Abstract
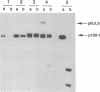
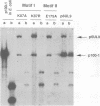
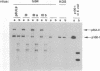
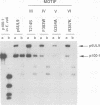
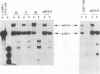
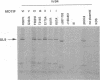
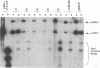
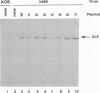
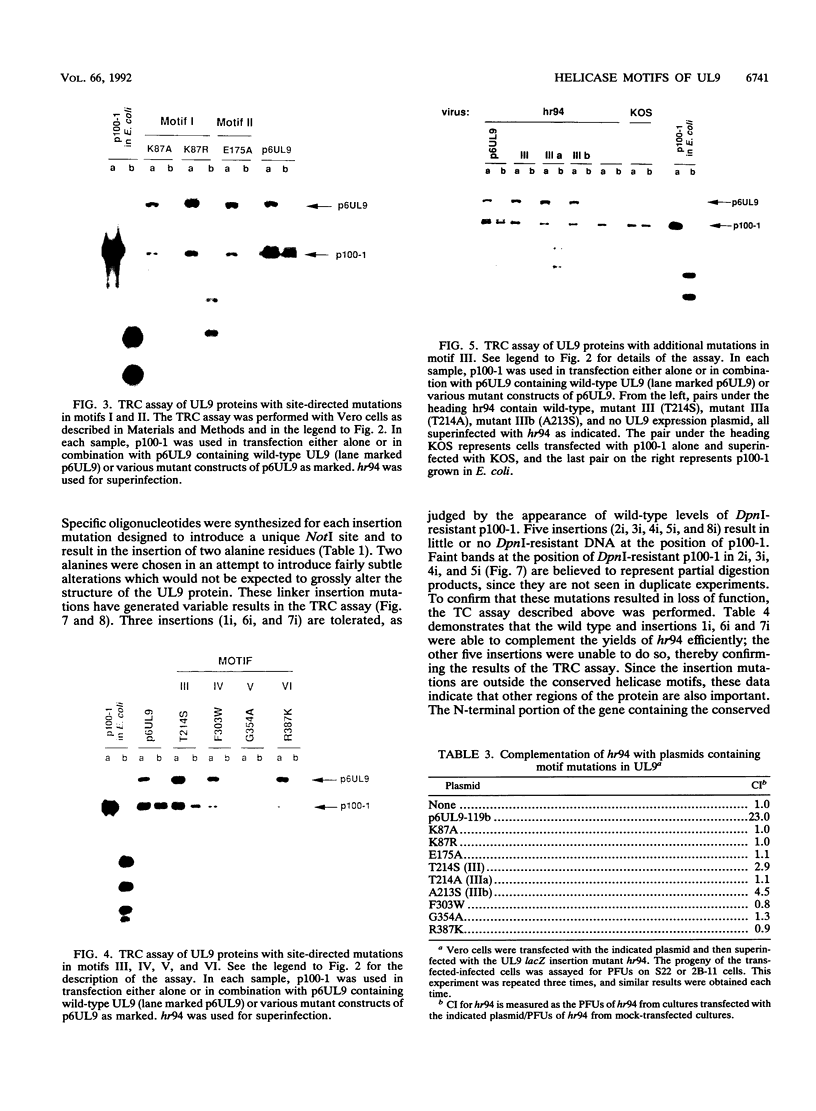
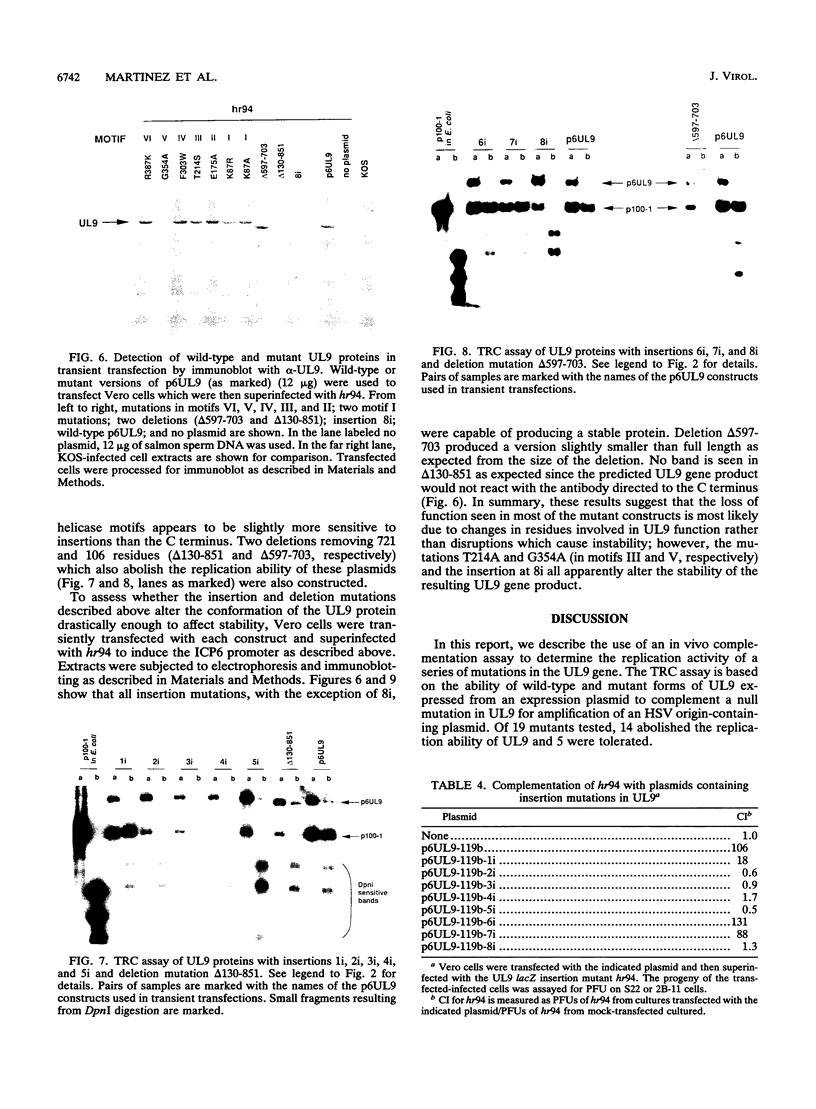
The UL9 gene of herpes simplex virus encodes a protein that specifically recognizes sequences within the viral origins of replication and exhibits helicase and DNA-dependent ATPase activities. The specific DNA binding domain of the UL9 protein was localized to the carboxy-terminal one-third of the molecule (H. M. Weir, J. M. Calder, and N. D. Stow, Nucleic Acids Res. 17:1409-1425, 1989). The N-terminal two-thirds of the UL9 gene contains six sequence motifs found in all members of a superfamily of DNA and RNA helicases, suggesting that this region may be important for helicase activity of UL9. In this report, we examined the functional significance of these six motifs for the UL9 protein through the introduction of site-specific mutations resulting in single amino acid substitutions of the most highly conserved residues within each motif. An in vivo complementation test was used to study the effect of each mutation on the function of the UL9 protein in viral DNA replication. In this assay, a mutant UL9 protein expressed from a transfected plasmid is used to complement a replication-deficient null mutant in the UL9 gene for the amplification of herpes simplex virus origin-containing plasmids. Mutations in five of the six conserved motifs inactivated the function of the UL9 protein in viral DNA replication, providing direct evidence for the importance of these conserved motifs. Insertion mutants resulting in the introduction of two alanines at 100-residue intervals in regions outside the conserved motifs were also constructed. Three of the insertion mutations were tolerated, whereas the other five abolished UL9 function. These data indicate that other regions of the protein, in addition to the helicase motifs, are important for function in vivo. Several mutations result in instability of the mutant products, presumably because of conformational changes in the protein. Taken together, these results suggest that UL9 is very sensitive to mutations with respect to both structure and function, perhaps reflecting its multifunctional character.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Al-Shawi M. K., Parsonage D., Senior A. E. Directed mutagenesis of the strongly conserved aspartate 242 in the beta-subunit of Escherichia coli proton-ATPase. J Biol Chem. 1988 Dec 25;263(36):19633–19639. [PubMed] [Google Scholar]
- Azzaria M., Schurr E., Gros P. Discrete mutations introduced in the predicted nucleotide-binding sites of the mdr1 gene abolish its ability to confer multidrug resistance. Mol Cell Biol. 1989 Dec;9(12):5289–5297. doi: 10.1128/mcb.9.12.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Bruckner R. C., Crute J. J., Dodson M. S., Lehman I. R. The herpes simplex virus 1 origin binding protein: a DNA helicase. J Biol Chem. 1991 Feb 5;266(4):2669–2674. [PubMed] [Google Scholar]
- Carmichael E. P., Kosovsky M. J., Weller S. K. Isolation and characterization of herpes simplex virus type 1 host range mutants defective in viral DNA synthesis. J Virol. 1988 Jan;62(1):91–99. doi: 10.1128/jvi.62.1.91-99.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael E. P., Weller S. K. Herpes simplex virus type 1 DNA synthesis requires the product of the UL8 gene: isolation and characterization of an ICP6::lacZ insertion mutation. J Virol. 1989 Feb;63(2):591–599. doi: 10.1128/jvi.63.2.591-599.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P., Crumpacker C. S., Schaffer P. A., Wilkie N. M. Physical and genetic analysis of the herpes simplex virus DNA polymerase locus. Virology. 1980 Jun;103(2):311–326. doi: 10.1016/0042-6822(80)90190-7. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Coen D. M., Aschman D. P., Gelep P. T., Retondo M. J., Weller S. K., Schaffer P. A. Fine mapping and molecular cloning of mutations in the herpes simplex virus DNA polymerase locus. J Virol. 1984 Jan;49(1):236–247. doi: 10.1128/jvi.49.1.236-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley A. J., Knipe D. M., Jones P. C., Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J Virol. 1981 Jan;37(1):191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute J. J., Tsurumi T., Zhu L. A., Weller S. K., Olivo P. D., Challberg M. D., Mocarski E. S., Lehman I. R. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Dean F. B., Bullock P., Murakami Y., Wobbe C. R., Weissbach L., Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci U S A. 1987 Jan;84(1):16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F. B., Dodson M., Echols H., Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., Deb S. P. A 269-amino-acid segment with a pseudo-leucine zipper and a helix-turn-helix motif codes for the sequence-specific DNA-binding domain of herpes simplex virus type 1 origin-binding protein. J Virol. 1991 Jun;65(6):2829–2838. doi: 10.1128/jvi.65.6.2829-2838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Dean F. B., Bullock P., Echols H., Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987 Nov 13;238(4829):964–967. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- Domier L. L., Franklin K. M., Shahabuddin M., Hellmann G. M., Overmeyer J. H., Hiremath S. T., Siaw M. F., Lomonossoff G. P., Shaw J. G., Rhoads R. E. The nucleotide sequence of tobacco vein mottling virus RNA. Nucleic Acids Res. 1986 Jul 11;14(13):5417–5430. doi: 10.1093/nar/14.13.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P., Gustafsson C. M., Hammarsten O. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oris. J Biol Chem. 1990 Oct 5;265(28):17167–17173. [PubMed] [Google Scholar]
- Elias P., Lehman I. R. Interaction of origin binding protein with an origin of replication of herpes simplex virus 1. Proc Natl Acad Sci U S A. 1988 May;85(9):2959–2963. doi: 10.1073/pnas.85.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fierer D. S., Challberg M. D. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J Virol. 1992 Jul;66(7):3986–3995. doi: 10.1128/jvi.66.7.3986-3995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M. L., Dorsky D. I., Crumpacker C. S., Parris D. S. The essential 65-kilodalton DNA-binding protein of herpes simplex virus stimulates the virus-encoded DNA polymerase. J Virol. 1989 Dec;63(12):5023–5029. doi: 10.1128/jvi.63.12.5023-5029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D. J., Weller S. K. An ICP6::lacZ insertional mutagen is used to demonstrate that the UL52 gene of herpes simplex virus type 1 is required for virus growth and DNA synthesis. J Virol. 1988 Aug;62(8):2970–2977. doi: 10.1128/jvi.62.8.2970-2977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D. J., Weller S. K. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology. 1988 Sep;166(1):41–51. doi: 10.1016/0042-6822(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. A conserved NTP-motif in putative helicases. Nature. 1988 May 5;333(6168):22–22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988 Aug 1;235(1-2):16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. One more conserved sequence motif in helicases. Nucleic Acids Res. 1988 Aug 11;16(15):7734–7734. doi: 10.1093/nar/16.15.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Gallagher M. P., Mimmack M. L., Pearce S. R. A family of closely related ATP-binding subunits from prokaryotic and eukaryotic cells. Bioessays. 1988 Apr;8(4):111–116. doi: 10.1002/bies.950080406. [DOI] [PubMed] [Google Scholar]
- Hloch P., Schiedner G., Stahl H. Complete cDNA sequence of the human p68 protein. Nucleic Acids Res. 1990 May 25;18(10):3045–3045. doi: 10.1093/nar/18.10.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman T. C. A new superfamily of replicative proteins. Nature. 1988 May 5;333(6168):22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Koff A., Schwedes J. F., Tegtmeyer P. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J Virol. 1991 Jun;65(6):3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A., Tegtmeyer P. Characterization of major recognition sequences for a herpes simplex virus type 1 origin-binding protein. J Virol. 1988 Nov;62(11):4096–4103. doi: 10.1128/jvi.62.11.4096-4103.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lane D. Enlarged family of putative helicases. Nature. 1988 Aug 11;334(6182):478–478. doi: 10.1038/334478a0. [DOI] [PubMed] [Google Scholar]
- Liu Q. Y., Summers W. C. Site-directed mutagenesis of a nucleotide-binding domain in HSV-1 thymidine kinase: effects on catalytic activity. Virology. 1988 Apr;163(2):638–642. doi: 10.1016/0042-6822(88)90308-x. [DOI] [PubMed] [Google Scholar]
- Marchetti M. E., Smith C. A., Schaffer P. A. A temperature-sensitive mutation in a herpes simplex virus type 1 gene required for viral DNA synthesis maps to coordinates 0.609 through 0.614 in UL. J Virol. 1988 Mar;62(3):715–721. doi: 10.1128/jvi.62.3.715-721.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz B., Subak-Sharpe J. H., Preston V. G. Physical mapping of temperature-sensitive mutations of herpes simplex virus type 1 using cloned restriction endonuclease fragments. J Gen Virol. 1983 Oct;64(Pt 10):2261–2270. doi: 10.1099/0022-1317-64-10-2261. [DOI] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Analysis of the essential and excision repair functions of the RAD3 gene of Saccharomyces cerevisiae by mutagenesis. Mol Cell Biol. 1986 Apr;6(4):1218–1227. doi: 10.1128/mcb.6.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo P. D., Nelson N. J., Challberg M. D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo P. D., Nelson N. J., Challberg M. D. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J Virol. 1989 Jan;63(1):196–204. doi: 10.1128/jvi.63.1.196-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula A. A., Sauer R. T. Genetic analysis of protein stability and function. Annu Rev Genet. 1989;23:289–310. doi: 10.1146/annurev.ge.23.120189.001445. [DOI] [PubMed] [Google Scholar]
- Parris D. S., Cross A., Haarr L., Orr A., Frame M. C., Murphy M., McGeoch D. J., Marsden H. S. Identification of the gene encoding the 65-kilodalton DNA-binding protein of herpes simplex virus type 1. J Virol. 1988 Mar;62(3):818–825. doi: 10.1128/jvi.62.3.818-825.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purifoy D. J., Lewis R. B., Powell K. L. Identification of the herpes simplex virus DNA polymerase gene. Nature. 1977 Oct 13;269(5629):621–623. doi: 10.1038/269621a0. [DOI] [PubMed] [Google Scholar]
- Purifoy D. J., Powell K. L. Temperature-sensitive mutants in two distinct complementation groups of herpes simplex virus type 1 specify thermolabile DNA polymerase. J Gen Virol. 1981 May;54(Pt 1):219–222. doi: 10.1099/0022-1317-54-1-219. [DOI] [PubMed] [Google Scholar]
- Rabkin S. D., Hanlon B. Nucleoprotein complex formed between herpes simplex virus UL9 protein and the origin of DNA replication: inter- and intramolecular interactions. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10946–10950. doi: 10.1073/pnas.88.23.10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Song J. M., Montelone B. A., Siede W., Friedberg E. C. Effects of multiple yeast rad3 mutant alleles on UV sensitivity, mutability, and mitotic recombination. J Bacteriol. 1990 Dec;172(12):6620–6630. doi: 10.1128/jb.172.12.6620-6630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Stahl H., Dröge P., Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986 Aug;5(8):1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg S. J., Kingsbury R. C., McKnight S. L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G., van Ham R. C., Vermeulen W., Bootsma D., van der Eb A. J., Hoeijmakers J. H. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990 Aug 24;62(4):777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- Weir H. M., Calder J. M., Stow N. D. Binding of the herpes simplex virus type 1 UL9 gene product to an origin of viral DNA replication. Nucleic Acids Res. 1989 Feb 25;17(4):1409–1425. doi: 10.1093/nar/17.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Carmichael E. P., Aschman D. P., Goldstein D. J., Schaffer P. A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987 Nov;161(1):198–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- Weller S. K., Lee K. J., Sabourin D. J., Schaffer P. A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983 Jan;45(1):354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988 Feb;62(2):435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohda M., Ohta S., Hisabori T., Kagawa Y. Site-directed mutagenesis of stable adenosine triphosphate synthase. Biochim Biophys Acta. 1988 Mar 30;933(1):156–164. doi: 10.1016/0005-2728(88)90065-5. [DOI] [PubMed] [Google Scholar]
- Zhu L. A., Weller S. K. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J Virol. 1992 Jan;66(1):469–479. doi: 10.1128/jvi.66.1.469-479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Weller S. K. UL5, a protein required for HSV DNA synthesis: genetic analysis, overexpression in Escherichia coli, and generation of polyclonal antibodies. Virology. 1988 Oct;166(2):366–378. doi: 10.1016/0042-6822(88)90507-7. [DOI] [PubMed] [Google Scholar]
- la Cour T. F., Nyborg J., Thirup S., Clark B. F. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO J. 1985 Sep;4(9):2385–2388. doi: 10.1002/j.1460-2075.1985.tb03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]