (e.g. HSP 70-kDa protein 5, HSP 8, HSP 9B, and HSC 70), molecular chaperone proteins (e.g. chaperonin containing TCP1 subunit 2, calreticulin-like protein, chaperone protein GP96, and retinoblastoma-binding protein 4), cell cycle proteins (e.g. cell division cycle gene CDC48 and prohibitin), and multiple forms of the yolk protein vitellogenin. Annotated proteins involved in organ functions included those specific to kidney (e.g. intraflagellar protein IFT81), skeletal muscle (e.g. creatine kinase), liver (e.g. basic fatty acid-binding protein and 6-phosphofructokinase), central nervous system (e.g. synaptosome-associated protein and brain-type fatty acid-binding protein), heart (e.g. ATPase 2A), and lens proteins (crystallin γN2 and βB1). Proteins regulating developmental processes included proteins such as β-catenins 1 and 2, staufen homolog 2, and kelch-like 1.
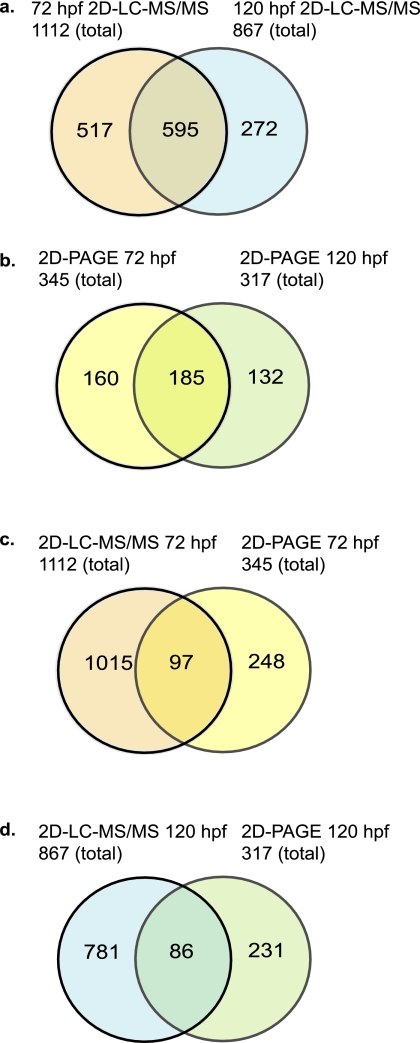
About 50% of all identified proteins were detected at both embryonic stages (Fig. 3). Interestingly a large fraction of proteins were exclusively identified by 2D PAGE (248 and 231 at 72 and 120 hpf, respectively) but not by 2D LC-MS/MS. Only a total of 97 proteins at 72 hpf and 86 at 120 hpf were detected by both 2D PAGE and 2D LC-MS/MS at either stage (Fig. 3). Thus, ∼70% of the proteins identified on gels were not detected by 2D LC-MS/MS. A potential explanation for this discrepancy is that the gel separation method may favor proteins that are not well digested in solution.
Fig. 3.
Comparison of proteins identified by 2D LC-MS/MS and from 2D PAGE. a and b, Venn diagram illustration of the overlap between proteins identified by 2D LC-MS/MS (a) or from 2D PAGE (b) at 72 and 120 hpf. c and d, overlap between 2D LC-MS/MS- and 2D PAGE-identified proteins at 72 hpf (c) and at 120 hpf (d).
Differential Protein Expression during Development—
Zebrafish embryonic development between the hatching period (72 hpf) and the larval stage (120 hpf) is characterized by rapid maturation of primal organs to form a viable organism. We assessed relative protein expression changes during this period by DIGE. Protein lysates of both stages were differentially fluorescently labeled, and an internal control was included to which protein spot intensities of both stages were normalized. A total of 148 of 789 resolved protein spots were in excess of 3-fold more abundant at 120 hpf than at 72 hpf. The expression intensity of 236 spots was below a third of that observed at 72 hpf. Sixty-one spots with a more than 3-fold increase and 103 spots with a more than 3-fold decrease in relative expression at 120 versus 72 hpf were identified with an error probability of less then 0.05 (Table I and supplemental Table 4).
Table I.
Protein spot identifications with 3-fold change or more in relative expression at 120 versus 72 hpf
h., heavy; l., light; mitochondr., mitochondrial; Eukar. transl. init. fact., eukaryotic translation initiation factor; Het., heterogeneous; contain., containing; prot., protein; APEX, apurinic/apyrimidinic endonuclease.
| Spota | Accessionb | Protein name | Scorec | Molecular weight | pI | ID Spec.d | Pep. count (n)e | Unmtch. (n)f | Cov.g | -Fold changeh |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| 1699 | IPI00503469 | Actin | 152 | 41947 | 5.22 | 590 | 11 | 61 | 40 | 24.1 |
| 2270 | IPI00551966 | Actin, α1, skeletal muscle | 90 | 41955 | 5.23 | 607 | 8 | 54 | 31 | 10.8 |
| 2124 | IPI00482295 | Actin, cytoplasmic 1 | 74 | 41739 | 5.3 | 580 | 6 | 71 | 18 | 10.3 |
| 1709 | IPI00503469 | Actin | 101 | 41947 | 5.22 | 601 | 7 | 54 | 23 | 10 |
| 2071 | IPI00503469 | Actin | 134 | 41947 | 5.22 | 593 | 12 | 57 | 36 | 8.5 |
| 2068 | IPI00503469 | Actin | 135 | 41947 | 5.22 | 587 | 10 | 56 | 35 | 7 |
| 1752 | IPI00503469 | Actin | 115 | 41947 | 5.22 | 571 | 10 | 64 | 38 | 5 |
| 2001 | IPI00503469 | Actin | 103 | 41947 | 5.22 | 606 | 9 | 53 | 32 | 4.7 |
| 2315 | IPI00503469 | Actin | 90 | 41947 | 5.22 | 568 | 7 | 64 | 21 | 4.6 |
| 2298 | IPI00503469 | Actin | 85 | 41947 | 5.22 | 608 | 7 | 52 | 21 | 4.4 |
| 2096 | IPI00503469 | Actin | 113 | 41947 | 5.22 | 564 | 7 | 69 | 27 | 4.2 |
| 1466 | IPI00503469 | Actin | 219 | 41947 | 5.22 | 569 | 12 | 60 | 41 | 3.3 |
| 2049 | IPI00503469 | Actin | 85 | 41947 | 5.22 | 577 | 9 | 66 | 32 | 3.1 |
| 1510 | IPI00551966 | Actin, α1, skeletal muscle | 103 | 41955 | 5.23 | 63 | 8 | 63 | 30 | −4.4 |
| 1392 | IPI00503469 | Actin | 89 | 41947 | 5.22 | 613 | 9 | 58 | 33 | −13.7 |
| 1379 | IPI00503469 | Actin | 102 | 41947 | 5.22 | 611 | 9 | 50 | 29 | −16.8 |
| 1393 | IPI00503469 | Actin | 222 | 41947 | 5.22 | 636 | 12 | 49 | 44 | −19.2 |
| 1919 | IPI00483287 | Fast skeletal myosin h. chain 3 | 94 | 51813 | 5.49 | 583 | 13 | 50 | 29 | 27.3 |
| 1713 | IPI00497758 | Fast myosin heavy chain 4 | 74 | 222067 | 5.54 | 566 | 12 | 40 | 9 | 14.5 |
| 1861 | IPI00483287 | Fast skeletal myosin h. chain 3 | 91 | 51813 | 5.49 | 572 | 13 | 63 | 31 | 11.6 |
| 1834 | IPI00483287 | Fast skeletal myosin h. chain 3 | 88 | 51813 | 5.49 | 603 | 14 | 58 | 31 | 7 |
| 1766 | IPI00497758 | Fast myosin heavy chain 4 | 76 | 222067 | 5.54 | 582 | 20 | 55 | 11 | 6.4 |
| 1803 | IPI00509014 | α-Tropomyosin | 226 | 32720 | 4.7 | 592 | 18 | 58 | 47 | 5.3 |
| 1822 | IPI00497758 | Fast myosin heavy chain 4 | 65 | 222067 | 5.54 | 610 | 18 | 57 | 12 | 3.7 |
| 1605 | IPI00509014 | α-Tropomyosin | 241 | 32720 | 4.7 | 117 | 20 | 51 | 49 | 3.3 |
| 1338 | IPI00483287 | Fast skeletal myosin h. chain 3 | 79 | 51813 | 5.49 | 638 | 13 | 58 | 29 | −3.2 |
| 1519 | IPI00497758 | Fast myosin heavy chain 4 | 165 | 222067 | 5.54 | 674 | 19 | 23 | 10 | −3.3 |
| 1975 | IPI00499941 | Fast skeletal myosin l. chain 1a | 138 | 20918 | 4.63 | 668 | 10 | 54 | 61 | −3.4 |
| 1467 | IPI00497758 | Fast myosin heavy chain 4 | 139 | 222067 | 5.54 | 657 | 21 | 33 | 10 | −3.4 |
| 1495 | IPI00483287 | Fast skeletal myosin h. chain 3 | 105 | 51813 | 5.49 | 666 | 14 | 41 | 30 | −3.7 |
| 1595 | IPI00483287 | Fast skeletal myosin h. chain 3 | 74 | 51813 | 5.49 | 667 | 9 | 23 | 19 | −4.8 |
| 1530 | IPI00509014 | α-Tropomyosin | 272 | 32720 | 4.7 | 81 | 21 | 53 | 53 | −4.8 |
| 1885 | IPI00509014 | α-Tropomyosin | 79 | 32720 | 4.7 | 663 | 11 | 57 | 31 | −6.3 |
| 1414 | IPI00497758 | Fast myosin heavy chain 4 | 71 | 222067 | 5.54 | 640 | 14 | 23 | 6 | −6.6 |
| 1491 | IPI00497758 | Fast myosin heavy chain 4 | 92 | 222067 | 5.54 | 664 | 20 | 28 | 10 | −7.6 |
| 1235 | PI00483287 | Fast skeletal myosin h. chain 3 | 117 | 51813 | 5.49 | 622 | 15 | 40 | 32 | −8.3 |
| 2158 | IPI00488085 | Myosin light chain 2 | 216 | 18853 | 4.68 | 654 | 12 | 45 | 69 | −8.6 |
| 1531 | IPI00497758 | Fast myosin heavy chain 4 | 92 | 222067 | 5.54 | 659 | 16 | 26 | 7 | −9 |
| 1109 | IPI00497758 | Fast myosin heavy chain 4 | 100 | 222067 | 5.54 | 649 | 27 | 43 | 17 | −11.1 |
| 1029 | IPI00497758 | Fast myosin heavy chain 4 | 131 | 222067 | 5.54 | 615 | 22 | 48 | 10 | −11.2 |
| 1392 | IPI00483287 | Fast skeletal myosin h. chain 3 | 184 | 51813 | 5.49 | 613 | 7 | 61 | 23 | −13.7 |
| 1133 | IPI00483287 | Fast skeletal myosin h. chain 3 | 103 | 51813 | 5.49 | 643 | 15 | 52 | 31 | −14.2 |
| 1104 | IPI00497758 | Fast myosin heavy chain 4 | 79 | 222067 | 5.54 | 629 | 22 | 52 | 14 | −15.4 |
| 1405 | IPI00500057 | Myosin, heavy polypeptide 2 | 66 | 221742 | 5.55 | 626 | 10 | 45 | 31 | −24.5 |
| 2022 | IPI00607465 | Vg1 protein | 276 | 36409 | 9.23 | 46 | 11 | 61 | 36 | −44.4 |
| 2026 | IPI00607465 | Vg1 protein | 179 | 36409 | 9.23 | 55 | 11 | 61 | 37 | −82.4 |
| 2028 | IPI00607465 | Vg1 protein | 264 | 36409 | 9.23 | 67 | 12 | 61 | 37 | −40.5 |
| 2040 | IPI00607465 | Vg1 protein | 225 | 36409 | 9.23 | 60 | 11 | 61 | 35 | −7.3 |
| 2046 | IPI00607465 | Vg1 protein | 161 | 36409 | 9.23 | 78 | 10 | 57 | 32 | −6.7 |
| 1539 | IPI00496717 | Vitellogenin 1 | 68 | 149452 | 8.68 | 64 | 11 | 50 | 10 | −10.6 |
| 1666 | IPI00513217 | Vitellogenin 1 | 88 | 21118 | 9 | 75 | 7 | 53 | 40 | −16.1 |
| 1695 | IPI00513217 | Vitellogenin 1 | 65 | 21118 | 9 | 34 | 8 | 54 | 52 | −5.2 |
| 1712 | IPI00513217 | Vitellogenin 1 | 130 | 21118 | 9 | 36 | 7 | 56 | 39 | −14.9 |
| 1734 | IPI00513217 | Vitellogenin 1 | 84 | 21118 | 9 | 50 | 6 | 59 | 32 | −192.6 |
| % | ||||||||||
| 2022 | IPI00496717 | Vitellogenin 1 | 240 | 149452 | 8.68 | 46 | 14 | 56 | 11 | −44.4 |
| 2026 | IPI00496717 | Vitellogenin 1 | 155 | 149452 | 8.68 | 55 | 16 | 56 | 14 | −82.4 |
| 2028 | IPI00496717 | Vitellogenin 1 | 228 | 149452 | 8.68 | 67 | 16 | 55 | 12 | −40.5 |
| 2040 | IPI00496717 | Vitellogenin 1 | 192 | 149452 | 8.68 | 60 | 15 | 56 | 12 | −7.3 |
| 2046 | IPI00496717 | Vitellogenin 1 | 148 | 149452 | 8.68 | 78 | 16 | 51 | 15 | −6.7 |
| 1624 | IPI00507087 | Muscle creatine kinase | 75 | 42797 | 6.32 | 65 | 9 | 55 | 27 | −5.7 |
| 1532 | IPI00495855 | l-Lactate dehydrogenase B | 130 | 36089 | 6.43 | 48 | 11 | 39 | 36 | −10.5 |
| 1423 | IPI00490850 | Aldolase c | 193 | 39234 | 6.21 | 54 | 16 | 55 | 55 | −34.1 |
| 1322 | IPI00485952 | Creatine kinase, mitochondr. 2 | 87 | 46265 | 6.49 | 19 | 9 | 33 | 24 | −24.1 |
| 1231 | IPI00490877 | Eno3 protein | 118 | 47402 | 6.2 | 20 | 13 | 57 | 45 | −50.3 |
| 1363 | IPI00508284 | Ribosomal protein SA | 74 | 33990 | 4.75 | 30 | 5 | 36 | 30 | −12.9 |
| 1285 | IPI00501593 | RNA-binding protein 4 | 100 | 46105 | 6.81 | 18 | 10 | 16 | 29 | −18 |
| 1285 | IPI00615024 | RNA binding motif protein 4 | 76 | 46089 | 6.81 | 17 | 8 | 16 | 23 | −18 |
| 1501 | IPI00496845 | Eukar. transl. init. fact. 3 | 87 | 38572 | 5.53 | 73 | 11 | 44 | 40 | −4 |
| 1495 | IPI00491050 | Het. nuclear ribonucleoprotein | 78 | 37087 | 5.76 | 35 | 7 | 51 | 22 | −3.7 |
| 1819 | IPI00480889 | Prohibitin | 126 | 29666 | 5.28 | 37 | 9 | 60 | 38 | −5 |
| 1029 | IPI00498630 | Chaperonin contain. TCP1 S5 | 125 | 59925 | 5.33 | 26 | 17 | 50 | 22 | −11.2 |
| 1064 | IPI00508003 | Hspd1 protein | 90 | 61157 | 5.56 | 32 | 12 | 30 | 29 | −10.4 |
| 2062 | IPI00495773 | Crystallin, γN2 | 194 | 21730 | 5.86 | 660 | 11 | 47 | 62 | −18.6 |
| 1921 | IPI00502990 | Crystallin, βB1 | 183 | 26797 | 6.44 | 52 | 10 | 58 | 46 | −4 |
| 2002 | IPI00490966 | βA4-Crystallin | 216 | 23013 | 6.25 | 40 | 13 | 58 | 85 | −8.4 |
| 1979 | IPI00504818 | βA1–2-Crystallin | 83 | 24521 | 6.4 | 77 | 9 | 62 | 67 | −6.4 |
| 2478 | IPI00513361 | Novel β-type globin | 111 | 11951 | 6.81 | 95 | 7 | 70 | 77 | −3.2 |
| 2478 | IPI00502256 | Similar to embryonic 1 | 124 | 16151 | 6.89 | 95 | 8 | 68 | 63 | −3.2 |
| 1531 | IPI00498781 | Similar to vertebrate APEX | 97 | 34874 | 5.77 | 68 | 9 | 65 | 32 | −9 |
| 2743 | IPI00510181 | Ubiquitin ribosomal prot. S27a | 71 | 17987 | 9.68 | 89 | 5 | 37 | 30 | 9.3 |
| 2743 | IPI00619743 | Ubiquitin C | 117 | 26486 | 7.85 | 89 | 5 | 37 | 40 | 9.3 |
| 2010 | IPI00483436 | NADH dehydrogenase | 66 | 23687 | 5.74 | 104 | 8 | 56 | 38 | 5.3 |
Gel spot number (also reference to supplemental Table 4).
IPI accession number.
MASCOT protein score.
Mass spectrum ID (as reference to view the annotated mass spectrum in supplemental Table 4).
Number of unique peptides matched to mass peaks.
Number of unmatched mass peaks.
Sequence coverage in percent.
-Fold change of normalized spot volume between embryonic stage (120 vs. 72 hpf).
Annotated proteins included primarily structural protein isoforms, which were resolved in multiple spots, such as actin, myosin, tropomyosin, and tubulin isoforms. For example, actin isoforms were resolved in 17 spots (13 increased and four decreased at 120 hpf); myosins were identified in 27 spots (eight increased and 19 decreased at 120 hpf) representing multiple distinct isoforms and post-translationally modified or partially truncated variants. Vitellogenin (IPI00496717 and IPI00607465) was identified in 10 spots, also indicative of the existence of multiple modified variants. As expected, this yolk protein was decreased at 120 hpf as it is consumed for energy and protein production during development.
Proteins involved in energy production and metabolism (muscle-specific creatine kinase, IPI00507087; l-lactate dehydrogenase B chain, IPI00495855; aldolase c fructose-bisphosphate, IPI00490850; creatine kinase mitochondrial 2, IPI00485952; and enolase 3 protein, IPI00490877) were between 5- and 50-fold less abundant at 120 hpf than at 72 hpf. Transcription/translation proteins (ribosomal protein SA, IPI00508284; RNA binding motif protein 4, IPI00615024; eukaryotic translation initiation factor 3, IPI00496845; and heterogeneous nuclear ribonucleoprotein that binds to nascent RNA polymerase II transcripts and plays a role in both transcript-specific packaging and alternative splicing of pre-mRNAs, IPI00491050) were 4–18-fold more abundant at 72 hpf than at 120 hpf, consistent with the faster synthesis of cellular proteins during organismal growth at the earlier developmental stage. Similarly prohibitin (IPI00480889), chaperonin containing TCP1 subunit 5 (IPI00498630), and heat shock protein Hspd1 (IPI00508003), which are all involved in cell cycle control, were more abundant (5–10-fold) at 72 hpf than at 120 hpf. All four lens proteins (crystallin γN2, IPI00495773; β-crystallin B1, IPI00502990; β-crystallin A4, IPI00490966; and β-crystallin A1–2, IPI00504818) were more prominent (6–19-fold) at the earlier stage relative to total protein, consistent with the relatively larger volume of the eyes in comparison with the whole organism at this stage. Three embryonic proteins, novel β-type globin (IPI00513361), novel protein similar to embryonic 1 (IPI00502256), and novel protein similar to vertebrate apurinic/apyrimidinic endonuclease (APEX) (IPI00498781), were also decreased at 120 hpf (3–9-fold).
Apart from the structural proteins, several proteins that were more abundant at 120 hpf than at 72 hpf fell into the hypothetical/predicted or unknown categories. Other up-regulated proteins were ubiquitin C (IPI00619743; 9-fold) and ribosomal protein S27a (IPI00510181; 9-fold), which are involved in targeting cellular proteins for degradation (Table I).
Pathway Membership of Detected Proteins—
We sought to categorize protein identifications using a pathway enrichment analysis based on a database that describes signaling pathways and network relationship (Ingenuity Systems). As this resource does not include D. rerio sequences, we performed a BLASTP search of all protein identifications against the RefSeq human and mouse databases. This resulted in 731 RefSeq cross-references for 120 hpf (83% of total identifications from IPI) and 883 for 72 hpf (80% of total identification from IPI). The network and pathway database contained functional annotations for 461 proteins at 120 hpf (55% of original protein identifications) and 561 proteins at 72 hpf (51% of original identifications). These were analyzed for their membership in collated canonical and metabolic signaling pathways. A total of 163 proteins for each time point, 120 and 72 hpf, were mapped to a canonical signaling pathway. Metabolic pathway information existed for 366 proteins for 120 hpf and 397 proteins for 72 hpf (supplemental Tables 5 and 6).
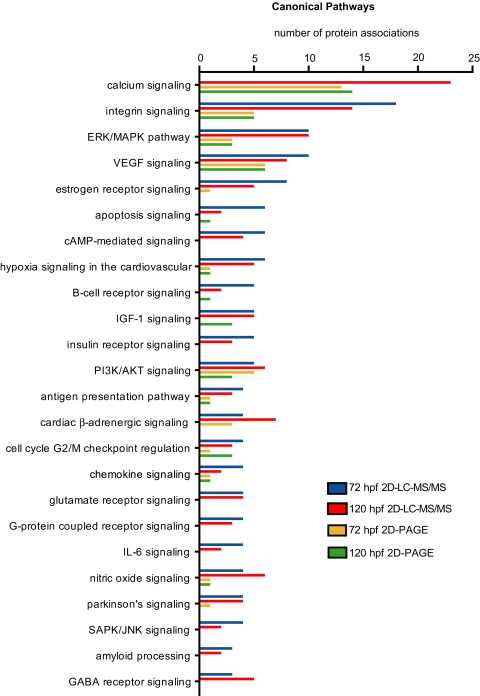
The distribution of proteins mapped to the canonical pathways is illustrated in Fig. 4. Pathways with the most contributing proteins were related to calcium, integrin, extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase, and vascular endothelial growth factor signaling. Proteins associated with morphogenesis such as the WNT/β-catenin pathway were less prominent but present at both 120 and 72 hpf (not shown). Indeed the developmental stages were relatively similar in their functional associations with the notable exception of the calcium signaling pathway, which was detected at 120 hpf (23 proteins) but was absent at 72 hpf.
Fig. 4.
Pathway analysis. The number of proteins associated with canonical signaling pathways as defined by Ingenuity Pathway analysis is shown. Only pathways with five or more protein associations are shown. ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol 3-kinase; VEGF, vascular endothelial growth factor; SAPK, stress-associated protein kinase; JNK, c-Jun NH2-terminal kinase; GABA, γ-aminobutyric acid.
Proteins associated with metabolic signaling pathways are represented in supplemental Fig. 1. Again the numbers of proteins classified as pathway members were similar for both embryo stages. More proteins identified at 72 hpf were grouped into the two metabolic pathways oxidative phosphorylation and ubiquinone biosynthesis than at the 120-hpf stage (12 and eight more proteins, respectively).
BLASTP analysis of protein sequences identified from 2D PAGE resulted in 235 RefSeq cross-references for 120 hpf (73%) and 262 cross-references for 72 hpf (75%). A total of 86 proteins for 120 hpf (26%) and 96 proteins at 72 hpf (27%) were annotated with canonical and signaling pathway information in the Ingenuity Systems pathway database. Fewer pathways were detected compared with 2D LC-MS/MS identifications. However, the pathway profile was again similar between the stages (Fig. 4 and supplemental Fig. 1).
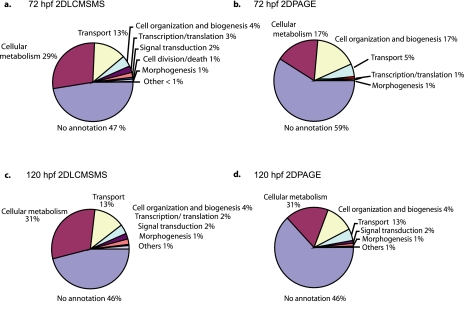
A second approach to the functional analysis of the embryonic zebrafish proteins was based on GO annotations. Annotated proteins were categorized into the broad GO classes biological process, molecular function, and cellular component. A graphical representation of these categories for each embryonic stage is shown in Fig. 5 and supplemental Fig. 2 for 72 hpf and supplemental Fig. 3 for 120 hpf. The protein identifications at both 120 and 72 hpf were categorized similarly using GO. Approximately 30% of proteins had GO annotation to cellular metabolism, 13% had GO annotation to transport, 4% had GO annotation to cell organization and biogenesis, and 2% had GO annotation to translation/transcription and signal transduction. About 1% of proteins were annotated with functions relating to morphogenesis, cell differentiation, and development. Structural molecule activity, a category that is often over-represented in proteomics analyses, was associated with 8% of the proteins at 120 hpf and 6% at 72 hpf. Enzyme inhibitor activity, signal transducer activity, and motor activity all had 1% or less associated proteins at both 120 and 72 hpf. Cellular component information was unavailable for 60% of proteins. Association to organelles was ∼20%, association to intracellular localization was 10%, and association to membrane localization was 8% at both stages.
Fig. 5.
Gene Ontology classification. Gene Ontology biological process classification of proteins identified at 72 and 120 hpf from 2D LC-MS/MS and 2D PAGE is shown.
Some of the GO categories were more frequently represented than expected from a random distribution. Such enrichment of functional groups points to protein classes that are either specifically expressed in embryos of the selected developmental stages or preferentially detected by the selected methodologies. Enriched GO classes are shown in supplemental Tables 7 and 8. GO categories with the most significant enrichment at 72 hpf were “development” (p = 0.009), “cellular metabolism” (p = 0.022), and “cell death” (p = 0.027). Cell differentiation, cell organization, biogenesis, and transport were less significantly enriched (0.05 > p > 0.027). GO categories significantly enriched at 120 hpf were cellular metabolism (p = 0.007), cell death (p = 0.021), and “growth” (p = 0.021). Others enriched with 0.05 > p > 0.025 were development, “transport,” “cell organization,” “biogenesis,” and “signal transduction.” In the GO category “molecular function,” “catalytic activity” (GO:0003824; p = 0.005) and “binding” (GO:0005488; p = 0.018) were most prominent at 120 hpf. Both “intracellular complex” (GO:0005622; p = 0.005 72 hpf) and “protein complex” (GO:0043234; p = 0.028) were most enriched at 72 hpf.
Zebrafish Proteomics Database—
A relational database was constructed by combination of IPI, GenBank, the NCBI taxonomy, GO assignments, and the experimental data. A Web application interface was developed for user-friendly ad hoc queries of the sequence annotation as well as perusal of the experimental and data mining results. The zebrafish proteomics database is available on line. The source code and associated database are also available for download at the site (see supplemental data for the URL database link under Instructions for Downloading).
DISCUSSION
Sequencing of the genome has fostered various initiatives to catalog the proteome of genetic model organisms such as yeast (37), Caenorhabditis elegans (38), Drosophila (39, 40), and the mouse (41). Similarly a collaborative program has been established to explore systematically the human proteome (42, 43). Such initiatives afford the opportunity to refine gene models created during the genome annotation process, recognize genes that may give rise to multiple proteins by means of alternative splicing or post-translational modification, assess tissue- and/or process-specific protein regulation, and avail a resource for the selection of specific proteins such as candidate disease biomarkers. The zebrafish has emerged as a tractable model organism for high throughput screening in vertebrate biology and genetics (6) due to its large clutch size, external development, transparency during development, and ease of husbandry. More recently, small chemical compound screens targeting organ systems, such as the cardiovascular system (44), central nervous system (45), and the blood-forming organs (46), have illustrated the utility of zebrafish in drug development. Its genome has been sequenced, and about 40% of its genes have been comprehensively annotated (Vertebrate Genome Annotation (VEGA) 49).
We used a bimodal strategy, shotgun proteomics and 2D PAGE, to analyze expressed zebrafish proteins during two advanced stages of development. The approach was informed by a number of considerations including (i) the selection of developmental stages that will likely be screened for dysregulated protein expression in large scale mutagenesis or chemical screens, (ii) the simplicity of sample preparation and analytical methodology (mindful of their possible adaptation to high throughput screening), (iii) the eminent quality of mass spectrometric protein identifications, (iv) the accessibility of the protein data in a fully searchable database and their integration with existing genomics and genetics resources provided to the zebrafish research community through the ZFIN database (32), and (v) the comprehensive categorization of proteins by functional classes to facilitate the selection of candidate proteins, for example for the design of targeted quantitative mass spectrometry assays (29). Our approach yielded 1384 unique proteins at 72 or 120 hpf by 2D LC-ESI-MS/MS and 477 unique proteins at 72 or 120 hpf by 2D PAGE-MALDI-TOF/TOF, which showed an overlap of about 30%. More unique proteins were identified by LC-MS/MS at 72 hpf (1112 proteins) than at 120 hpf (867 proteins), although equal amounts of protein were analyzed. While the precise cause for this disparity remains unknown, this may reflect developmental differences in the complexity of the protein samples favoring a larger number of high confidence identifications in the less developed embryo.
Shotgun proteomics studies produce hundreds of thousands of mass spectra derived from fragmented peptide ions that include the amino acid sequence information. These sequences are typically inferred automatically by matching the fragmentation ion spectra to theoretical or empirical spectra in peptide sequence databases, a process that may result in the generation of large numbers of false identifications (31) even if the error rate is small. Abundant proteins are generally detected with a high degree of confidence, whereas many lower abundance protein identifications have low reproducibility (31). Combining multiple biological and/or technical replicates improves markedly the sensitivity of protein identifications from MS/MS data, and both sensitivity and specificity are enhanced further by a combined spectra analysis with complementary database search algorithms, such as SEQUEST plus MASCOT (30, 47). These algorithms use distinct assessments of the plausibility of the inferred peptide sequences. SEQUEST uses heuristic metrics of the fit between the measured and theoretical spectra; MASCOT estimates a probability that the fragment ions in the spectrum could be generated by chance from sequences in the database. Thus, we integrated data from both biological replicates and such “orthogonal” dual data base searches to maximize the confidence in the protein identifications. We used an algorithm, EBP, that estimates accurately sensitivity and the false identification rate for complex protein samples and uses a function to combine data from replicate samples and multiple database search algorithms (30). This strategy identified from 327,906 potential peptide sequence calls proteins at both developmental stages with false identification rates of less than 1%.
Roughly 10% of the identified proteins were hypothetical, predicted, or unannotated proteins (based on Ensembl Assembly Zv7, April 2007 (13)). Thus, our analysis provides evidence in support of high quality annotations for numerous additional zebrafish genes. Comprehensive annotation of protein-coding genes remains challenging (48). Indeed most annotation pipelines, including the automated Ensembl pipeline (13) and the manual Vertebrate Genome Annotation (VEGA) pipeline (49) used for zebrafish, require confirmation of computationally predicted genes by independent evidence and/or manual validation for highest quality annotation. The additional evidence can take the form of experimentally documented transcription within the species (such as expressed sequence tags) or conservation across distant organisms. Indeed computational gene finding increasingly incorporates cross-species homology between closely related genomes to produce improved gene models (50). However, this evidence may not be sufficient as conservation across species is not limited to protein-coding regions (12). Similarly alternative splicing and overlapping genes present particularly complex annotation problems, and indeed, some estimates suggest that the majority of genes undergo alternative splicing (51, 52). Direct mass spectrometric identification of peptide sequences can resolve such ambiguities (40) as it generates an independent line of evidence at the translational level with error sources distinct from nucleotide-based approaches. Here we provided rigorous peptide level evidence for ∼1500 genes of the zebrafish genome. Given an estimated total number of zebrafish genes of about 22,000–23,000, our absolute coverage is in the range of 7%. As such peptide level information adds an important element to the repertoire of available annotation evidence (18) a larger collaborative effort, similar to the human and mouse proteome organizations, to enhance markedly the coverage of the zebrafish proteome seems warranted. Indeed one might expect that proteomics studies might accompany vertebrate genome annotation projects routinely in the future just like microbial genome annotation projects have been accelerated by proteomics investigations (14, 15).
The applicability of protein mass spectrometry in zebrafish has been explored in 2D-PAGE based investigations, which have detected small sets of zebrafish proteins mass spectrometrically with variable measures of error control (53–57). A more comprehensive shotgun proteomics investigation focused on the identification of protein in livers of adult fish as a resource for toxicological studies (58). Here we provide comprehensive 2D PAGE and shotgun proteomics identifications obtained from whole zebrafish embryos. A likely application of these data is the design of targeted quantitative mass spectrometry assays that might be used in mutagenesis or chemical screens. Targeted quantitative proteomics approaches are based on stable isotope dilution LC/multiple reaction monitoring-MS methods (59–64) and allow quantitation of compounds with high specificity and precision (29, 63). Although the sensitivity of immunoassays for protein quantitation still exceeds most mass spectrometry assays, stable isotope analogs normalize for selective losses of analytes as well as act as carriers for trace amounts of analytes subjected to complex isolation procedures (29). The development of such assays, however, requires detailed prior knowledge of (i) which proteins are expressed in the sample and are reliably detectable, (ii) which peptides are uniquely diagnostic for the targeted proteins, and (iii) under which experimental conditions they can be detected (i.e. in which strong cation exchange chromatography fractions do they elute). Our data set provides this information.
Analysis of known or predicted protein functions within the data set revealed a similar representation of protein classes relevant for cell function at both developmental stages, including proteins related to structure, transcription/translation, cell cycle, nucleotide metabolism, ion transport, carbohydrate, energy, and lipid metabolism. Proteins associated with organ systems such as central nervous system, heart, and skeletal muscle were represented in both stages. Analysis of relative expression changes revealed that proteins involved in energy production, transcription/translation, and cell cycle control were relatively more abundant at 72 hpf, consistent with the faster synthesis of cellular proteins during organismal growth at this time compared with 120 hpf. A large fraction, greater than 50% for both data sets, lacked functional information such as Gene Ontology classifications. More than 40 and 60% had no information relating to “molecular and biological function” and “cellular processes,” respectively. Thus, all protein assignments at both stages were aligned with sequences in the human or mouse RefSeq protein databases. This revealed alignment of 83% at 120 hpf and 80% at 72 hpf. However, these homologous sequences also had poor annotation in Ingenuity Pathway analysis. Thus, many of the identified proteins may represent candidates for the exploration of their protein functions.
Our large scale proteome analysis of embryonic zebrafish tissue revealed expression of previously uncharacterized proteins and detected developmentally regulated functional protein classes. The data are accessible on line in a fully searchable database that links protein identifications to existing resources including the ZFIN database (32). This new resource should allow the selection of candidate proteins for targeted quantitation (29) in mutagenesis and chemical screens and may refine systematic genetic network analysis in vertebrate development and biology.
Supplementary Material
Footnotes
Published, MCP Papers in Press, January 22, 2008, DOI 10.1074/mcp.M700382-MCP200
The abbreviations used are: ZFIN, Zebrafish Information Network; EBP, Empirical Bayes Protein Identifier; GO, Gene Ontology; hpf, hours postfertilization; HSP, heat shock protein; IPI, International Protein Index; 2D, two-dimensional; PMF, peptide mass fingerprint; ACTH, adrenocorticotropic hormone; BLAST, Basic Local Alignment Search Tool.
This work was supported, in whole or in part, by National Institutes of Health Grant HL 62250 (to G. A. F.). This work was also supported by the American Heart Association (National Scientist Development Grant 0430148N to T. G.) and the Higher Education Authority of Ireland (to M. B. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Raw mass spectral data have been submitted to the Proteomics Identifications (PRIDE) database (1) under accession numbers 2030–2045, 2047–2128, 2154–2353, 2383–2413.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Jones, P., Cote, R. G., Martens, L., Quinn, A. F., Taylor, C. F., Derache, W., Hermjakob, H., and Apweiler, R. ( 2006) PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 34, D659–D663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimmel, C. B. ( 1989) Genetics and early development of zebrafish. Trends Genet. 5, 283–288 [DOI] [PubMed] [Google Scholar]
- 3.Brownlie, A., Donovan, A., Pratt, S. J., Paw, B. H., Oates, A. C., Brugnara, C., Witkowska, H. E., Sassa, S., and Zon, L. I. ( 1998) Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat. Genet. 20, 244–250 [DOI] [PubMed] [Google Scholar]
- 4.Grosser, T., Yusuff, S., Cheskis, E., Pack, M. A., and FitzGerald, G. A. ( 2002) Developmental expression of functional cyclooxygenases in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 99, 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pini, B., Grosser, T., Lawson, J. A., Price, T. S., Pack, M. A., and FitzGerald, G. A. ( 2005) Prostaglandin E synthases in zebrafish. Arterioscler. Thromb. Vasc. Biol. 25, 315–320 [DOI] [PubMed] [Google Scholar]
- 6.Amsterdam, A., and Hopkins, N. ( 2006) Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 22, 473–478 [DOI] [PubMed] [Google Scholar]
- 7.Grunwald, D. J., and Streisinger, G. ( 1992) Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet. Res. 59, 103–116 [DOI] [PubMed] [Google Scholar]
- 8.Gaiano, N., Amsterdam, A., Kawakami, K., Allende, M., Becker, T., and Hopkins, N. ( 1996) Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature 383, 829–832 [DOI] [PubMed] [Google Scholar]
- 9.Wienholds, E., Schulte-Merker, S., Walderich, B., and Plasterk, R. H. ( 2002) Target-selected inactivation of the zebrafish rag1 gene. Science 297, 99–102 [DOI] [PubMed] [Google Scholar]
- 10.Jekosch, K. ( 2004) The zebrafish genome project: sequence analysis and annotation. Methods Cell Biol. 77, 225–239 [DOI] [PubMed] [Google Scholar]
- 11.Woods, I. G., Kelly, P. D., Chu, F., Ngo-Hazelett, P., Yan, Y. L., Huang, H., Postlethwait, J. H., and Talbot, W. S. ( 2000) A comparative map of the zebrafish genome. Genome Res. 10, 1903–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., Antonarakis, S. E., Attwood, J., Baertsch, R., Bailey, J., Barlow, K., Beck, S., Berry, E., Birren, B., Bloom, T., Bork, P., Botcherby, M., Bray, N., Brent, M. R., Brown, D. G., Brown, S. D., Bult, C., Burton, J., Butler, J., Campbell, R. D., Carninci, P., Cawley, S., Chiaromonte, F., Chinwalla, A. T., Church, D. M., Clamp, M., Clee, C., Collins, F. S., Cook, L. L., Copley, R. R., Coulson, A., Couronne, O., Cuff, J., Curwen, V., Cutts, T., Daly, M., David, R., Davies, J., Delehaunty, K. D., Deri, J., Dermitzakis, E. T., Dewey, C., Dickens, N. J., Diekhans, M., Dodge, S., Dubchak, I., Dunn, D. M., Eddy, S. R., Elnitski, L., Emes, R. D., Eswara, P., Eyras, E., Felsenfeld, A., Fewell, G. A., Flicek, P., Foley, K., Frankel, W. N., Fulton, L. A., Fulton, R. S., Furey, T. S., Gage, D., Gibbs, R. A., Glusman, G., Gnerre, S., Goldman, N., Goodstadt, L., Grafham, D., Graves, T. A., Green, E. D., Gregory, S., Guigo, R., Guyer, M., Hardison, R. C., Haussler, D., Hayashizaki, Y., Hillier, L. W., Hinrichs, A., Hlavina, W., Holzer, T., Hsu, F., Hua, A., Hubbard, T., Hunt, A., Jackson, I., Jaffe, D. B., Johnson, L. S., Jones, M., Jones, T. A., Joy, A., Kamal, M., Karlsson, E. K., Karolchik, D., Kasprzyk, A., Kawai, J., Keibler, E., Kells, C., Kent, W. J., Kirby, A., Kolbe, D. L., Korf, I., Kucherlapati, R. S., Kulbokas, E. J., Kulp, D., Landers, T., Leger, J. P., Leonard, S., Letunic, I., Levine, R., Li, J., Li, M., Lloyd, C., Lucas, S., Ma, B., Maglott, D. R., Mardis, E. R., Matthews, L., Mauceli, E., Mayer, J. H., McCarthy, M., McCombie, W. R., McLaren, S., McLay, K., McPherson, J. D., Meldrim, J., Meredith, B., Mesirov, J. P., Miller, W., Miner, T. L., Mongin, E., Montgomery, K. T., Morgan, M., Mott, R., Mullikin, J. C., Muzny, D. M., Nash, W. E., Nelson, J. O., Nhan, M. N., Nicol, R., Ning, Z., Nusbaum, C., O'Connor, M. J., Okazaki, Y., Oliver, K., Overton-Larty, E., Pachter, L., Parra, G., Pepin, K. H., Peterson, J., Pevzner, P., Plumb, R., Pohl, C. S., Poliakov, A., Ponce, T. C., Ponting, C. P., Potter, S., Quail, M., Reymond, A., Roe, B. A., Roskin, K. M., Rubin, E. M., Rust, A. G., Santos, R., Sapojnikov, V., Schultz, B., Schultz, J., Schwartz, M. S., Schwartz, S., Scott, C., Seaman, S., Searle, S., Sharpe, T., Sheridan, A., Shownkeen, R., Sims, S., Singer, J. B., Slater, G., Smit, A., Smith, D. R., Spencer, B., Stabenau, A., Stange-Thomann, N., Sugnet, C., Suyama, M., Tesler, G., Thompson, J., Torrents, D., Trevaskis, E., Tromp, J., Ucla, C., Ureta-Vidal, A., Vinson, J. P., Von Niederhausern, A. C., Wade, C. M., Wall, M., Weber, R. J., Weiss, R. B., Wendl, M. C., West, A. P., Wetterstrand, K., Wheeler, R., Whelan, S., Wierzbowski, J., Willey, D., Williams, S., Wilson, R. K., Winter, E., Worley, K. C., Wyman, D., Yang, S., Yang, S. P., Zdobnov, E. M., Zody, M. C., and Lander, E. S. ( 2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 [DOI] [PubMed] [Google Scholar]
- 13.Flicek, P., Aken, B. L., Beal, K., Ballester, B., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cunningham, F., Cutts, T., Down, T., Dyer, S. C., Eyre, T., Fitzgerald, S., Fernandez-Banet, J., Graf, S., Haider, S., Hammond, M., Holland, R., Howe, K. L., Howe, K., Johnson, N., Jenkinson, A., Kahari, A., Keefe, D., Kokocinski, F., Kulesha, E., Lawson, D., Longden, I., Megy, K., Meidl, P., Overduin, B., Parker, A., Pritchard, B., Prlic, A., Rice, S., Rios, D., Schuster, M., Sealy, I., Slater, G., Smedley, D., Spudich, G., Trevanion, S., Vilella, A. J., Vogel, J., White, S., Wood, M., Birney, E., Cox, T., Curwen, V., Durbin, R., Fernandez-Suarez, X. M., Herrero, J., Hubbard, T. J., Kasprzyk, A., Proctor, G., Smith, J., Ureta-Vidal, A., and Searle, S. ( 2008) Ensembl 2008. Nucleic Acids Res. 36, D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaffe, J. D., Berg, H. C., and Church, G. M. ( 2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 [DOI] [PubMed] [Google Scholar]
- 15.Jaffe, J. D., Stange-Thomann, N., Smith, C., DeCaprio, D., Fisher, S., Butler, J., Calvo, S., Elkins, T., FitzGerald, M. G., Hafez, N., Kodira, C. D., Major, J., Wang, S., Wilkinson, J., Nicol, R., Nusbaum, C., Birren, B., Berg, H. C., and Church, G. M. ( 2004) The complete genome and proteome of Mycoplasma mobile. Genome Res. 14, 1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalume, D. E., Peri, S., Reddy, R., Zhong, J., Okulate, M., Kumar, N., and Pandey, A. ( 2005) Genome annotation of Anopheles gambiae using mass spectrometry-derived data. BMC Genomics 6, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savidor, A., Donahoo, R. S., Hurtado-Gonzales, O., Verberkmoes, N. C., Shah, M. B., Lamour, K. H., and McDonald, W. H. ( 2006) Expressed peptide tags: an additional layer of data for genome annotation. J. Proteome Res. 5, 3048–3058 [DOI] [PubMed] [Google Scholar]
- 18.Tanner, S., Shen, Z., Ng, J., Florea, L., Guigo, R., Briggs, S. P., and Bafna, V. ( 2007) Improving gene annotation using peptide mass spectrometry. Genome Res. 17, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong, T. P., Rosenberg, M., Mohideen, M. A., Weinstein, B., and Fishman, M. C. ( 2000) gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science 287, 1820–1824 [DOI] [PubMed] [Google Scholar]
- 20.Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S., and Hopkins, N. ( 2004) A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093 [DOI] [PubMed] [Google Scholar]
- 21.Matthews, R. P., Plumb-Rudewiez, N., Lorent, K., Gissen, P., Johnson, C. A., Lemaigre, F., and Pack, M. ( 2005) Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development 132, 5295–5306 [DOI] [PubMed] [Google Scholar]
- 22.Wallace, K. N., Dolan, A. C., Seiler, C., Smith, E. M., Yusuff, S., Chaille-Arnold, L., Judson, B., Sierk, R., Yengo, C., Sweeney, H. L., and Pack, M. ( 2005) Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev. Cell 8, 717–726 [DOI] [PubMed] [Google Scholar]
- 23.Farber, S. A., Pack, M., Ho, S. Y., Johnson, I. D., Wagner, D. S., Dosch, R., Mullins, M. C., Hendrickson, H. S., Hendrickson, E. K., and Halpern, M. E. ( 2001) Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science 292, 1385–1388 [DOI] [PubMed] [Google Scholar]
- 24.Poss, K. D., Nechiporuk, A., Hillam, A. M., Johnson, S. L., and Keating, M. T. ( 2002) Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development 129, 5141–5149 [DOI] [PubMed] [Google Scholar]
- 25.Furthauer, M., Reifers, F., Brand, M., Thisse, B., and Thisse, C. ( 2001) sprouty4 acts in vivo as a feedback-induced antagonist of FGF signaling in zebrafish. Development 128, 2175–2186 [DOI] [PubMed] [Google Scholar]
- 26.Rolinski, B., Arnecke, R., Dame, T., Kreischer, J., Olgemoller, B., Wolf, E., Balling, R., Hrabe de Angelis, M., and Roscher, A. A. ( 2000) The biochemical metabolite screen in the Munich ENU Mouse Mutagenesis Project: determination of amino acids and acylcarnitines by tandem mass spectrometry. Mamm. Genome 11, 547–551 [DOI] [PubMed] [Google Scholar]
- 27.Wu, J. Y., Kao, H. J., Li, S. C., Stevens, R., Hillman, S., Millington, D., and Chen, Y. T. ( 2004) ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J. Clin. Investig. 113, 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss, A. W. ( 2004) Tandem mass spectrometry in discovery of disorders of the metabolome. J. Clin. Investig. 113, 354–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oe, T., Ackermann, B. L., Inoue, K., Berna, M. J., Garner, C. O., Gelfanova, V., Dean, R. A., Siemers, E. R., Holtzman, D. M., Farlow, M. R., and Blair, I. A. ( 2006) Quantitative analysis of amyloid β peptides in cerebrospinal fluid of Alzheimer's disease patients by immunoaffinity purification and stable isotope dilution liquid chromatography/negative electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20, 3723–3735 [DOI] [PubMed] [Google Scholar]
- 30.Price, T. S., Lucitt, M. B., Wu, W., Austin, D. J., Pizarro, A., Yocum, A. K., Blair, I. A., FitzGerald, G. A., and Grosser, T. ( 2007) EBP, a program for protein identification using multiple tandem mass spectrometry datasets. Mol. Cell. Proteomics 6, 527–536 [DOI] [PubMed] [Google Scholar]
- 31.States, D. J., Omenn, G. S., Blackwell, T. W., Fermin, D., Eng, J., Speicher, D. W., and Hanash, S. M. ( 2006) Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat. Biotechnol. 24, 333–338 [DOI] [PubMed] [Google Scholar]
- 32.Sprague, J., Bayraktaroglu, L., Clements, D., Conlin, T., Fashena, D., Frazer, K., Haendel, M., Howe, D. G., Mani, P., Ramachandran, S., Schaper, K., Segerdell, E., Song, P., Sprunger, B., Taylor, S., Van Slyke, C. E., and Westerfield, M. ( 2006) The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 34, D581–D585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westerfield, M. 1995. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), University of Oregon Press, Eugene, OR
- 34.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 35.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. ( 2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draghici, S., Khatri, P., Martins, R. P., Ostermeier, G. C., and Krawetz, S. A. ( 2003) Global functional profiling of gene expression. Genomics 81, 98–104 [DOI] [PubMed] [Google Scholar]
- 37.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. ( 2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 38.Gillette, W. K., Esposito, D., Frank, P. H., Zhou, M., Yu, L. R., Jozwik, C., Zhang, X., McGowan, B., Jacobowitz, D. M., Pollard, H. B., Hao, T., Hill, D. E., Vidal, M., Conrads, T. P., Veenstra, T. D., and Hartley, J. L. ( 2005) Pooled ORF expression technology (POET): using proteomics to screen pools of open reading frames for protein expression. Mol. Cell. Proteomics 4, 1647–1652 [DOI] [PubMed] [Google Scholar]
- 39.Beller, M., Riedel, D., Jansch, L., Dieterich, G., Wehland, J., Jackle, H., and Kuhnlein, R. P. ( 2006) Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics 5, 1082–1094 [DOI] [PubMed] [Google Scholar]
- 40.Brunner, E., Ahrens, C. H., Mohanty, S., Baetschmann, H., Loevenich, S., Potthast, F., Deutsch, E. W., Panse, C., de Lichtenberg, U., Rinner, O., Lee, H., Pedrioli, P. G., Malmstrom, J., Koehler, K., Schrimpf, S., Krijgsveld, J., Kregenow, F., Heck, A. J., Hafen, E., Schlapbach, R., and Aebersold, R. ( 2007) A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 25, 576–583 [DOI] [PubMed] [Google Scholar]
- 41.Cutillas, P., and Vanhaesebroeck, B. ( 2007) Quantitative profile of five murine core proteomes using label-free functional proteomics. Mol. Cell. Proteomics 6, 1560–1573 [DOI] [PubMed] [Google Scholar]
- 42.Omenn, G. S., States, D. J., Adamski, M., Blackwell, T. W., Menon, R., Hermjakob, H., Apweiler, R., Haab, B. B., Simpson, R. J., Eddes, J. S., Kapp, E. A., Moritz, R. L., Chan, D. W., Rai, A. J., Admon, A., Aebersold, R., Eng, J., Hancock, W. S., Hefta, S. A., Meyer, H., Paik, Y. K., Yoo, J. S., Ping, P., Pounds, J., Adkins, J., Qian, X., Wang, R., Wasinger, V., Wu, C. Y., Zhao, X., Zeng, R., Archakov, A., Tsugita, A., Beer, I., Pandey, A., Pisano, M., Andrews, P., Tammen, H., Speicher, D. W., and Hanash, S. M. ( 2005) Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics 5, 3226–3245 [DOI] [PubMed] [Google Scholar]
- 43.Hamacher, M., Apweiler, R., Arnold, G., Becker, A., Bluggel, M., Carrette, O., Colvis, C., Dunn, M. J., Frohlich, T., Fountoulakis, M., van Hall, A., Herberg, F., Ji, J., Kretzschmar, H., Lewczuk, P., Lubec, G., Marcus, K., Martens, L., Palacios Bustamante, N., Park, Y. M., Pennington, S. R., Robben, J., Stuhler, K., Reidegeld, K. A., Riederer, P., Rossier, J., Sanchez, J. C., Schrader, M., Stephan, C., Tagle, D., Thiele, H., Wang, J., Wiltfang, J., Yoo, J. S., Zhang, C., Klose, J., and Meyer, H. E. ( 2006) HUPO Brain Proteome Project: summary of the pilot phase and introduction of a comprehensive data reprocessing strategy. Proteomics 6, 4890–4898 [DOI] [PubMed] [Google Scholar]
- 44.Peterson, R. T., Shaw, S. Y., Peterson, T. A., Milan, D. J., Zhong, T. P., Schreiber, S. L., MacRae, C. A., and Fishman, M. C. ( 2004) Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 22, 595–599 [DOI] [PubMed] [Google Scholar]
- 45.Mendelsohn, B. A., Yin, C., Johnson, S. L., Wilm, T. P., Solnica-Krezel, L., and Gitlin, J. D. ( 2006) Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell Metab. 4, 155–162 [DOI] [PubMed] [Google Scholar]
- 46.North, T., Goessling, W., Walkley, C., Lengerke, C., Kopani, K., Lord, A., Weber, G., Jang, I., Grosser, T., FitzGerald, G. A., Daley, G., Orkin, S., and Zon, L. ( 2007) Prostaglandin E2 regulates vertebrate hematopoietic stem cell homeostasis. Nature 447, 1007–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deutsch, E. W., Eng, J. K., Zhang, H., King, N. L., Nesvizhskii, A. I., Lin, B., Lee, H., Yi, E. C., Ossola, R., and Aebersold, R. ( 2005) Human Plasma PeptideAtlas. Proteomics 5, 3497–3500 [DOI] [PubMed] [Google Scholar]
- 48.Mueller, M., Martens, L., and Apweiler, R. ( 2007) Annotating the human proteome: beyond establishing a parts list. Biochim. Biophys. Acta 1774, 175–191 [DOI] [PubMed] [Google Scholar]
- 49.Loveland, J. ( 2005) VEGA, the genome browser with a difference. Brief. Bioinformatics 6, 189–193 [DOI] [PubMed] [Google Scholar]
- 50.Korf, I., Flicek, P., Duan, D., and Brent, M. R. ( 2001) Integrating genomic homology into gene structure prediction. Bioinformatics 17, Suppl. 1, S140–S148 [DOI] [PubMed] [Google Scholar]
- 51.Mironov, A. A., Fickett, J. W., and Gelfand, M. S. ( 1999) Frequent alternative splicing of human genes. Genome Res. 9, 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Florea, L., Di Francesco, V., Miller, J., Turner, R., Yao, A., Harris, M., Walenz, B., Mobarry, C., Merkulov, G. V., Charlab, R., Dew, I., Deng, Z., Istrail, S., Li, P., and Sutton, G. ( 2005) Gene and alternative splicing annotation with AIR. Genome Res. 15, 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Damodaran, S., Dlugos, C. A., Wood, T. D., and Rabin, R. A. ( 2006) Effects of chronic ethanol administration on brain protein levels: a proteomic investigation using 2-D DIGE system. Eur. J. Pharmacol. 547, 75–82 [DOI] [PubMed] [Google Scholar]
- 54.Goishi, K., Shimizu, A., Najarro, G., Watanabe, S., Rogers, R., Zon, L. I., and Klagsbrun, M. ( 2006) αA-crystallin expression prevents γ-crystallin insolubility and cataract formation in the zebrafish cloche mutant lens. Development 133, 2585–2593 [DOI] [PubMed] [Google Scholar]
- 55.Knoll-Gellida, A., Andre, M., Gattegno, T., Forgue, J., Admon, A., and Babin, P. J. ( 2006) Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals. BMC Genomics 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Link, V., Carvalho, L., Castanon, I., Stockinger, P., Shevchenko, A., and Heisenberg, C. P. ( 2006) Identification of regulators of germ layer morphogenesis using proteomics in zebrafish. J. Cell Sci. 119, 2073–2083 [DOI] [PubMed] [Google Scholar]
- 57.Tay, T. L., Lin, Q., Seow, T. K., Tan, K. H., Hew, C. L., and Gong, Z. ( 2006) Proteomic analysis of protein profiles during early development of the zebrafish, Danio rerio. Proteomics 6, 3176–3188 [DOI] [PubMed] [Google Scholar]
- 58.Wang, N., Mackenzie, L., De Souza, A. G., Zhong, H., Goss, G., and Li, L. ( 2007) Proteome profile of cytosolic component of zebrafish liver generated by LC-ESI MS/MS combined with trypsin digestion and microwave-assisted acid hydrolysis. J. Proteome Res. 6, 263–272 [DOI] [PubMed] [Google Scholar]
- 59.Barr, J. R., Maggio, V. L., Patterson, D. G., Jr., Cooper, G. R., Henderson, L. O., Turner, W. E., Smith, S. J., Hannon, W. H., Needham, L. L., and Sampson, E. J. ( 1996) Isotope dilution-mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin. Chem. 42, 1676–1682 [PubMed] [Google Scholar]
- 60.Wu, S. L., Amato, H., Biringer, R., Choudhary, G., Shieh, P., and Hancock, W. S. ( 2002) Targeted proteomics of low-level proteins in human plasma by LC/MSn: using human growth hormone as a model system. J. Proteome Res. 1, 459–465 [DOI] [PubMed] [Google Scholar]
- 61.Kuhn, E., Wu, J., Karl, J., Liao, H., Zolg, W., and Guild, B. ( 2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 62.Beynon, R. J., Doherty, M. K., Pratt, J. M., and Gaskell, S. J. ( 2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2, 587–589 [DOI] [PubMed] [Google Scholar]
- 63.Anderson, L., and Hunter, C. L. ( 2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 64.Ciccimaro, E., Hevko, J., and Blair, I. A. ( 2006) Analysis of phosphorylation sites on focal adhesion kinase using nanospray liquid chromatography/multiple reaction monitoring mass spectrometry. Rapid Commun. Mass Spectrom. 20, 3681–3692 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.