Abstract
Facultative phototrophic bacterium Rhodobacter capsulatus DsbA-null mutants are proficient in photosynthesis but are defective in respiration especially in enriched growth medium at 35 °C. They also exhibit severe pleiotropic phenotypes extending from motility defects to osmofragility and oxidative stresses. In this work, using a combined proteomics and molecular genetics approach, we demonstrated that the respiratory defect of R. capsulatus DsbA-null mutants originates from the overproduction of the periplasmic protease DegP, which renders them temperature-sensitive for growth. The DsbA-null mutants reverted frequently to overcome this growth defect by decreasing, but not completely eliminating, their DegP activity. In agreement with these findings, we showed that overproduction of DegP abolishes the newly restored respiratory growth ability of the revertants in all growth media. Structural localizations of the reversion mutations in DegP revealed the regions and amino acids that are important for its protease-chaperone activity. Remarkably although R. capsulatus DsbA-null or DegP-null mutants were viable, DegP-null DsbA-null double mutants were lethal at all growth temperatures. This is unlike Escherichia coli, and it indicates that in the absence of DsbA some DegP activity is required for survival of R. capsulatus. Absence of a DegQ protease homologue in some bacteria together with major structural variations among the DegP homologues, including a critical disulfide bond-bearing region, correlates well with the differences seen between various species like R. capsulatus and E. coli. Our findings illustrate the occurrence of two related but distinct periplasmic protease families in bacterial species.
In both prokaryotes and eukaryotes, proteins that are targeted to the extracytoplasmic environments often require oxidative protein folding to attain their biologically active three-dimensional (3D)1 structures. Disulfide bond formation plays a key role in this fundamental cellular process. In the presence of molecular oxygen, disulfide bonds can occur spontaneously, but the non-catalyzed reaction is inefficient and error-prone (1), requiring enzymatic catalysis in vivo.
The disulfide bond-forming dithiol:disulfide oxidoreductases are ubiquitous in prokaryotes and eukaryotes. They possess a thioredoxin fold with a common structural motif (Cys-Xaa-Xaa-Cys) forming a catalytic disulfide bond at the N terminus of an α-helix (2). In eukaryotic cells, the major oxidative folding catalyst is the protein-disulfide isomerase (EC 5.3.4.1) located in the endoplasmic reticulum where it receives oxidizing equivalents from the membrane-bound endoplasmic reticulum oxidoreductase I (3–6). In prokaryotic cells the major disulfide bond catalyst is the dithiol:disulfide oxidoreductase DsbA (7, 8). In Gram-negative bacteria, it is located in the periplasm where most of the disulfide bond formation occurs. DsbA is maintained in its oxidized state by the inner membrane protein DsbB, which is functionally similar to the endoplasmic reticulum oxidoreductase I of eukaryotes (9, 10).
DsbA homologues have been found in many bacteria (11–15), including the purple, non-sulfur, facultative phototrophic bacterium Rhodobacter capsulatus (16), and disulfide bond formation has been studied extensively in Escherichia coli. Upon translocation of a polypeptide into the periplasm, DsbA rapidly and nonspecifically oxidizes pairs of cysteine thiols into disulfide bonds by transferring its active site disulfide (7, 17). In the absence of DsbA, oxidation of secreted proteins ceases, and unfolded or misfolded proteins accumulate. These inactive proteins are either repaired via the thioreductive pathways (18) or degraded by periplasmic proteases like DegP (19, 20) that are important for cell survival.
Use of global approaches indicated that many proteins are DsbA substrates in vivo (17, 21, 22). Consequently DsbA-null mutants exhibit highly pleiotropic phenotypes, extending from the lack of motility, osmofragility, and mucoid colony morphology to increased sensitivity to various molecules like reducing agents (e.g. dithiothreitol), antibiotics (e.g. benzylpenicillin), heavy metals and oxyanions (e.g. Hg2+, Cd2+, and TeO32−), biofilm formation, and reduced virulence (18). In an earlier study we found that, unlike in E. coli (23–25), DsbA-null mutants of R. capsulatus produce c-type cytochromes, indicating that DsbA is not essential for the cytochrome c maturation process (16). However, we observed that DsbA-null mutants of R. capsulatus are severely impaired for respiratory (Res) growth, especially in enriched growth medium (mineral-peptone-yeast extract (MPYE)), although they are proficient for photosynthetic (Ps) growth. The Res growth defect, which was not observed in E. coli, was surprising as R. capsulatus Res electron transport chain is branched with a cytochrome oxidase and a quinol oxidase acting as redundant terminal oxidases (26, 27). In addition, R. capsulatus DsbA-null mutants reverted frequently (at a frequency of roughly 10−6) bypassing the Res growth impairment (16). The broad scope of the phenotypes observed led us to undertake a combined proteomics and molecular genetics approach to investigate their molecular bases.
In the present study, we conducted a three-way comparative survey of the extracytoplasmic subproteome of R. capsulatus from the wild type, a DsbA-null mutant, and a revertant of a DsbA-null mutant using two-dimensional gel electrophoresis (2D-GE) coupled to mass spectrometric identifications. The data led us to the major extracytoplasmic stress response protein DegP (also called HtrA or protease Do), which was apparently the only protein highly overproduced in the DsbA mutant and which decreased back to the wild-type levels in a respiration-proficient DsbA revertant. This suggested that overproduction of DegP might be the culprit for the Res growth deficiency of the DsbA-null mutants of R. capsulatus, and we demonstrated this to be the case using molecular genetics approaches. Our data also indicated that the Res growth defect of the DsbA-null mutant could be aggravated or alleviated by decreasing or increasing the protease activity of DegP, respectively. Moreover R. capsulatus DsbA-null DegP-null double mutants were lethal, showing that in this species, which lacks a second DegQ-like periplasmic protease unlike E. coli, some DegP activity was necessary for cell survival in the absence of DsbA. These species differences correlated well with a DsbA-controlled disulfide bond-bearing domain, which is present in E. coli but absent in R. capsulatus DegP homologues, and illustrated the occurrence of two interrelated but distinct extracytoplasmic protease families in bacterial species.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions—
The bacterial strains and plasmids used in this work are described in Table I. E. coli strains were grown aerobically at 37 °C in Luria-Bertani broth (LB medium) supplemented with appropriate antibiotics, ampicillin, kanamycin, spectinomycin, or tetracycline at a final concentrations of 100, 50, 50, or 12.5 μg/ml, respectively. R. capsulatus strains were grown chemoheterotrophically at either 35 or 25 °C as appropriate either in Sistrom's minimal medium A (MedA; containing succinate and NH4+ as carbon and nitrogen sources, respectively) (28) or in enriched medium (MPYE; containing complex carbon and nitrogen sources provided by mineral-peptone-yeast extract) (29). Kanamycin or spectinomycin at a final concentration of 10 μg/ml was used as needed (30).
Table I.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Relevant phenotypea,b | Source or Ref. |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| HB101 | F− Δ(gpt-proA)62 leuB6supE44ara-14 galK2lacY1Δ(mcrC-mrr) rpsL20 xyl-5mtl-1 recA13 | StrR, rB− mB− | 31 |
| XL1-Blue | F′::Tn10 proA+B+lacIq Δ(lacZ)M15/recA1 endA1 gyrA96(NalR) thi hsdR17(rK− mK+) supE44 relA1 lac | Stratagene | |
| R. capsulatus | |||
| MT1131c | crtD121 RifR | Wild type, Nadi+ Ps+ | 86 |
| Y262 | GTA overproducer | 32 | |
| MD20 | Δ(dsbA::kan) | Res+ Ps+ on Med A Res− Ps+ on MPYE | 16 |
| ST20 | Δ(dsbA::spe) | Res+ Ps+ on Med A Res− Ps+ on MPYE | This work |
| MD20Ri (i = 1–12) | MD20 revertants selected on MPYE under respiratory growth conditions | Res+ Ps+ on MPYE | This work |
| OZL2 | Δ(degP::spe) | Res+ Ps+ on MPYE Res−/+ Psslow on MedA | This work |
| Plasmids | |||
| pRK2013 | tra+ (RK2) | KanR, helper | 87 |
| pRK415 | Broad host range vector | TetR | 35 |
| pBluescript | Cloning vector, pBluescript II KS(+) | AmpR | Stratagene |
| pHP45Ω-Spe | Ωspe in pHP45 vector | SpeR, AmpR | 33 |
| pTC4-1K | 4.2-kb XbaI-KpnI insert of the dsbA::kanallele cloned into the corresponding sites of pRK415 | TetR, KanR | 16 |
| pBS-degPWT | 2.5-kb XbaI and KpnI fragment of pRK-degPWT cloned into the corresponding sites of pBluescript | AmpR | This work |
| pRK-degPWT | 2.5-kb PCR product containing degP, with 500 bp up- and downstream, cloned into the XbaI-KpnI sites of pRK415 | TetR | This work |
| pOZL1 | The SpeR cartridge of pHP45ΩSpe inserted in the unique EcoRI site of pBS-degPWT | SpeR, AmpR | This work |
| pOZL2 | XbaI and KpnI fragment of pOZL1 cloned into sites of pRK415 | SpeR, TetR | This work |
| pBS-degPS234A | 2.5-kb XbaI and KpnI fragment of pRK-degPS234A cloned into the corresponding sites of pBluescript | AmpR | This work |
| pRK-degPS234A | S234A mutation of pRK-degPWT | TetR | This work |
| pRK-degPR3 | 2.5-kb PCR product containing degP-R3, with 500 bp up- and downstream, cloned into the XbaI-KpnI sites of pRK415 | TetR | This work |
| pRK-degPR9 | 2.5-kb PCR product containing degP-R9, with 500 bp up- and downstream, cloned into the XbaI-KpnI sites of pRK415 | TetR | This work |
MedA and MPYE refer to minimal and enriched growth media, respectively.
Ps and Nadi refer to photosynthetic growth and cytochrome c oxidase-dependent catalysis of α-naphthol to indophenol blue, respectively at 35 °C. Abbreviations of antibiotic resistances are as follows: Amp, ampicillin; Kan, kanamycin; Spe, spectinomycin; Tet, tetracycline. GTA, gene transfer agent.
R. capsulatusMT1131 is referred to as “wild type” because it is the wild type with respect to its growth properties. All R. capsulatusstrains, except Y262, are derivatives of MT1131, which was originally isolated as a green derivative of R. capsulatusSB1003 (86).
Molecular Genetics Techniques—
Standard molecular genetics techniques were performed according to Sambrook and Russell (31). Conjugal transfer of plasmids from E. coli to R. capsulatus and interposon mutagenesis via the gene transfer agent (32) using the spectinomycin resistance (SpeR) cassette from pHP45Ω (33) were performed as described earlier (34). A 2.5-kb DNA fragment containing the entire degP gene was PCR-amplified using wild-type chromosomal DNA isolated with the Qiagen DNeasy kit (Valencia, CA) from R. capsulatus strain MT1131 and the primers DegP-500u-F (5′-CTC TCT AGA GCC GAA GCT TAC GGC AAG GAT GAA A-3′) and DegP-500d-R (5′-GAG GGT ACC CAT GTG CTT GTC GAA CCC GAA GA-3′). These primers contained the XbaI and KpnI sites engineered at their 5′-ends, allowing the PCR products thus digested to be cloned into the respective sites of the plasmids pRK415 and pBSII to yield pRK-degPWT and pBS-degPWT, respectively. Using similar PCR experiments with chromosomal DNA isolated from the dsbA revertants MD20R3 and MD20R9 as templates, the degPR3 and degPR9 alleles of degP were also cloned into pRK415 to yield pRK-degPR3 and pRK-degPR9, respectively. For construction of pBS-degPS234A, the catalytic center serine 234 of degP was mutated to alanine using the QuikChange XL site-directed mutagenesis kit from Stratagene (La Jolla, CA) with pBS-degPWT DNA as a template and the mutagenic primers DegP-S234A-F (5′-CGA TCA ACC GCG GCA ACG CGG GCG GGC CCT TGT TC-3′) and DegP-S234A-R (5′-GAA CAA GGG CCC GCC CGC GTT GCC GCG GTT GAT CG-3′). The XbaI-KpnI fragment of pBS-degPS234A was then transferred into the respective sites of pRK415 to yield pRK-degPS234A. To create a deletion-insertion allele of degP, its 576-bp EcoRI fragment on pBS-degPWT was replaced with the 2-kb EcoRI fragment of pHP45ΩSpe carrying an SpeR cartridge (33), yielding plasmid pOZL1 (Table I). The 3.8-kb XbaI-KpnI fragment of pOZL1 with the degP::Spe allele was first cloned into the corresponding sites of pRK415 to yield pOZL2. As needed, appropriate plasmids were conjugated into the gene transfer agent over producer strain Y262 (32), and the desired alleles were introduced into appropriate strains selecting for antibiotic resistance as described earlier (34).
DNA Sequence Analysis—
All PCR products and plasmids pRK415 and pBSII derivatives were cleaned with the Qiagen PCR purification and plasmid isolation kits, respectively, and sequenced by using appropriate primers for verification. As needed, chromosomal DNA isolated from the desired R. capsulatus strains (MT1131, MD20, MD20R3, MD20R4, MD20R7, MD20R8, MD20R9, MD20R10, MD20R12, and OZL2) with the DNeasy kit was used as a template, and the degP locus was amplified using the primers DegP-500u-F and DegP-500d-R to generate a 2.5-kb PCR product and sequenced after purification by automated DNA sequencing with the Big-Dye terminator cycle sequencing kit (Applied Biosystems). The sequencing primers DegP-500u-F, DegP-23u-F (5′-GCT TTT ATA GAA GGC CAG CCG GAG CAC-3′), DegP-23d-F (5′-GAA GGG TGC CTT TCT CCG TTC AGC-3′), DegP-1000m-R (5′-CTG ATG ATG ACG TCG CCC GAT TTC AT-3′), and DegP-500d-R were used to define the degP mutations. DNA analyses were done by using MacVector (Accelrys, San Diego, CA).
Preparation of Periplasmic Protein Extracts Using Polymyxin B Sulfate—
Periplasmic extracts were prepared by using polymyxin B sulfate (36) with the following modifications. Bacterial cells were harvested from 1-liter overnight cultures by centrifugation at 4 °C (5000 × g for 20 min), washed with ice-cold 20 mm Tris-HCl, pH 8.0, and gently resuspended in cold extraction buffer consisting of 1 mg/ml polymyxin B sulfate, 20 mm Tris-HCl, 250 mm NaCl (pH 8.0) (5 ml/g of wet cells). The suspension was gently stirred for 1 h at 4 °C and centrifuged at 10,000 × g for 20 min at 4 °C. The supernatant was transferred into a clean tube, recentrifuged at 150,000 × g for 2 h at 4 °C, saved as the periplasmic fractions, and stored at −20 °C.
Sample Preparation and 2D-GE—
Sample preparation and 2D-GE were performed essentially as described earlier (37). Briefly periplasmic proteins were precipitated with TCA, acetone (20%, w/v); washed twice with ice-cold acetone to remove residual TCA; and dried under vacuum using a SpeedVac. Pellets were resuspended in 2D-GE sample solubilization buffer (8 m urea, 4% CHAPS, 40 mm Tris, 0.2% Bio-Lyte (pH 3–10), and 65 mm DTT) and mixed gently at room temperature until complete solubilization. Insoluble materials were removed by centrifugation at 25,000 × g at room temperature, and protein concentrations of the supernatants were determined by using the Bradford method (Bio-Rad).
For 2D-GE, samples containing 300 μg of solubilized proteins were applied to 18-cm, pH 4–7 IPG strips (Bio-Rad); following a 12-h passive rehydration, IEF was carried out by using PROTEAN IEF cell (Bio-Rad) at 20 °C at a maximum of 7000 V for 15–18 h; and the strips thus prepared were kept frozen at −20 °C until use. For the second dimension SDS-PAGE, IPG strips were reduced with 1% (w/v) DTT and alkylated with 2.5% (w/v) iodoacetamide at room temperature both prepared in equilibration buffer consisting of 50 mm Tris-HCl, pH 8.8, 6 m urea, 30% (v/v) glycerol, 2% SDS, and 0.02% bromphenol blue. After equilibration, the IPG strips were layered on top of the second dimension resolving gel slabs and overlaid with a solution of molten 0.5% agarose in SDS electrophoresis buffer. The second dimension Laemmli-type SDS-PAGE was carried out using 11% gels without any stacking (38) at 40 mA/gel in a PROTEAN II XL cell (Bio-Rad), and gels were stained with colloidal Coomassie Brilliant Blue (39).
2D Gel Image Analyses—
Following staining, gels were digitized with SilverFast scan software (Epson Corporate, Long Beach, CA) using a flatbed scanner (Epson Expression 1680), and the PDQuest 2D gel analysis software (version 7.1.0, Bio-Rad) was used to analyze gel images. At least three independently run gels using periplasmic fractions prepared from appropriate strains were used for data collection. Following automatic detection mode, spots were manually edited to exclude those that were not present on all replica gels. For quasiquantitative comparisons, protein spots observed in appropriate strains were normalized for the total density of each gel after calibration by manual indication of the lowest and highest density spots. Normalized density values were then used for comparisons, and spots exhibiting at least 3-fold increases or decreases were retained for further studies. Database construction and annotation were carried out using PDQuest.
Sample Preparation and Mass Spectrometry Analyses—
Protein spots were manually excised from the gels and subjected to in-gel trypsin digestions as described previously (37). Tryptic peptide extracts were analyzed either by MALDI-TOF-MS using a Micromass M@LDI Reflectron mass spectrometer (Waters/Micromass, Milford, MA) or by nano-LC-MS/MS using an LCQ Deca XP Plus Thermo Finnigan mass spectrometer coupled to an Ultimate Nano liquid chromatography system (Thermo Finnigan, San Jose, CA).
Analysis by MALDI-TOF-MS—
For the MALDI-TOF-MS, a sample droplet (1.2 μl) was applied onto a 96-well stainless steel target plate between two layers of matrix (α-cyano-4-hydroxycinnamic acid). As needed, prior to MALDI-TOF-MS, reversed-phase ZipTips (Millipore, Bedford, MA) were used according to the manufacturer's instructions for enhanced sample clean-up, and mass calibration was done before each run as described earlier (37). Spectra were obtained using the reflectron mode with an acceleration voltage of 20 kV for the mass range 700–3500 Da, and data collected using 200 laser shots generated by a pulsed nitrogen laser (337 nm; pulse width, 4 ns) hitting the target spot at several positions were combined to generate a peptide mass fingerprint. Raw spectra were analyzed by using the MassLynx (version 4.0, Waters/Micromass) software with the intensities of all peaks normalized to that of the most intense peak of the selected m/z region. The resulting mass spectra were calibrated using internal calibration with the ACTH-(18–39) clip peak at m/z 2465.1989 in the lock mass solution, and the calibrated spectra were used for database searches via the Micromass Protein-Lynx Global Server (version 2.0, Waters/Micromass) with an m/z tolerance of 40 ppm. Protein identifications were assigned by comparing peak lists, generated from peptide mass fingerprinting, with a database containing theoretical tryptic digests of R. capsulatus ORFs (release date, January 2001; 3717 entries) obtained from Integrated Genomics Inc., Chicago, IL. Criteria for positive identification of proteins were set as follows. (i) To distinguish a valid match from a false positive, a minimum of four measured peptide masses must match tryptic peptide masses calculated for an individual protein in the database with 40 ppm or better mass accuracy. (ii) The matched peptides must provide at least 15% sequence coverage of the identified protein. (iii) The protein must exhibit a significant difference in the number of matched peptides compared with the next potential hit. (iv) The similarity in the molecular weight and pI of the identified protein compared with the estimated values obtained from the image analysis was also considered. For each sample, spectra acquisition and annotation were repeated at least twice both automatically and manually to minimize identification errors.
Analysis by LC-MS/MS—
For the nano-LC-MS/MS, autosampling and chromatography were performed essentially as described earlier (37). Tryptic peptide mixtures were first loaded onto a μ-precolumn (C18, 5 μm, 100 Å, 300-μm inner diameter × 5 mm) (LC Packings), washed for 4 min at a flow rate of 0.25 μl/min with LC buffer A (5% acetonitrite, 0.1% formic acid) (37), and then transferred onto an analytical C18 nanocapillary HPLC column using a nanocolumn switching device (Switchos, LC Packings) to direct the flow either to waste or to the analytical column. The peptide mixture was fractioned on an LC Packings PepMap C18 column (75-μm inner diameter × 150 mm) with a 3-μm particle size and a 100-Å pore diameter. A fused silica needle with 8-μm aperture (New Objective, Woburn, MA) was used for ionization of peptides. Mass spectra were measured with an LCQ Deca XP Plus ion trap mass spectrometer (Thermo Finnigan). Mass spectrometry scans as well as HPLC solvent gradients were controlled via the Xcalibur software (version 1.3, Thermo Finnigan). Peak lists were generated using Extract-msn in Bioworks 3.1 software (Thermo Finnigan). From raw files, MS/MS spectra were exported as individual files in .dta format under the following settings: peptide mass range, 500–3500 Da; minimal total ion intensity threshold, 100,000; minimal number of fragment ions, 15; precursor mass tolerance, 1.4 Da; group scan, 25; group count, 1. The mass spectra were filtered for common contaminants such as keratin during the .dta file creation process. The resulting .dta files from each analysis were automatically combined into a single text file. The resulting peak lists were searched against the R. capsulatus protein database (release date, January 2001; 3717 entries) obtained from Integrated Genomics Inc. using SEQUEST software (version 3.1) on a local server by comparison with the theoretical spectra of all possible peptide fragments from the SEQUEST database of choice (i.e. R. capsulatus annotated protein database in this case). The following parameters were used: trypsin was selected as the enzyme, for proteolytic cleavage only trypsin cleavage after arginine and lysine was allowed, and the number of maximal internal (missed) cleavage sites was set to 2. Mass tolerance for precursor and fragment ions was 2.5 and 1.0 Da, respectively. No modification was considered. Matching peptides were filtered according to correlation scores (XCorr at least 1.5, 2.0, and 3.0 for +1, +2, and +3 charged peptides, respectively) to give high confidence protein identification. The default setting was used for all other variables.
Zymogram Gel Analysis—
Periplasmic fractions and whole cell extracts of R. capsulatus strains were solubilized in zymogram sample buffer containing 62.5 mm Tris-HCl, pH 6.8, 25% glycerol, 4% SDS, and 0.01% bromophenol blue without any reducing agent and loaded in a 12.5% zymogram gel containing 1 mg/ml casein as a substrate. Electrophoresis was carried out for 2 h at 4 °C and 150 V under non-reducing conditions using TGS buffer consisting of 25 mm Tris-HCl, pH 8.3, 192 mm glycine, and 0.1% SDS. Following electrophoresis, the gels were shaken gently for 60 min at room temperature in zymogram renaturation buffer containing 2.5% (v/v) Triton X-100 (Sigma); incubated overnight at 35 °C in a buffer containing 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 5 mm CaCl2, and 0.02% Brij-35; stained with Coomassie Brilliant Blue; and destained to reveal zones of protease activity.
Bioinformatics Tools and Homology Modeling Softwares—
Prediction softwares SignalP (version 3.0) (40–42) and PSORTb (version 2.0) (43, 44) were used to predict the likely subcellular localization of identified proteins, and the transmembrane helical domains in proteins were predicted with the TMHMM Server (version 2.0) (45). A three-dimensional structural model of R. capsulatus DegP was achieved using the crystal structure of E. coli DegP (Protein Data Bank code 1KY9) as a template for the biopolymer homology modeling software of SWISS-MODEL/Deep View (version 3.7) (Swiss-Prot, Basel, Switzerland) (46). Because R. capsulatus and E. coli DegP are highly homologous (36% identical and 71% similar based on ClustalW (version 1.82) analysis) no structural optimization was performed.
Chemicals—
All chemicals were purchased from commercial sources as high purity standards, and the solvents used were HPLC spectral grade.
RESULTS
Pleiotropic Phenotypes of R. capsulatus DsbA-null Mutants and Their Revertants—
R. capsulatus DsbA-null mutants (e.g. MD20 Δ(dsbA::kan)) exhibit severe pleiotropic phenotypes extending from motility defects to increased osmosensitivity and oxidative stresses. These mutants grow well under anoxygenic Ps conditions but are unable to do so under Res conditions especially in enriched MPYE growth medium at 35 °C (16). They exhibit improved Res growth in MedA (which contains Cu2SO4) or upon supplementation of enriched medium with redox-active chemicals like the cysteine/cystine couple. Addition of Cu2SO4 (10–20 μm) to enriched MPYE medium or its omission from MedA restored or abolished, respectively, the Res growth abilities of these mutants (Table II). As R. capsulatus contains two independent Res branches and terminal oxidases (26, 27), the growth defect suggested that multiple components might be affected by the absence of DsbA. In addition, DsbA-null mutants reverted readily on enriched medium at 35 °C (at a frequency of about 10−6) to regain Res growth ability. A number of such Ps+, Res+ revertants (MD20Ri with i from 1 to 12) were retained, and one of them (MD20R3) was characterized (Table II) and used for further studies.
Table II.
Phenotypes of various R. capsulatus strains
P, R, and Cox refer to photosynthetic growth, respiratory growth, and ability to exhibit cytochrome c oxidase activity; + and − indicate proficiency and deficiency, respectively, s and vs correspond to slow and very slow growth, respectively; and na is not applicable.
| 35 °C
|
25 °C
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MPYE | MPYE + Cu2+ | MedA | MedA-Cu2 | MPYE | MPYE + Cu2+ | MedA | MedA − Cu2+ | |
| MT1131 (wild type) | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ |
| MD20 Δ(dsbA::kan) | P+,R−,Coxna | P+,R+,Cox+ | P+,R+,Cox+ | P+,R−,Coxna | P+,R+,Cox− | P+,R+,Cox+ | P+,R+,Cox+ | P+,Rs,Coxna |
| MD20R3 | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ |
| OZL2 Δ(degP::spe) | P+,R+,Cox+ | P+,R+,Cox+ | P+,Rvs,Cox+ | P+,Rvs,Cox+ | P+,R+,Cox+ | P+,R+,Cox+ | P+,Rvs,Cox+ | P+,Rvs,Cox+ |
| pRK-degPR3 or pRK-degPR9/ Δ(degP::spe dsbA::kan) | Ps,R+,Cox− | P+,R+,Cox− | P−,R−,Coxna | P−,R−,Coxna | Ps,Rs,Cox− | Ps,Rs,Cox− | P−,R−,Coxna | P−,R−,Coxna |
Periplasmic Subproteomes of a DsbA-null Mutant and Its Revertant MD20R3—
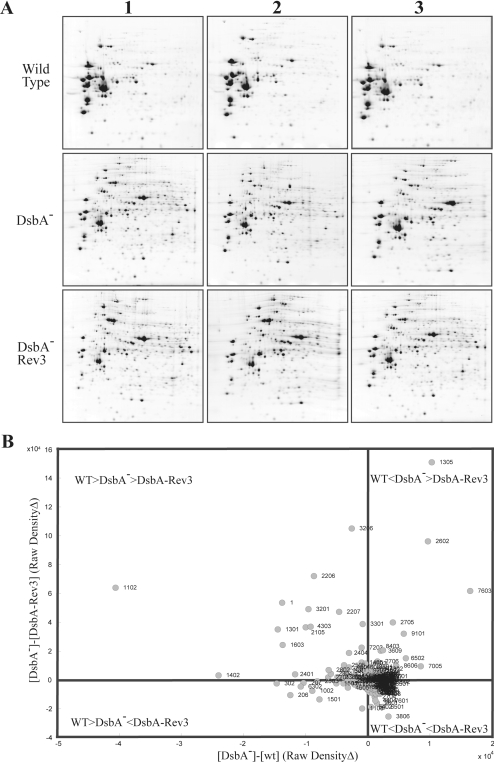
The highly intricate phenotypes of R. capsulatus DsbA-null mutants (Table II) led us to choose a global proteomics approach to investigate the molecular basis underlying the Res growth defects. Given that DsbA is a periplasmic protein, analyses were focused on extracytoplasmic subproteomes as a first step to reduce biological sample complexity. The wild-type (MT1131), DsbA-null mutant (MD20), and its revertant (MD20R3) strains were grown by respiration in the permissive MedA, and cells were harvested at similar growth stages. Periplasmic fractions were prepared using polymyxin B sulfate, separated by 2D-GE, and analyzed using the PDQuest software as described under “Experimental Procedures.” For each strain, at least triplicate cultures and multiple gels per culture were performed to ensure statistical reproducibility (Fig. 1A). For all protein spots, intensity variations between the wild-type, DsbA-null mutant, and its revertant MD20R3 strains were monitored (Fig. 1B).
Fig. 1.
A, 2D gel analysis of periplasmic proteins in various R. capsulatus strains. Periplasmic protein fractions prepared from the wild type (MT1131), a DsbA− mutant (MD20), and a revertant of the DsbA− mutant (MD20R3) were separated by isoelectric focusing and SDS-polyacrylamide gel electrophoresis. Gels were stained with colloidal Coomassie, scanned, and analyzed using PDQuest 2D gel analysis software (Bio-Rad) as described under “Experimental Procedures.” Each study group contained at least three independent replicas, variations in the amounts of the spots were calculated using PDQuest software (see supplemental Table 1 for magnitudes of changes), and chosen protein spots were analyzed by MALDI-TOF-MS or nLC-MS/MS as described under “Experimental Procedures.” B, comparison of the changes in the abundance of different protein spots. Normalized raw density values calculated by PDQuest analysis software were used to compare the abundance of the protein spots in different strains described in A. Spot numbers refer to those attributed by the image analysis program PDQuest. The orders of magnitude of relative abundance of each spot for wild type (WT), DsbA-null mutant (DsbA−), and its revertant (DsbA-Rev3) are indicated on the graph. Protein spots that are at, or near, the origin represent those of unchanged abundance as compared with the wild type.
The analyses visualized at least 340 individual protein spots of which 225 showed less than and the remaining 115 showed more than 3-fold variations in abundance between the wild-type, DsbA-null mutant, and the revertant strains. The latter spots were taken as affected differentially by the absence of DsbA and identified by MALDI-TOF-MS or nano-LC-MS/MS together with 158 of the 225 additional spots (corresponding to 118 distinct proteins) as a protein identification validity check (data not shown). Among the identified proteins those lacking a signal sequence, as indicated by SignalP version 3.0 (40–42) and PSORTb version 2.0 (43, 44) programs, were not pursued further. As expected, R. capsulatus DsbA (RRC03149) was detected only in the periplasmic fractions from the wild-type strain MT1131 and was absent in both the DsbA-null mutant MD20 and its revertant MD20R3. In total, the abundance of 52 bona fide extracytoplasmic protein spots corresponding to 38 distinct ORFs either decreased (25 ORFs) or increased (13 ORFs) by at least 3-fold in the DsbA-null mutant MD20 and its revertant MD20R3 as compared with the wild-type strain MT1131 (Table III).
Table III.
Summary of decreased and increased protein spots in R. capsulatus DsbA-null mutant and revertant compared with the wild-type strain MT1131
Note that all proteins listed except PgaA, which is an integral membrane protein, have a putative signal sequence recognized by the program SignalP.
| ORF no. | Protein name | SSP no.a | Gene name | Cysb | Theoretical
|
Experimental
|
MD20/MT1131c | MD20R3/MT1131c | MS identification method | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mr | pI | Mr | pI | ||||||||
| Decreased proteins | |||||||||||
| RRC03149 | Thiol:disulfide interchange protein DsbA | 8003 | dsbA | 4 | 23.38 | 6.00 | 19.27 | 5.64 | 0.00 | 0.00 | nLC-MS/MS + MALDI-TOF |
| RRC00091 | Spermidine/putrescine-binding periplasmic protein | 1402 | potD | 2 | 38.43 | 4.60 | 40.26 | 4.41 | 0.18 | 0.17 | nLC-MS/MS + MALDI-TOF |
| RRC01245 | Spermidine/putrescine-binding periplasmic proteind | 1302 | 2 | 37.56 | 4.49 | 38.31 | 4.48 | 0.01 | 0.01 | nLC-MS/MS + MALDI-TOF | |
| 2301 | 2 | 37.56 | 4.49 | 38.33 | 4.52 | 0.24 | 0.26 | nLC-MS/MS + MALDI-TOF | |||
| RRC01333 | Periplasmic oligopeptide-binding protein (OppA) | 2702 | oppA | 2 | 57.80 | 4.50 | 57.11 | 4.55 | 0.37 | 0.01 | nLC-MS/MS |
| RRC02807 | Oligopeptide-binding protein AppAd | 3802 | appA | 8 | 61.57 | 4.53 | 61.31 | 4.56 | 0.34 | 0.27 | nLC-MS/MS + MALDI-TOF |
| 2703 | 8 | 61.57 | 4.53 | 60.46 | 4.55 | 0.37 | 0.33 | nLC-MS/MS + MALDI-TOF | |||
| RRC00775 | Lysine-arginine-ornithine-binding protein | 101 | argT | 2 | 25.18 | 5.10 | 27.63 | 4.26 | 0.27 | 0.23 | nLC-MS/MS |
| RRC03799 | Glutamate/glutamine/aspartate/asparagine-binding protein BZTA | 2201 | yhdW | 5 | 35.53 | 4.49 | 34.79 | 4.54 | 0.47 | 0.46 | nLC-MS/MS + MALDI-TOF |
| RRC04018 | Glutamine-binding periplasmic protein precursor | 1001 | glnH | 1 | 27.63 | 4.50 | 25.99 | 4.37 | 0.44 | 0.33 | nLC-MS/MS |
| RRC04050 | Periplasmic leucine-binding proteins | 4605 | 3 | 37.59 | 6.40 | 48.41 | 5.11 | 0.39 | 0.49 | nLC-MS/MS + MALDI-TOF | |
| RRC00677 | Sorbitol-binding periplasmic proteind | 2502 | 3 | 47.76 | 4.85 | 46.74 | 4.55 | 0.13 | 0.14 | nLC-MS/MS | |
| 2503 | 3 | 47.76 | 4.85 | 46.74 | 4.56 | 0.12 | 0.13 | nLC-MS/MS | |||
| RRC00724 | d-Xylose-binding periplasmic protein | 1301 | xylF | 2 | 35.03 | 4.29 | 36.88 | 4.38 | 0.61 | 0.51 | nLC-MS/MS + MALDI-TOF |
| RRC01127 | d-Ribose-binding periplasmic proteind | 102 | rbsB | 1 | 32.94 | 4.38 | 32.38 | 4.32 | 0.00 | 0.00 | nLC-MS/MS + MALDI-TOF |
| 1102 | 1 | 32.94 | 4.38 | 30.45 | 4.36 | 0.38 | 0.28 | nLC-MS/MS + MALDI-TOF | |||
| RRC00984 | ABC transporter substrate-binding protein | 2401 | 2 | 35.20 | 4.70 | 41.75 | 4.55 | 0.23 | 0.21 | nLC-MS/MS + MALDI-TOF | |
| RRC01044 | Fe(III)-binding periplasmic proteind | 5302 | 2 | 35.50 | 5.24 | 38.00 | 5.35 | 0.20 | 0.23 | nLC-MS/MS + MALDI-TOF | |
| 6302 | 2 | 35.50 | 5.24 | 37.85 | 5.43 | 0.18 | 0.22 | nLC-MS/MS + MALDI-TOF | |||
| RRC02708 | ABC transporter substrate-binding proteind | 1504 | motA | 2 | 41.97 | 4.84 | 46.71 | 4.51 | 0.00 | 0.00 | nLC-MS/MS |
| 1502 | 2 | 41.97 | 4.84 | 46.72 | 4.40 | 0.44 | 0.52 | nLC-MS/MS | |||
| 1603 | 2 | 41.97 | 4.84 | 46.78 | 4.41 | 0.69 | 0.64 | nLC-MS/MS | |||
| RRC03395 | Phosphate-binding periplasmic protein | 2404 | pstS | 5 | 37.01 | 4.70 | 40.90 | 4.56 | 0.57 | 0.31 | nLC-MS/MS + MALDI-TOF |
| RRC03762 | Molybdate-binding periplasmic protein | 4001 | modA | 3 | 30.76 | 5.19 | 26.06 | 4.94 | 0.01 | 0.01 | nLC-MS/MS |
| RRC04244 | Nucleoside-binding proteind | 4302 | 1 | 38.59 | 5.10 | 38.48 | 4.87 | 0.35 | 0.22 | nLC-MS/MS | |
| 4303 | 1 | 38.59 | 5.10 | 38.59 | 5.08 | 0.62 | 0.47 | nLC-MS/MS + MALDI-TOF | |||
| RRC00804 | Acriflavin resistance protein E | 9202 | acrA | 1 | 36.36 | 8.55 | 36.60 | 5.94 | 0.61 | 0.01 | nLC-MS/MS |
| RRC02879 | Periplasmic component of efflux system | 5404 | 1 | 34.68 | 5.04 | 39.90 | 5.36 | 0.00 | 0.00 | nLC-MS/MS + MALDI-TOF | |
| RRC02811 | Glutathione peroxidase (EC 1.11.1.9) | 5002 | 2 | 19.41 | 5.90 | 19.56 | 5.36 | 0.05 | 0.00 | nLC-MS/MS + MALDI-TOF | |
| RRC01089 | 5′-Nucleotidase precursor (EC 3.1.3.5) | 3602 | yunD | 4 | 54.37 | 4.59 | 54.5 | 4.61 | 0.67 | 0.57 | nLC-MS/MS |
| RRC04139 | Acyl-CoA thioesterase I precursor (EC 3.1.2.1) | 1004 | tesA | 1 | 24.16 | 5.30 | 22.70 | 4.51 | 0.14 | 0.01 | nLC-MS/MS + MALDI-TOF |
| RRC03417 | Flagellin | 404 | fliC | 0 | 40.01 | 4.20 | 40.01 | 4.20 | 0.00 | 0.00 | nLC-MS/MS + MALDI-TOF |
| RRC03489 | Peptidyl-prolyl cis-trans isomerase | 1002 | ppiB | 0 | 20.32 | 4.60 | 19.30 | 4.43 | 0.25 | 0.31 | nLC-MS/MS + MALDI-TOF |
| Increased proteins | |||||||||||
| RRC03107 | Zinc protease (EC 3.4.99.5) | 2601 | ymfH | 0 | 49.75 | 4.70 | 49.56 | 4.56 | >10 | >10 | nLC-MS/MS + MALDI-TOF |
| RRC03108 | Zinc proteased | 2501 | 0 | 46.69 | 4.54 | 46.43 | 4.55 | 10 | 14 | nLC-MS/MS + MALDI-TOF | |
| 1506 | 0 | 46.69 | 4.54 | 46.70 | 4.55 | >10 | >10 | MALDI-TOF | |||
| RRC03327 | Protease (transglutaminase-like protein) | 601 | 10 | 67.05 | 8.11 | 51.41 | 9.50 | 4 | 5 | nLC-MS/MS | |
| RRC00485 | Outer membrane lipoprotein carrier proteind | 7003 | lolA | 0 | 21.97 | 5.96 | 24.05 | 5.53 | 4 | 4 | nLC-MS/MS + MALDI-TOF |
| 7005 | 0 | 21.97 | 5.96 | 22.84 | 5.53 | 2 | 2 | nLC-MS/MS | |||
| RRC04580 | Peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) | 3207 | 0 | 30.84 | 5.00 | 31.08 | 4.77 | 5 | 8 | nLC-MS/MS | |
| RRC02959 | CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase | 2107 | pgsA | 0 | 23.02 | 10.0 | 29.23 | 4.55 | 6 | 5 | nLC-MS/MS |
| RRC04027 | Periplasmic glucan biosynthesis protein MdoG | 4806 | mdoG | 1 | 62.62 | 5.38 | 58.48 | 5.27 | 5 | 7 | nLC-MS/MS |
| RRC00951 | Outer membrane protein | 7003 | yiaD | 2 | 23.35 | 5.96 | 24.05 | 5.53 | 4 | 4 | nLC-MS/MS |
| RRC03455 | Outer membrane protein/DsbA-like thioredoxin domaind | 2106 | 2 | 26.79 | 4.57 | 27.03 | 4.54 | 5 | 7 | nLC-MS/MS | |
| 2102 | 2 | 26.79 | 4.57 | 28.35 | 4.55 | 3 | 3 | nLC-MS/MS | |||
| RRC01299 | Taurine-binding periplasmic protein | 208 | 2 | 34.91 | 4.60 | 35.20 | 4.37 | >10 | >10 | MALDI-TOF | |
| RRC01191 | Mannitol-binding periplasmic protein | 4404 | 3 | 39.82 | 5.43 | 39.59 | 5.30 | >10 | >10 | nLC-MS/MS + MALDI-TOF | |
| RRC01654 | Hypothetical exported protein | 2704 | 2 | 56.26 | 4.50 | 58.83 | 4.56 | >10 | >10 | nLC-MS/MS + MALDI-TOF | |
| RRC01208 | DegP (EC 3.4.21.2)d | 2602 | degP | 0 | 51.57 | 4.84 | 51.42 | 4.66 | >10 | 1.6 | nLC-MS/MS + MALDI-TOF |
| 2705 | 0 | 51.57 | 4.84 | 51.91 | 4.58 | >10 | 1.7 | nLC-MS/MS + MALDI-TOF | |||
| 2706 | 0 | 51.54 | 4.84 | 52.40 | 4.56 | >10 | 1.7 | nLC-MS/MS + MALDI-TOF | |||
| 3609 | 0 | 51.54 | 4.84 | 52.85 | 4.61 | >10 | 1.7 | nLC-MS/MS + MALDI-TOF | |||
SSP no. corresponds to the spot number given by the image analysis program PDQuest.
Cys indicates the number of cysteine residues found.
MD20/MT1131 and MD20R3/MT1131 indicate the ratio of a given spot between these strains.
These proteins ran as multiple spots on 2D-GE, and each spot was identified independently.
Periplasmic Proteins Decreased in a DsbA-null Mutant and in Its Revertant—
In E. coli, the abundance of extracytoplasmic proteins changes in the absence of DsbA (10, 47, 48). In R. capsulatus DsbA-null mutant MD20 and its revertant MD20R3, at least 33 protein spots corresponding to 25 periplasmic proteins also showed decreases in their abundance (Table III). As the same set of proteins remained affected in both MD20 and its Res+ revertant MD20R3, apparently reversion did not reinstate disulfide bond formation capability. A majority of the decreased proteins belonged to the ATP-binding cassette (ABC) transporter family (49–51). This group included two spermidines/putrescines (i.e. R. capsulatus PotD homologues RRC00091 and RRC01245); two oligopeptide-binding proteins, OppA (BS-oppA/RRC01333) and AppA (BS-appA/RRC02807); and an additional spot identified as a different form of AppA. OppA and AppA are periplasmic oligopeptide permeases involved in the internalization of peptides for nutrition, recycling of muropeptides, and sensing the external environment via specific or nonspecific peptides (52, 53). Additional periplasmic proteins of decreased abundance corresponded to the components of various ABC-type transporters involved in transport of amino acids (EC-argT/RRC00775, EC-yhdW/RRC03799, EC-glnH/RRC04018, and RRC04050), sugars (RRC00677, EC-xylF/RRC00724, and EC-rbsB/RRC01127), or extracellular solutes (RRC00984, RRC01044, RRC02708, EC-pstS/RRC03395, EC-modA/RRC03762, and RRC04244). The latter two proteins (RRC03762 and RRC01044) are homologues of ModA related to molybdate and iron uptake, respectively. In E. coli and Salmonella typhimurium ModA levels also decrease in DsbA− mutants (21, 54, 55). Moreover the acriflavin resistance protein AcrA (RRC00804), believed in E. coli to affect septum formation during cell division and cell membrane permeability, and the periplasmic component of efflux system YhiI (EC-yhiI/RRC02879) of unknown substrate specificity also decreased (Table III).
Another group of decreased extracytoplasmic proteins included glutathione peroxidase (RRC02811), 5′-nucleotidase (BS-yunD/RRC01089), and acyl-CoA thioesterase I (EC 3.1.2.1)/lysophospholipase (3.1.1.5) (EC-tesA/RRC04139). Glutathione peroxidase is an antioxidant enzyme found in many organisms, including humans and other mammals, plants, parasitic worms (56–58), and prokaryotes like E. coli, Neisseria meningitidis, and Streptococcus pyogenes (59–61). These proteins are involved in protection against reactive oxygen species, redox regulation of metabolic processes, peroxynitrite scavenging, and modulation of inflammatory processes (62). 5′-Nucleotidase YunD is a disulfide-linked homodimeric enzyme that hydrolyzes extracellular nucleotides into membrane-permeable nucleosides, and TesA is a multifunctional enzyme with thioesterase, esterase, arylesterase, protease, and lysophospholipase activities (55).
Interestingly the levels of flagellin (EC-fliC/RRC03417) and peptidyl-prolyl cis-trans isomerase (RRC03489) also decreased in the absence of DsbA (Table III). The flagellar assembly component flagellin (FliC) is known to be affected by the absence of DsbA and becomes protease-accessible (63). The peptidyl-prolyl cis-trans isomerases accelerate folding of proteins, and their decrease in the absence of DsbA appears at first counterintuitive. However, several peptidyl-prolyl cis-trans isomerases exist in R. capsulatus, and their relationship(s) to disulfide bond formation processes are currently unknown.
Periplasmic Proteins Increased in a DsbA-null Mutant and in Its Revertant—
In Gram-negative bacteria, accumulation of abnormal envelope proteins induces stress response components as a compensatory effect, eliminating non-native proteins (64). In R. capsulatus DsbA-null mutant MD20 and its revertant MD20R3 the abundance of several periplasmic proteases, chaperones, and folding catalysts were also increased (Table III). This list included two zinc proteases of the insulinase family (BS-yfmH/RRC03107 and RRC03108), a hypothetical protein annotated as transglutaminase-like protease (RRC03327), the outer membrane lipoprotein carrier protein (EC-lolA/RRC00485), and a peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (RRC04580). Additional periplasmic proteins of increased abundance included PgsA (RRC02959), which is an integral membrane protein involved in phospholipid biosynthesis, and MdoG (EC-mdoG/RRC04027), which is related to membrane-derived oligosaccharides synthesis (65, 66). In E. coli, PgsA encodes phosphatidyl-glycerophosphate synthase (EC 2.7.8.5) (CDP-1,2-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase), which catalyzes the production of phosphatidylglycerol and cardiolipin (67–70), and MdoG is involved in the synthesis of osmoregulated periplasmic glucans (71). Osmoregulated periplasmic glucans are a family of anionic and highly branched oligosaccharides found in the periplasm of Gram-negative bacteria that accumulate in response to low osmolarity of the medium (72).
In addition, the amounts of two putative outer membrane proteins (EC-yiaD/RRC00951 and RRC03455) that are associated with the peptidoglycan-associated (lipo)protein OmpA family, thought to be regulated by the EnvZ-OmpR signal transduction system (73), also increased in the DsbA-null mutant and its revertant. Interestingly RRC03455 has a DsbA-like thioredoxin domain, suggesting an ability to catalyze disulfide bond formation. Moreover in the absence of DsbA the abundance of the osmoprotectants taurine- (i.e. 2-aminoethanesulfonate) (RRC01299) and mannitol (RRC01191)-binding proteins increased, reflecting that osmosensing mechanisms are activated. Finally the abundance of a hypothetical protein, RRC01654 of unknown function, also increased indicating that it was expressed in R. capsulatus.
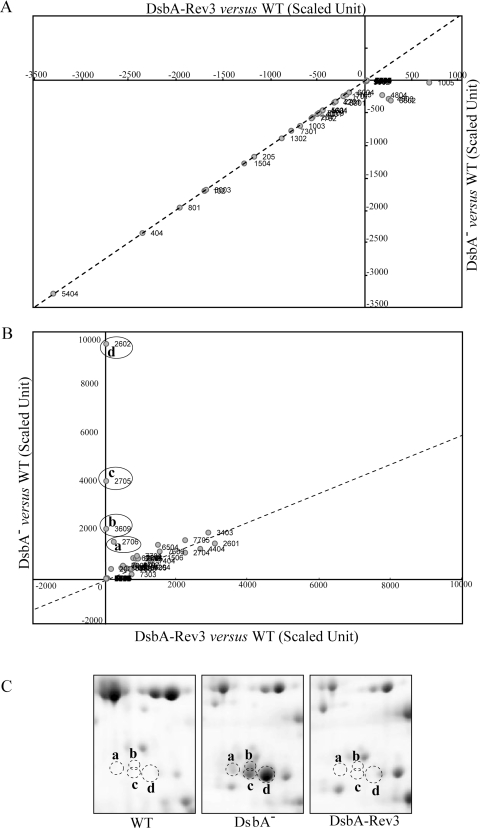
Comparative Analysis of the DsbA-null Mutant Versus Its Revertant—
The next level of data analysis focused on the proteins that exhibited differences between the DsbA-null mutant MD20 and its revertant MD20R3 after normalization of the intensities of protein spots against the wild-type levels. This comparison indicated that the decreased or increased abundance of most proteins followed similar trends in both DsbA-null mutant and in its revertant (Fig. 2, A and B). If the abundance of a protein was increased or decreased in the DsbA-null mutant MD20, it also increased or decreased in its revertant MD20R3 and vice versa with the exception of four spots (Fig. 2B, circled spots a, b, c, and d). These four spots increased significantly in the DsbA mutant but returned to the wild-type levels in the revertant (Fig. 2C). Mass spectrometric identification revealed that all four protein spots corresponded to R. capsulatus DegP (also known as protease Do or HtrA) homologue, a major periplasmic stress response protease, and led us to examine the role of this protein in R. capsulatus.
Fig. 2.
Relative ratios of protein spots in the periplasmic fractions of a DsbA− mutant versus its revertant. Comparative analyses of the scaled data (i.e. mutant or revertant data minus wild-type data) indicated that the ratio of most of the proteins in the DsbA-null mutant and in its revertant remained unchanged. A, protein spots that are decreased in their abundance in the DsbA-null (DsbA−) and its revertant (DsbA-Rev3) as compared with the WT strain. B, protein spots that are increased in their abundance in the DsbA-null (DsbA−) and/or its revertant (DsbA-Rev3) as compared with the WT strain. Note that the amounts of four protein spots (circled and labeled a, b, c, and d) increased in the DsbA− mutant but not in its revertant DsbA-Rev3 were identified to be the same periplasmic protease, DegP. C, 2D gel portions where the protein spots corresponding to a, b, c, and d in the WT, DsbA−, and DsbA-Rev3 (left, middle, and right panels, respectively) strains are shown (see supplemental Table 1 for magnitudes of changes deduced by the PDQuest software).
Overproduction or Absence of DegP Is Deleterious in the Absence of DsbA—
As no information was available on R. capsulatus degP, first a DegP− derivative of the wild-type strain MT1131 was constructed by interposon mutagenesis using a Δ(degP::spe) allele and selecting for SpeR on enriched medium (see “Experimental Procedures”). The DegP-null mutant thus obtained, OZL2, was Res+ and Ps+ on enriched medium at 35 °C but grew poorly in minimal medium under similar growth conditions (Table II), and the growth defect was less pronounced at lower (i.e. 25 °C) temperatures. OZL2 also reverted frequently for better growth in minimal medium and was fully complemented by a plasmid carrying a wild-type copy of degP. These phenotypes were intriguing but not pursued further as degP was not required for Res growth and for cytochrome c oxidase production on enriched medium of an otherwise wild-type R. capsulatus strain such as OZL2.
We thought that if the culprit for the Res− phenotype of a DsbA-null mutant were DegP overproduction then its elimination would alleviate the growth defect. Conversely the introduction of a plasmid carrying a wild-type copy of degP into a Res+ revertant of a DsbA-null mutant might overproduce DegP and hinder its growth ability. However, despite repeated efforts on both minimal and enriched media at either 25 or 35 °C under Res or Ps growth conditions neither the introduction of Δ(degP::spe) into the DsbA-null mutant MD20 nor the introduction of Δ(dsbA::kan) into the DegP-null mutant OZL2 was successful, suggesting that a DegP-null DsbA-null mutant might be lethal. Indeed this was confirmed by the isolation of a chromosomal (Δ(dsbA::kan) Δ(degP::spe)) double mutant using a DegP-null mutant that was rendered diploid for dsbA via a plasmid-borne copy of it like OZL2/pdsbAWT (Table I). In contrast, a similar experiment using a protease-inactive mutant of DegP, carrying the S234A substitution at the catalytic triad in its active site, yielded no such mutant. This indicated that in R. capsulatus some DegP activity might be necessary in the absence of DsbA. Thus, a chromosomal dsbA::kan degP::spe double mutant, complemented by an allele of DegP carried by a revertant of a DsbA-null mutant, was sought to demonstrate that indeed this was the case. Thus, the degPR3 or degPR9 alleles (see below) were cloned into the plasmid pRK415 (Table I, pRK-degPR3 and pRK-degPR9) and conjugated into OZL2 (Δ(degP::spe)) followed by the introduction of a null allele (Δ(dsbA::kan)) by interposon mutagenesis to produce such mutants successfully. Moreover we also observed that although a multicopy plasmid carrying degP (pRK-degPWT) had no effect on the Res growth of wild-type cells it abolished readily the Res growth of the DsbA-null mutant revertant MD20R3 on all media. Overproduction of DegP hampered the Res growth abilities of a DsbA-null mutant even on minimal medium. These findings established that in the absence of DsbA either the overproduction or the absence of DegP was deleterious in R. capsulatus.
DegP Protease Activities of the DsbA-null Mutant Revertants—
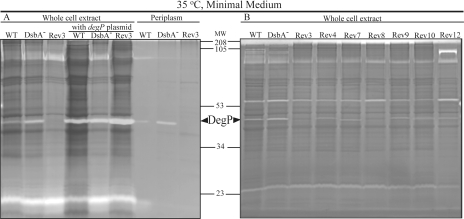
The above data predicted that the Res+ revertants of the DsbA-null mutant, like MD20R3, must have decreased DegP activity. Gel-based protease activities (zymograms) using periplasmic fractions or whole cell extracts of appropriate strains were examined using gel-incorporated casein as a substrate as described under “Experimental Procedures.” These zymograms revealed various protease activities present in R. capsulatus, and among them the band corresponding to DegP was recognized using strains containing a multicopy plasmid carrying degP (pRK-degPWT) (Fig. 3A). The data confirmed that indeed the DsbA-null mutant had high whereas its revertant MD20R3 had low levels of DegP activity compared with the wild-type R. capsulatus strain MT1131. Similarly several independently isolated Res+ revertants of the DsbA-null mutant MD20 (MD20R4, -7, -8, -9, -10, and -12) also exhibited decreased amounts of DegP protease activity as compared with their DsbA-null parent MD20 (Fig. 3B). The overall data correlated well the Res growth defect of a DsbA-null mutant with its highly increased DegP protease activity.
Fig. 3.
Gel-based protease activity assays (zymograms) of various R. capsulatus protein extracts. In each case, periplasmic or whole cell lysates (10 μg of total proteins) prepared from cells grown by respiration in minimal medium at 35 °C were resolved in 12.5% casein zymogram gels under non-reducing conditions. After electrophoresis, gels were incubated overnight in a renaturation buffer and stained with Coomassie Blue. WT, DsbA−, and Rev3 correspond to the wild-type MT1131, DsbA-null mutant MD20, and its revertant MD20R3, respectively. Rev4, Rev7, Rev8, Rev9, Rev10, and Rev12 indicate the lysates prepared from the revertants MD20R4, -7, -8, -9, -10, and -12, respectively, of the DsbA-null mutant MD20 of R. capsulatus as listed in Table I. The band corresponding to DegP is indicated by arrows, and molecular weight markers (MW) are shown between A and B.
DsbA-null Revertants Carry Mutations in degP—
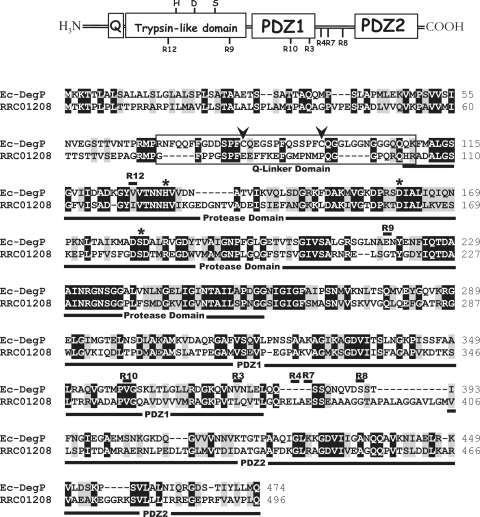
The DNA sequences of the degP loci of the DsbA-null revertants MD20Ri (Table I) were determined to define whether their decreased protease activities were due to mutations in degP. Indeed the single missense mutations L373S, L382P, E384K, G392C, G217S, V356M, and I120F were found in the degP locus of the revertants MD20R3, -R4, -R7, -R8, -R9, -R10, and -R12, respectively (Fig. 4).
Fig. 4.
Amino acid sequence alignment between the E. coli (Ec) DegP and its R. capsulatus homologue RRC01208. The Q-linker domain of DegP is boxed, and the trypsin-like protease, PDZ1, and PDZ2 domains are underlined. The two conserved cysteine residues in the Q-linker domain and the Asp-His-Ser catalytic triad of the trypsin-like domain are indicated with arrows and *, respectively. The point mutations found in the revertants MD20R3, -4, -7, -8, -9, -10, and -12 (Table I) of the DsbA-null mutant MD20 are indicated by R3, R4, R7, R8, R9, R10, and R12, respectively. Black and gray boxes correspond to identical and similar amino acid residues, respectively.
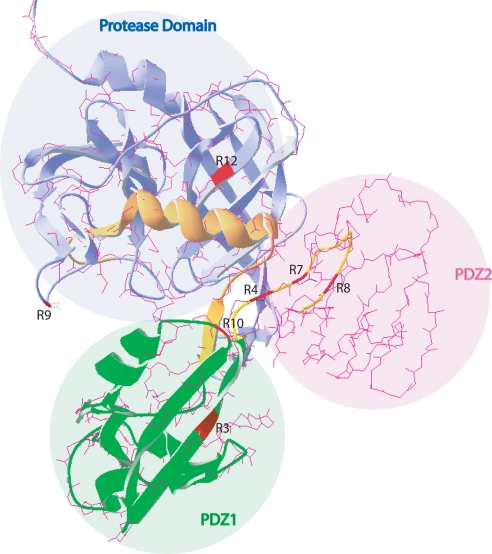
E. coli DegP is a well studied protein with a known 3D structure (74). It is a hexameric protein with each monomer formed of a trypsin-like protease domain containing a His-Asp-Ser catalytic triad, which is characteristic of serine proteases, and two PDZ (named after three proteins, PSD-95, Disk-large, and ZO-1) domains (Fig. 4) (74–77). The mobile side walls of the central cavity in the hexameric DegP is formed by the PDZ domains, composed of 80–100-amino acid-long sequences, implicated in the entry of the proteins into the proteolytic sites to be degraded (78–80). As R. capsulatus and E. coli DegP proteins are highly homologous (36% identity and 71% similarity) (Fig. 4), building a 3D structural model using the available E. coli structure as a template was attempted (Fig. 5). Although it was possible to derive reliable 3D models for the protease and the PDZ1 domains, this was not the case for the PDZ2 domain. Localization on this structural model of the degP mutations found in the revertants of MD20 indicated that two of them (MD20R9 and -R12) were in the protease domain, two (MD20R3 and -R10) were in the C-terminal region of the PDZ1, and the remaining three (MD20R4, -R7, and -R8) were clustered within a 29-amino acid-long linker region between the PDZ1 and the PDZ2 domains (Figs. 4 and 5). The locations of these mutations were consistent with decreased protease activities of the corresponding mutant DegP proteins as deletion of either or both of the PDZ domains is known to affect severely the protease but much less the chaperone function of DegP (81).
Fig. 5.
Homology modeling of R. capsulatus DegP-like protein using the crystal structure of E. coli DegP. Homology modeling was performed by using the SWISS-MODEL automated protein structure homology modeling server. The amino acid sequence alignment of R. capsulatus and E. coli DegP shown in Fig. 4 was loaded into the server, and the resulting .pdb files containing superimposed structures of E. coli DegP (Protein Data Bank code 1KY9) and hypothetical R. capsulatus DegP-like protein were visualized by using the Deep View/Swiss-Pdb Viewer program. Protease, PDZ1, and PDZ2 domains are colored in blue, green, and pink, respectively, and the mutations found in the revertants (MD20R3, -4, -7, -8, -9, -10, and -12 (R3, R4, R7, R8, R9, R10, and R12) described in Fig. 4) are shown in red on the structure. Note that only the protease and the PDZ1 domains could be modeled reliably.
R. capsulatus DsbA-null Mutants Are Temperature-sensitive for Respiratory Growth—
The facts that the E. coli DegP acts mainly as a protease at high temperatures and as a chaperone at low temperatures (81) led us to test the Res growth ability on enriched medium of R. capsulatus DsbA-null mutants at temperatures between 35 and 20 °C. When the growth temperature was lowered to 25–28 °C DsbA-null mutants regained Res growth ability on enriched medium. Like the protease activity of purified E. coli DegP, which is less active at 28 than 42 °C, zymogram data also indicated that wild-type R. capsulatus cells grown on enriched medium at 25 °C exhibited lower DegP activity than those grown at 35 °C (data not shown), suggesting that lowering the growth temperature possibly lowered the protease and increased the chaperone activity of DegP to allow growth.
DISCUSSION
In an earlier study on cyt c biogenesis, we had observed that R. capsulatus DsbA-null mutants lacking the major periplasmic dithiol:disulfide oxidoreductase were unable to grow by respiration especially on enriched medium (16). Here we investigated further this intriguing observation and found that the defect could be alleviated by supplementing the growth medium with redox-active molecules such as Cu2+ ions or by frequent reversion events. We first attempted genetic complementation of a DsbA-null mutant with a transferable chromosomal library constructed using DNA isolated from one of its revertants, MD20R3. However, despite repeated efforts this approach was unsuccessful and raised the possibility that the reversion event might be due to the loss of a wild-type function, which turned out to be the case.
Next we compared the periplasmic subproteomes of a DsbA-null mutant and its parent and found several noteworthy differences. The comparative proteomics approach was very informative because it confirmed the absence of DsbA in the appropriate mutants and revealed drastic changes in the amounts of several extracytoplasmic proteins, which could be associated with pleiotropic phenotypes of DsbA-null mutants. For example, periplasmic glucan biosynthesis protein MdoG increased and flagellin decreased in agreement with the mucoid colony morphology and motility defects of the DsbA− mutants, respectively. However, due to the large number of differential changes observed (Table III), it was impossible to pinpoint the molecular basis of the Res growth deficiency seen in the absence of DsbA. Thus, we undertook a three-way comparison between a Res-deficient DsbA-null mutant, a revertant regaining the Res growth ability, and their parental strain. This second order of analysis was more incisive as it yielded only four protein spots that were highly increased in the DsbA-null mutant and returned back to the wild-type levels in the revertant strain. Interestingly mass spectrometry analyses identified all four spots to be the product of the same R. capsulatus RRC01208 gene. This protein turned out to be a homologue of the E. coli DegP and DegQ periplasmic proteases also called HtrA or protease Do in various genome annotations. Correlating the overproduction of this DegP-like periplasmic protease with the Res defect of a R. capsulatus DsbA-null mutant led us to hypothesize that this might be the culprit. Rigorous testing of this hypothesis required a direct molecular genetics approach. Indeed overproduction of R. capsulatus DegP-like periplasmic protease in a revertant of a DsbA-null mutant conferred back the Res defect, which was even more severe than that in the DsbA-null parent as it arrested Res growth on both enriched and minimal media. Clearly without a three-ways proteomics comparison coupled to multilevel data analyses, it would have been impossible to uncover any clue about the molecular basis of the Res defect associated with the absence of DsbA.
Perhaps even more remarkable was the finding that R. capsulatus mutants lacking both DsbA and the DegP-like protease were lethal. This is unlike E. coli where DegP is a known substrate of DsbA (82). In R. capsulatus, absence of DsbA induces overproduction of an active DegP-like protease and renders Res growth defective, and low amounts of this protease are required for growth. Indeed Res-proficient revertants of the DsbA-null mutant contained no null allele of degP only point mutations that lowered, but not completely eliminated, the DegP-like protease activity. Moreover the chromosomal copy of degP can be readily deleted if a DsbA-null mutant harbored a plasmid-borne copy (i.e. pRK-degPR3 or pRK-degPR9) of DegP point mutants. It is noteworthy that the lethality of a DegP-null mutant in a DsbA-null background provides a facile selection for point mutations decreasing the activity of R. capsulatus DegP-like protease. Although the structure of R. capsulatus DegP is unknown, its significant sequence similarity to the E. coli protein allows structural mapping of these mutations via homology modeling. Based on the crystal structures of human HtrA2 and E. coli DegP, the PDZ domains are thought to be involved in modulating the protease activity (77) and mediating the initial binding of substrates (74). If this is also the case for R. capsulatus DegP-like protease, then the mutations isolated using this approach could define easily the regions of DegP affecting its protease activity. Thus, detailed structure-function studies of R. capsulatus periplasmic DegP-like protease can now be undertaken.
Why the DsbA-DegP interactions are different in different strains is intriguing. E. coli DegP has a disulfide bond between two conserved cysteine residues in a structurally highly flexible region called the “Q-linker.” The three-dimensional structure of E. coli DegP (74, 83) indicates that the active enzyme is a hexamer constituted of two loosely bound dimers of tightly associated trimers. Apparently reduction of the conserved disulfide bridge in the Q-linker induces self-digestion of DegP via dissociation of its oligomeric structure (84). Although the R. capsulatus DegP-like protease discovered here is highly homologous to the E. coli DegP (36% amino acid identity and 71% similarity), a major difference between them lies in their Q-linker domains. Unlike the E. coli DegP, the R. capsulatus DegP-like homologue is devoid of the conserved cysteines and is not a substrate of DsbA. Hence its accumulation in the absence of DsbA is consistent with its structural dissimilarity, possibly rationalizing the Res differences seen between the R. capsulatus and E. coli DsbA-null mutants.
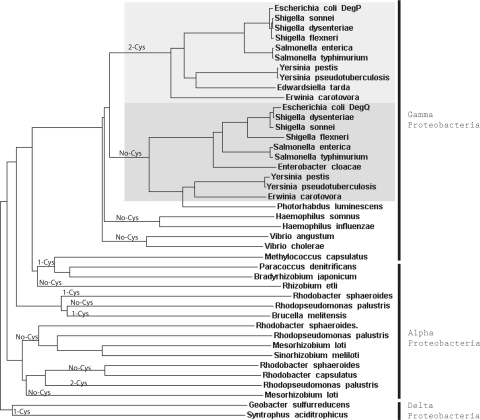
Homologues of the envelope stress protease DegP/HtrA, which was first identified in E. coli (19, 20), are found in a wide range of bacteria, fungi, plants, and mammals. Genomes of many eukaryotic and prokaryotic organisms are annotated to contain multiple DegP/HtrA homologues, which are sometimes referred to as heat shock proteases as well. Interestingly E. coli contains another periplasmic serine protease, DegQ, which is also highly homologous to DegP (55% amino acid identity and 83% similarity). The function of DegQ is less well understood, although it can act as a functional substitute for DegP when overexpressed. Remarkably the Q-linker of E. coli DegQ is distinct from that of DegP and, like the R. capsulatus DegP-like protease, is devoid of the cysteine pair. The Q-linker of R. capsulatus DegP-like protease has 31 and 25% identities and 55 and 52% similarities to those of E. coli DegP and DegQ, respectively. Recently based on sequence homologies between the Q-linker regions, Kim and Kim (80) have proposed that most of the DegP/HtrA homologues found in many bacteria should be renamed as DegQ homologues. Phylogenomics comparisons indicate that a number of bacteria (including R. capsulatus) are unlike enteric bacteria (including E. coli) in having a single periplasmic serine protease with a Q-linker structure in between those of E. coli DegP and DegQ (Fig. 6). In these species, a single DegP-like protease apparently fulfills both the DegP and DegQ functions together, suggesting that it would be essential under high envelope stress conditions like the absence of DsbA. In addition, unlike enteric bacteria many other species (including Rhodobacter sphaeroides) have various DegP-like proteases devoid of the conserved cysteine pair in their Q-linker domains even if they contain some cysteine residues in other parts of the protein (Fig. 6). Why a Q-linker with a conserved pair of cysteines is only seen in enteric bacteria and why such species always possess an additional DegP homologue (i.e. E. coli DegQ) devoid of such cysteines are intriguing questions. A possibility is that these species need to keep a DegQ-like protease activity at all times while having the ability to overproduce a DegP-like protease in a DsbA-dependent manner as part of their envelope stress response. This more evolved dual system then renders both DegP and DegQ nonessential for growth and disallows the dangerous overproduction of DegP in the absence of DsbA. Considering that the optimal growth temperature for enteric bacteria like E. coli is around 37 °C whereas that for the free living soil species like R. capsulatus is lower (about 30 °C), the occurrence of a second protease activity might reflect an adaptive response of bacteria to higher physiological growth temperatures.
Fig. 6.
Phylograms of DegP family members. Trees were generated by comparing via the ClustalW program the full-length sequences of DegP homologues from various bacterial species of available genome sequences. Note that the DegP homologues that have a conserved pair of cysteines in their Q-linker domains are clustered mainly among the enteric bacteria, like Escherichia, Shigella, Salmonella, Yersinia, and Erwinia species belonging to the Gamma Proteobacteria (in the top part of the figure). Note that in all other species the one or two (e.g. Rhodopseudomonas palustris) cysteine residues found are not located in the Q-linker domains of their DegP or DegQ homologues. Remarkably all species that have a DegP homologue with two Q-linker cysteines also have a DegQ homologue without any cysteine, illustrating the occurrence of two interrelated but distinct DegP-like and DegQ-like periplasmic protease families in bacterial species.
The E. coli DegP and its homologues also exhibit molecular chaperone activities in addition to their protease function. As a component of the heat shock response, the protease function of DegP/HtrA is more pronounced at higher growth temperatures, whereas at lower temperatures apparently its chaperone function dominates (81, 85). In the case of R. capsulatus, growth at lower temperatures alleviates the respiratory defect of DsbA-null mutants, and cells exhibit lower DegP-like protease activity as compared with those grown at normal temperature. Apparently the deleterious protease activity is decreased at lower temperatures to alleviate its detrimental effect on growth.
Interestingly, however, DsbA-null mutants grown at low temperature do not contain an active cyt cbb3 oxidase unlike their revertants, indicating that absence of DsbA somehow affects indirectly the production of an active cyt cbb3 oxidase possibly via some deleterious consequence(s) of the overproduction of DegP-like protease. Searching for revertants of DsbA-null mutants that regain the Res growth ability but still lack an active cyt cbb3 oxidase at normal growth temperature or isolating mutants that regain the ability to produce an active cyt cbb3 oxidase at lower temperature might be informative. Although the phenotypes of the DsbA-null mutants are complex and the mechanisms underlying its phenotypes are multifaceted, their studies continue to elucidate the physiological roles of disulfide bond formation in Res growth of the R. capsulatus.
Supplementary Material
Footnotes
Published, MCP Papers in Press, January 2, 2008, DOI 10.1074/mcp.M700433-MCP200
The abbreviations used are: 3D, three-dimensional; MPYE, mineral-peptone-yeast extract; MedA, Sistrom's minimal medium A; Res, respiratory; Ps, photosynthetic; 2D, two-dimensional; GE, gel electrophoresis; ACTH, adrenocorticotropin; nLC, nano-LC; ABC, ATP-binding cassette; cyt, cytochrome; WT, wild type.
This work was supported, in whole or in part, by National Institutes of Health Grant GM38237 (to F. D.). This work was also supported by United States Department of Energy Grant ER 9120053 (to F. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Anfinsen, C. B., Haber, E., Sela, M., and White, F. H., Jr. ( 1961) The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl. Acad. Sci. U. S. A. 47, 1309–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin, J. L. ( 1995) Thioredoxin—a fold for all reasons. Structure (Lond.) 3, 245–250 [DOI] [PubMed] [Google Scholar]
- 3.LaMantia, M. L., and Lennarz, W. J. ( 1993) The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell 74, 899–908 [DOI] [PubMed] [Google Scholar]
- 4.Pollard, M. G., Travers, K. J., and Weissman, J. S. ( 1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1, 171–182 [DOI] [PubMed] [Google Scholar]
- 5.Frand, A. R., and Kaiser, C. A. ( 1999) Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 4, 469–477 [DOI] [PubMed] [Google Scholar]
- 6.Gruber, C. W., Cemazar, M., Heras, B., Martin, J. L., and Craik, D. J. ( 2006) Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem. Sci. 31, 455–464 [DOI] [PubMed] [Google Scholar]
- 7.Bardwell, J. C., McGovern, K., and Beckwith, J. ( 1991) Identification of a protein required for disulfide bond formation in vivo. Cell 67, 581–589 [DOI] [PubMed] [Google Scholar]
- 8.Kamitani, S., Akiyama, Y., and Ito, K. ( 1992) Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messens, J., and Collet, J. F. ( 2006) Pathways of disulfide bond formation in Escherichia coli. Int. J. Biochem. Cell Biol. 38, 1050–1062 [DOI] [PubMed] [Google Scholar]
- 10.Kadokura, H., Katzen, F., and Beckwith, J. ( 2003) Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72, 111–135 [DOI] [PubMed] [Google Scholar]
- 11.Peek, J. A., and Taylor, R. K. ( 1992) Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 89, 6210–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomb, J. F. ( 1992) A periplasmic protein disulfide oxidoreductase is required for transformation of Haemophilus influenzae Rd. Proc. Natl. Acad. Sci. U. S. A. 89, 10252–10256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishihara, T., Tomita, H., Hasegawa, Y., Tsukagoshi, N., Yamagata, H., and Udaka, S. ( 1995) Cloning and characterization of the gene for a protein thiol-disulfide oxidoreductase in Bacillus brevis. J. Bacteriol. 177, 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng, T. C., Kwik, J. F., and Maier, R. J. ( 1997) Cloning and expression of the gene for a protein disulfide oxidoreductase from Azotobacter vinelandii: complementation of an Escherichia coli dsbA mutant strain. Gene (Amst.) 188, 109–113 [DOI] [PubMed] [Google Scholar]
- 15.Tinsley, C. R., Voulhoux, R., Beretti, J. L., Tommassen, J., and Nassif, X. ( 2004) Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J. Biol. Chem. 279, 27078–27087 [DOI] [PubMed] [Google Scholar]
- 16.Deshmukh, M., Turkarslan, S., Astor, D., Valkova-Valchanova, M., and Daldal, F. ( 2003) The dithiol:disulfide oxidoreductases DsbA and DsbB of Rhodobacter capsulatus are not directly involved in cytochrome c biogenesis, but their inactivation restores the cytochrome c biogenesis defect of CcdA-null mutants. J. Bacteriol. 185, 3361–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadokura, H., Tian, H., Zander, T., Bardwell, J. C., and Beckwith, J. ( 2004) Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303, 534–537 [DOI] [PubMed] [Google Scholar]
- 18.Nakamoto, H., and Bardwell, J. C. ( 2004) Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta 1694, 111–119 [DOI] [PubMed] [Google Scholar]
- 19.Strauch, K. L., and Beckwith, J. ( 1988) An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc. Natl. Acad. Sci. U. S. A. 85, 1576–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipinska, B., Sharma, S., and Georgopoulos, C. ( 1988) Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 16, 10053–10067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiniker, A., and Bardwell, J. C. ( 2004) In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 279, 12967–12973 [DOI] [PubMed] [Google Scholar]
- 22.Leichert, L. I., and Jakob, U. ( 2004) Protein thiol modifications visualized in vivo. PLoS Biol. 2, e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metheringham, R., Griffiths, L., Crooke, H., Forsythe, S., and Cole, J. ( 1995) An essential role for DsbA in cytochrome c synthesis and formate-dependent nitrite reduction by Escherichia coli K-12. Arch. Microbiol. 164, 301–307 [DOI] [PubMed] [Google Scholar]
- 24.Sambongi, Y., and Ferguson, S. J. ( 1996) Mutants of Escherichia coli lacking disulphide oxidoreductases DsbA and DsbB cannot synthesise an exogenous monohaem c-type cytochrome except in the presence of disulphide compounds. FEBS Lett. 398, 265–268 [DOI] [PubMed] [Google Scholar]
- 25.Allen, J. W., Barker, P. D., and Ferguson, S. J. ( 2003) A cytochrome b562 variant with a c-type cytochrome CXXCH heme-binding motif as a probe of the Escherichia coli cytochrome c maturation system. J. Biol. Chem. 278, 52075–52083 [DOI] [PubMed] [Google Scholar]
- 26.La Monica, R. F., and Marrs, B. L. ( 1976) The branched respiratory system of photosynthetically grown Rhodopseudomonas capsulata. Biochim. Biophys. Acta 423, 431–439 [DOI] [PubMed] [Google Scholar]
- 27.Zannoni, D., Melandri, B. A., and Baccarini-Melandri, A. ( 1976) Energy transduction in photosynthetic bacteria. X. Composition and function of the branched oxidase system in wild type and respiration deficient mutants of Rhodopseudomonas capsulata. Biochim. Biophys. Acta 423, 413–430 [DOI] [PubMed] [Google Scholar]
- 28.Sistrom, W. R. ( 1960) A requirement for sodium in the growth of Rhodopseudomonas spheroides. J. Gen. Microbiol. 22, 778–785 [DOI] [PubMed] [Google Scholar]
- 29.Myllykallio, H., Jenney, F. E., Jr., Moomaw, C. R., Slaughter, C. A., and Daldal, F. ( 1997) Cytochrome c(y) of Rhodobacter capsulatus is attached to the cytoplasmic membrane by an uncleaved signal sequence-like anchor. J. Bacteriol. 179, 2623–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daldal, F., Mandaci, S., Winterstein, C., Myllykallio, H., Duyck, K., and Zannoni, D. ( 2001) Mobile cytochrome c2 and membrane-anchored cytochrome cy are both efficient electron donors to the cbb3- and aa3-type cytochrome c oxidases during respiratory growth of Rhodobacter sphaeroides. J. Bacteriol. 183, 2013–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and Russell, D. W. ( 2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 32.Yen, H. C., Hu, N. T., and Marrs, B. L. ( 1979) Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131, 157–168 [DOI] [PubMed] [Google Scholar]
- 33.Prentki, P., and Krisch, H. M. ( 1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene (Amst.) 29, 303–313 [DOI] [PubMed] [Google Scholar]
- 34.Daldal, F., Cheng, S., Applebaum, J., Davidson, E., and Prince, R. C. ( 1986) Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. U. S. A. 83, 2012–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keen, N. T., Tamaki, S., Kobayashi, D., and Trollinger, D. ( 1988) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene (Amst.) 70, 191–197 [DOI] [PubMed] [Google Scholar]
- 36.Ren, Q., and Thony-Meyer, L. ( 2001) Physical interaction of CcmC with heme and the heme chaperone CcmE during cytochrome c maturation. J. Biol. Chem. 276, 32591–32596 [DOI] [PubMed] [Google Scholar]
- 37.Onder, O., Yoon, H., Naumann, B., Hippler, M., Dancis, A., and Daldal, F. ( 2006) Modifications of the lipoamide-containing mitochondrial subproteome in a yeast mutant defective in cysteine desulfurase. Mol. Cell. Proteomics 5, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 38.Laemmli, U. K. ( 1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 39.Neuhoff, V., Stamm, R., Pardowitz, I., Arold, N., Ehrhardt, W., and Taube, D. ( 1990) Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11, 101–117 [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. ( 1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8, 581–599 [DOI] [PubMed] [Google Scholar]
- 41.Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. ( 1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 42.Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. ( 2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 43.Gardy, J. L., Laird, M. R., Chen, F., Rey, S., Walsh, C. J., Ester, M., and Brinkman, F. S. ( 2005) PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617–623 [DOI] [PubMed] [Google Scholar]
- 44.Gardy, J. L., Spencer, C., Wang, K., Ester, M., Tusnady, G. E., Simon, I., Hua, S., deFays, K., Lambert, C., Nakai, K., and Brinkman, F. S. ( 2003) PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31, 3613–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. ( 2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 46.Guex, N., and Peitsch, M. C. ( 1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 47.Collet, J. F., and Bardwell, J. C. ( 2002) Oxidative protein folding in bacteria. Mol. Microbiol. 44, 1–8 [DOI] [PubMed] [Google Scholar]
- 48.Missiakas, D., and Raina, S. ( 1997) Protein folding in the bacterial periplasm. J. Bacteriol. 179, 2465–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linton, K. J., and Higgins, C. F. ( 1998) The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28, 5–13 [DOI] [PubMed] [Google Scholar]
- 50.Higgins, C. F. ( 2001) ABC transporters: physiology, structure and mechanism–an overview. Res. Microbiol. 152, 205–210 [DOI] [PubMed] [Google Scholar]
- 51.Jones, P. M., and George, A. M. ( 2002) Mechanism of ABC transporters: a molecular dynamics simulation of a well characterized nucleotide-binding subunit. CMLS Cell. Mol. Life Sci. 99, 12639–12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monnet, V. ( 2003) Bacterial oligopeptide-binding proteins. CMLS Cell. Mol. Life Sci. 60, 2100–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levdikov, V. M., Blagova, E. V., Brannigan, J. A., Wright, L., Vagin, A. A., and Wilkinson, A. J. ( 2005) The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J. Mol. Biol. 345, 879–892 [DOI] [PubMed] [Google Scholar]
- 54.Li, M. S., Kroll, J. S., and Yu, J. ( 2001) Influence of the yihE gene of Shigella flexneri on global gene expression: on analysis using DNA arrays. Biochem. Biophys. Res. Commun. 288, 91–100 [DOI] [PubMed] [Google Scholar]
- 55.Lo, Y. C., Lin, S. C., Shaw, J. F., and Liaw, Y. C. ( 2003) Crystal structure of Escherichia coli thioesterase I/protease I/lysophospholipase L1: consensus sequence blocks constitute the catalytic center of SGNH-hydrolases through a conserved hydrogen bond network. J. Mol. Biol. 330, 539–551 [DOI] [PubMed] [Google Scholar]
- 56.Mills, G. C. ( 1957) Hemoglobin catabolism. I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J. Biol. Chem. 229, 189–197 [PubMed] [Google Scholar]
- 57.Callahan, H. L., Crouch, R. K., and James, E. R. ( 1988) Helminth anti-oxidant enzymes: a protective mechanism against host oxidants? Parasitol. Today 4, 218–225 [DOI] [PubMed] [Google Scholar]
- 58.Criqui, M. C., Jamet, E., Parmentier, Y., Marbach, J., Durr, A., and Fleck, J. ( 1992) Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol. Biol. 18, 623–627 [DOI] [PubMed] [Google Scholar]
- 59.Moore, T. D., and Sparling, P. F. ( 1995) Isolation and identification of a glutathione peroxidase homolog gene, gpxA, present in Neisseria meningitidis but absent in Neisseria gonorrhoeae. Infect. Immun. 63, 1603–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeVeaux, L. C., and Kadner, R. J. ( 1985) Transport of vitamin B12 in Escherichia coli: cloning of the btuCD region. J. Bacteriol. 162, 888–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brenot, A., King, K. Y., Janowiak, B., Griffith, O., and Caparon, M. G. ( 2004) Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72, 408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Avissar, N., Whitin, J. C., Allen, P. Z., Wagner, D. D., Liegey, P., and Cohen, H. J. ( 1989) Plasma selenium-dependent glutathione peroxidase. Cell of origin and secretion. J. Biol. Chem. 264, 15850–15855 [PubMed] [Google Scholar]
- 63.Dailey, F. E., and Berg, H. C. ( 1993) Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 90, 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raivio, T. L., and Silhavy, T. J. ( 2001) Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55, 591–624 [DOI] [PubMed] [Google Scholar]
- 65.Kennedy, E. P., Rumley, M. K., Schulman, H., and Van Golde, L. M. ( 1976) Identification of sn-glycero-1-phosphate and phosphoethanolamine residues linked to the membrane-derived oligosaccharides of Escherichia coli. J. Biol. Chem. 251, 4208–4213 [PubMed] [Google Scholar]
- 66.van Golde, L. M. ( 1973) Metabolism of membrane phospholipids and its relation to a novel class of oligosaccharides in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 70, 1368–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kikuchi, S., Shibuya, I., and Matsumoto, K. ( 2000) Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J. Bacteriol. 182, 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heacock, P. N., and Dowhan, W. ( 1987) Construction of a lethal mutation in the synthesis of the major acidic phospholipids of Escherichia coli. J. Biol. Chem. 262, 13044–13049 [PubMed] [Google Scholar]
- 69.Gopalakrishnan, A. S., Chen, Y. C., Temkin, M., and Dowhan, W. ( 1986) Structure and expression of the gene locus encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J. Biol. Chem. 261, 1329–1338 [PubMed] [Google Scholar]
- 70.Usui, M., Sembongi, H., Matsuzaki, H., Matsumoto, K., and Shibuya, I. ( 1994) Primary structures of the wild-type and mutant alleles encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J. Bacteriol. 176, 3389–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bohin, J. P. ( 2000) Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol. Lett. 186, 11–19 [DOI] [PubMed] [Google Scholar]
- 72.Lequette, Y., Odberg-Ferragut, C., Bohin, J. P., and Lacroix, J. M. ( 2004) Identification of mdoD, an mdoG paralog which encodes a twin-arginine-dependent periplasmic protein that controls osmoregulated periplasmic glucan backbone structures. J. Bacteriol. 186, 3695–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wurtzel, E. T., Chou, M. Y., and Inouye, M. ( 1982) Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J. Biol. Chem. 257, 13685–13691 [PubMed] [Google Scholar]
- 74.Krojer, T., Garrido-Franco, M., Huber, R., Ehrmann, M., and Clausen, T. ( 2002) Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416, 455–459 [DOI] [PubMed] [Google Scholar]
- 75.Faccio, L., Fusco, C., Chen, A., Martinotti, S., Bonventre, J. V., and Zervos, A. S. ( 2000) Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J. Biol. Chem. 275, 2581–2588 [DOI] [PubMed] [Google Scholar]
- 76.Gray, C. W., Ward, R. V., Karran, E., Turconi, S., Rowles, A., Viglienghi, D., Southan, C., Barton, A., Fantom, K. G., West, A., Savopoulos, J., Hassan, N. J., Clinkenbeard, H., Hanning, C., Amegadzie, B., Davis, J. B., Dingwall, C., Livi, G. P., and Creasy, C. L. ( 2000) Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur. J. Biochem. 267, 5699–5710 [DOI] [PubMed] [Google Scholar]
- 77.Li, W., Srinivasula, S. M., Chai, J., Li, P., Wu, J. W., Zhang, Z., Alnemri, E. S., and Shi, Y. ( 2002) Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9, 436–441 [DOI] [PubMed] [Google Scholar]
- 78.Maurizi, M. R. ( 2002) Love it or cleave it: tough choices in protein quality control. Nat. Struct. Biol. 9, 410–412 [DOI] [PubMed] [Google Scholar]
- 79.Schlieker, C., Mogk, A., and Bukau, B. ( 2004) A PDZ switch for a cellular stress response. Cell 117, 417–419 [DOI] [PubMed] [Google Scholar]
- 80.Kim, D. Y., and Kim, K. K. ( 2005) Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38, 266–274 [DOI] [PubMed] [Google Scholar]
- 81.Spiess, C., Beil, A., and Ehrmann, M. ( 1999) A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97, 339–347 [DOI] [PubMed] [Google Scholar]
- 82.Skorko-Glonek, J., Sobiecka-Szkatula, A., and Lipinska, B. ( 2006) Characterization of disulfide exchange between DsbA and HtrA proteins from Escherichia coli. Acta Biochim. Pol. 53, 585–589 [PubMed] [Google Scholar]
- 83.Clausen, T., Southan, C., and Ehrmann, M. ( 2002) The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10, 443–455 [DOI] [PubMed] [Google Scholar]
- 84.Skorko-Glonek, J., Zurawa, D., Tanfani, F., Scire, A., Wawrzynow, A., Narkiewicz, J., Bertoli, E., and Lipinska, B. ( 2003) The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochim. Biophys. Acta 1649, 171–182 [DOI] [PubMed] [Google Scholar]
- 85.Rizzitello, A. E., Harper, J. R., and Silhavy, T. J. ( 2001) Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183, 6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scolnik, P. A., Walker, M. A., and Marrs, B. L. ( 1980) Biosynthesis of carotenoids derived from neurosporene in Rhodopseudomonas capsulata. J. Biol. Chem. 255, 2427–2432 [PubMed] [Google Scholar]
- 87.Ditta, G., Schmidhauser, T., Yakobson, E., Lu, P., Liang, X. W., Finlay, D. R., Guiney, D., and Helinski, D. R. ( 1985) Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13, 149–153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.