Abstract
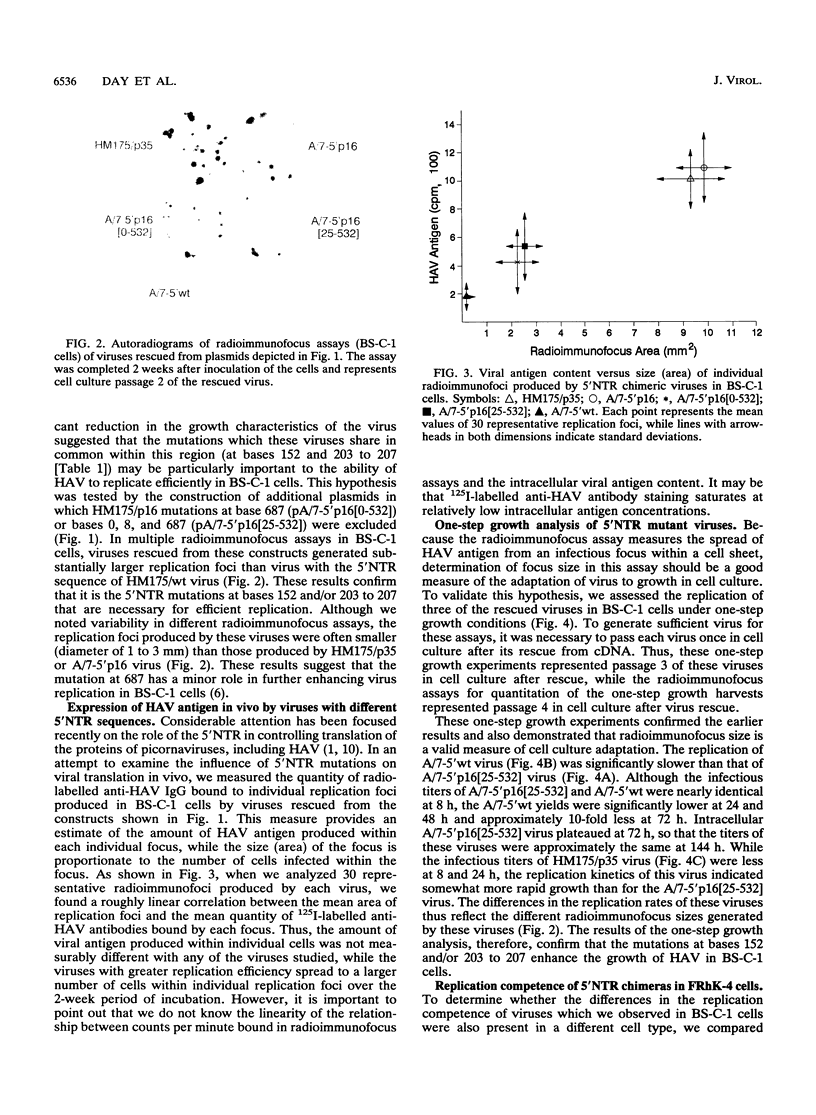
Passage of human hepatitis A virus (HAV) in cell culture results in attenuation of the virus as well as progressive increases in the efficiency of virus replication in cell culture. Because the presence of identical mutations within the 5' nontranslated regions (5'NTRs) of several independently isolated cell culture-adapted HAV variants suggests that the 5'NTR may play a role in determining this change in virus host range, we constructed chimeric infectious cDNA clones in which portions of the 5'NTR of cell culture-adapted HM175/p35 virus were replaced with cDNA from either wild-type virus (HM175/wt) or a second independently isolated, but closely related cell culture-adapted virus (HM175/p16). Substitution of the complete 5'NTR of HM175/p35 with the 5'NTR of HM175/wt resulted in virus with very small replication foci in continuous African green monkey kidney (BS-C-1) cells, indicating that 5'NTR mutations in HM175/p35 virus are required for optimal growth in these cells. A chimera with the 5'NTR sequence of HM175/p16 retained the large foci of HM175/p35 virus, while the growth properties of other viruses having chimeric 5'NTR sequences indicated that mutations at bases 152 and/or 203 to 207 enhance replication in BS-C-1 cells. These findings were confirmed in one-step growth experiments, which also indicated that radioimmunofocus size is a valid measure of virus replication competence in cell culture. An additional mutation at base 687 of HM175/p16 had only a minor role in enhancing growth. In contrast to their effect in BS-C-1 cells, these 5'NTR mutations did not enhance replication in continuous fetal rhesus monkey kidney (FRhK-4) cells. Thus, mutations at bases 152 and/or 203 to 207 enhance the replication of HAV in a highly host cell-specific fashion.
Full text
PDF


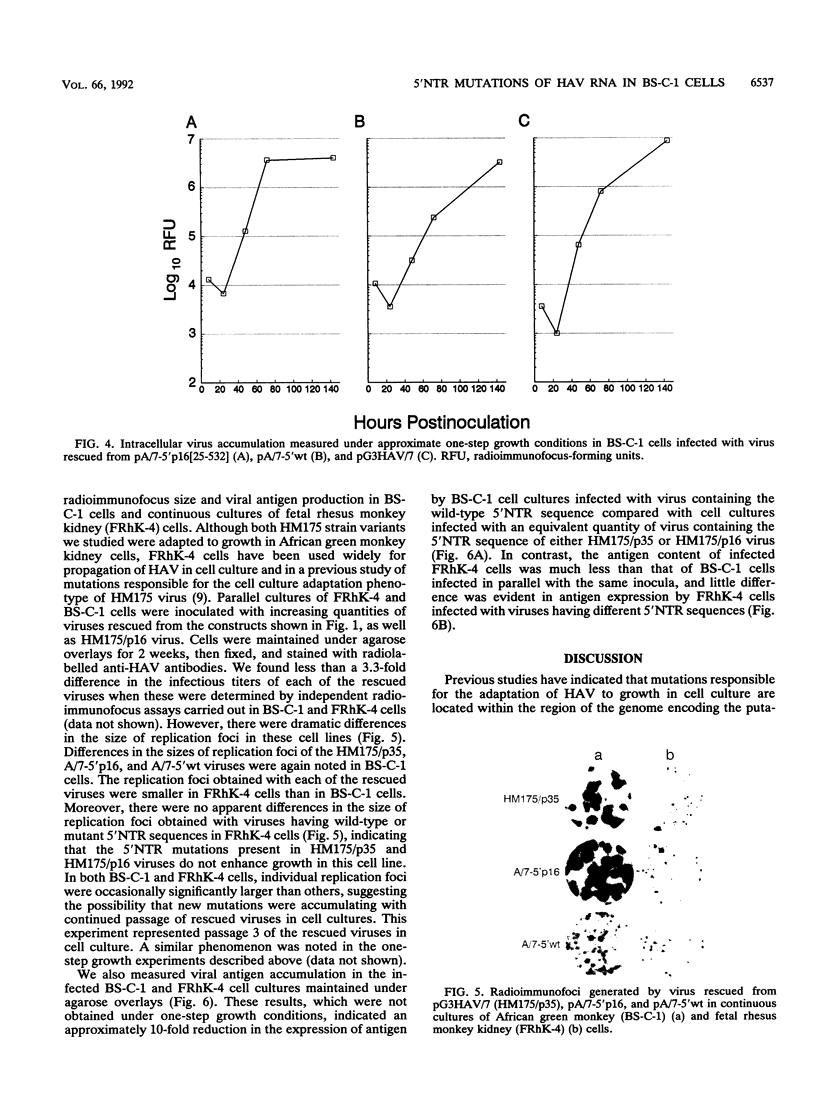


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown E. A., Day S. P., Jansen R. W., Lemon S. M. The 5' nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991 Nov;65(11):5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Rosenblum B., Ticehurst J. R., Daemer R. J., Feinstone S. M., Purcell R. H. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Feinstone S. M., Rosenblum B., Purcell R. H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987 Oct;61(10):3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., Huang Y. K., McRill C., Lewis M., Purcell R. H. Mutations in both the 2B and 2C genes of hepatitis A virus are involved in adaptation to growth in cell culture. J Virol. 1992 Feb;66(2):650–654. doi: 10.1128/jvi.66.2.650-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S. U., McRill C., Rosenblum B., Feinstone S., Purcell R. H. Mutations responsible for adaptation of hepatitis A virus to efficient growth in cell culture. J Virol. 1991 Sep;65(9):4882–4886. doi: 10.1128/jvi.65.9.4882-4886.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. K., Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990 Sep;4(9):1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- Jansen R. W., Newbold J. E., Lemon S. M. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology. 1988 Apr;163(2):299–307. doi: 10.1016/0042-6822(88)90270-x. [DOI] [PubMed] [Google Scholar]
- Lemon S. M., Binn L. N., Marchwicki R. H. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J Clin Microbiol. 1983 May;17(5):834–839. doi: 10.1128/jcm.17.5.834-839.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon S. M. Type A viral hepatitis. New developments in an old disease. N Engl J Med. 1985 Oct 24;313(17):1059–1067. doi: 10.1056/NEJM198510243131706. [DOI] [PubMed] [Google Scholar]
- Meerovitch K., Pelletier J., Sonenberg N. A cellular protein that binds to the 5'-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989 Jul;3(7):1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- Midthun K., Ellerbeck E., Gershman K., Calandra G., Krah D., McCaughtry M., Nalin D., Provost P. Safety and immunogenicity of a live attenuated hepatitis A virus vaccine in seronegative volunteers. J Infect Dis. 1991 Apr;163(4):735–739. doi: 10.1093/infdis/163.4.735. [DOI] [PubMed] [Google Scholar]
- Provost P. J., Banker F. S., Giesa P. A., McAleer W. J., Buynak E. B., Hilleman M. R. Progress toward a live, attenuated human hepatitis A vaccine. Proc Soc Exp Biol Med. 1982 May;170(1):8–14. doi: 10.3181/00379727-170-41387. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl G., Lemon S. M. Recent advances in hepatitis A vaccine development. Virus Res. 1990 Oct;17(2):75–92. doi: 10.1016/0168-1702(90)90070-r. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Maslova S. V., Agol V. I. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology. 1985 Dec;147(2):243–252. doi: 10.1016/0042-6822(85)90127-8. [DOI] [PubMed] [Google Scholar]
- Ticehurst J. R., Racaniello V. R., Baroudy B. M., Baltimore D., Purcell R. H., Feinstone S. M. Molecular cloning and characterization of hepatitis A virus cDNA. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5885–5889. doi: 10.1073/pnas.80.19.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Angel R. M., Papavassiliou A. G., Fernández-Tomás C., Silverstein S. J., Racaniello V. R. Cell proteins bind to multiple sites within the 5' untranslated region of poliovirus RNA. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8299–8303. doi: 10.1073/pnas.86.21.8299. [DOI] [PMC free article] [PubMed] [Google Scholar]





