Abstract
Molecular switching and ligand-based modulation of the 90-kDa heat-shock protein (Hsp90) chaperone activity may ultimately facilitate conformational coupling to the ATPase cycle along with activation and recruitment of the broad range of client proteins. We present an atomic resolution analysis of the Hsp90 N-terminal domain (NTD) binding energy landscape by simulating protein dynamics with a range of binding partners. We show that the activity of the molecular chaperone may be linked to (i) local folding-unfolding transitions and conformational switching of the “active site lid” upon binding and (ii) differences in the underlying protein dynamics as a function of the binding partner. This study suggests that structural plasticity of the Hsp90 NTD can be exploited by the molecular chaperone machinery to modulate enhanced structural rigidity during ATP binding and increased protein flexibility as a consequence of the inhibitor binding. The present study agrees with the experimental structural data and provides a plausible molecular model for understanding mechanisms of modulation of molecular chaperone activities by binding partners.
Keywords: allosteric regulation, binding energy landscapes, protein dynamics, protein folding
Heat-shock protein 90 (Hsp90) is an ATPase-directed molecular chaperone required for the correct conformational development, stability, and function of >100 key cellular proteins (1–4). The repertoire of Hsp90 client proteins entails mainly growth-regulatory and signaling molecules, including kinases; transcription factors; and multiple mutated, chimeric, and overexpressed signaling proteins that promote cancer cell growth and survival (5–7). Hsp90 plays a pivotal role at the cross-roads of multiple signaling pathways associated with cell proliferation and cell viability, where up-regulation of Hsp90 activity may help promote tumor cell adaptation (8). Consequently, inhibition of the molecular chaperone's function has become a prominent strategy in the development of rational cancer therapeutics, in which targeted suppression of the ATPase activity with small molecule inhibitors (9–12) demonstrated anticancer activity in preclinical models and promising safety profile in humans (8, 11–13). Conformational coupling to the ATPase cycle, which requires N-terminal dimerization for ATP hydrolysis, necessitates functionally important conformational transitions of a contiguous segment of the NTD structure (residues 94–125 in yeast Hsp90) known as the “ATP-lid” or “active site lid,” a segment composed of two helices and the intervening loop located immediately adjacent to the ATP binding site (14). The crystal structures of the N domain and middle segment of Echerichia coli HtpG with the bound ADP [Protein DataBank (PDB) entries 1Y4S and 1Y4U] and the crystal structures of human Hsp90 complexes with ADP (PDB entry 1BYQ) (15) and geldanamycin (1YET) (16), used to model the complexes in this study, have unequivocally revealed that the “lid” segment projects out of the N-domain, in the “open” or “lid-up” conformation (17, 18). In contrast, the crystal structure of the ATP-bound conformation of full-length Hsp90 (PDB entry 2CG9) has unveiled that the lid segment of Hsp90 can be displaced from its position in the isolated Hsp90 NTD structure and folds over the nucleotide pocket to interact with the bound ATP, yielding the “closed” or “lid-down” conformation (14). The mechanism of conformational coupling to the ATPase cycle results in a “tense,” structurally rigid conformational state of Hsp90 upon ATP binding, whereas a subsequent hydrolysis to ADP leads to a more “relaxed,” structurally flexible state of Hsp90 (4, 17). Here, we used all-atom molecular dynamics (MD) simulations in explicit water on a long time scale to directly investigate the mechanism of ligand-based modulation of the Hsp90 NTD conformational dynamics at atomic resolution. A comparative analysis of the Hsp90 NTD dynamics and binding mechanisms has been performed based on simulations of the unbound Hsp90 NTD in solution (apo structure), and a range of Hsp90 complexes with different binding partners, including two natural substrates ATP and ADP [Fig. 1 a and b and supporting information (SI) Fig. S1], complexes with the inhibitors Shepherdin (19) (Fig. 1c), the minimal active sequence Shepherdin[79–83] (20), and the rationally identified small molecule inhibitor 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (AICAR) (Fig. S2) (21). We demonstrate that the molecular basis of the Hsp90 function and inhibition is largely determined by the mechanism in which the diversity of binding partners can modulate the conformational dynamics of Hsp90, linked to the functional activities of the molecular chaperone. This study provides atomic-level insights into the ligand-based mechanisms modulating molecular chaperone activity, which is important in regulation of the chaperone cycle (14).
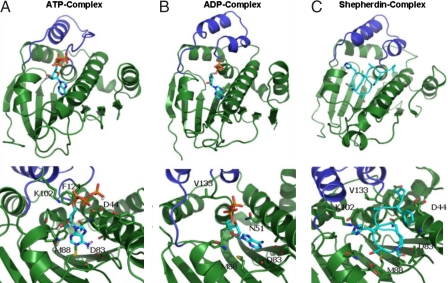
Fig. 1.
The representative structures of the most populated conformational clusters of Hsp90 NTD in complexes with different ligands emerged from simulations. (a) The Hsp90 NTD complex with ATP. (Upper) General view of the complex. (Lower) Close-up and a detailed view of the ligand interactions with the receptor. The stacking, attractive and repulsive interactions with F124, K102 and D44 are evident. (b) The Hsp90 NTD complex with ADP. (c) The Hsp90 NTD complex with Shepherdin. Dark blue, ATP-lid; light blue, ligand carbon atoms; red, oxygens; blue, nitrogens; orange, phosphorous.
Results and Discussion
Structural and Dynamical Analysis of the Hsp90 NTD Complexes.
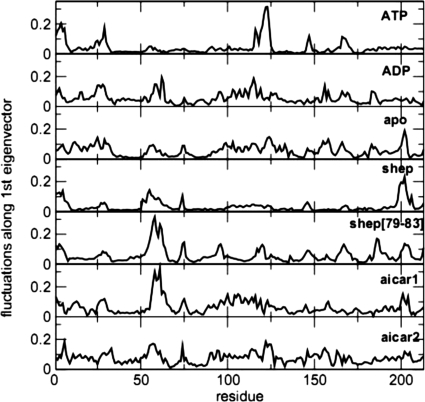
Structural and dynamic analysis of the MD trajectories obtained for the Hsp90 NTD complexes on a long time scale reveals that the most significant conformational variations involve the active site lid. A range of significant structural changes with respect to the initial open or lid-up conformation of the lid segment are discovered in simulations with the bound ATP (Fig. 1 and Fig. S3). Conformational transitions of the lid segment in the ATP bound complex result in an appreciable decrease of its α-helical content, featuring evolution toward a new configuration in which the lid plane lies perpendicular to its initial position. We find that the transition of the ATP-lid is induced by (i) the electrostatic interactions between the β-phosphate and the side chain of K102 (original PDB entry K112); (ii) the anion-π staking interactions (22, 23) between the ligand's α-phosphate oxygens and the aromatic ring of F124 (F134) (Fig. 1a). K102 and F124 are located respectively at the N and C termini of the lid, and the establishment of specific interactions with the natural ligand forces the rest of the lid to undergo extensive conformational restructuring, with a consequent increase of its flexibility. The ATP β and γ - phosphate groups determine electrostatic repulsion with the D44 side chain, located on the long helix 2. This interaction is, in turn, counterbalanced by an increase in the number of intradomain stabilizing interactions involving the Hsp90 helices 2 and 8. This analysis suggests that specific interactions that facilitate the loss of secondary structure in the lid are accompanied by a considerable concomitant consolidation of the Hsp90 NTD, stabilizing the domain in a more constrained, tense state. Essential dynamics (ED) analysis (24) was then used to further investigate the influence of this local conformational change on the global flexibility of the domain. ED identifies functionally relevant displacements of groups of residues and emphasizes the amplitude and direction of dominant protein motions by concentrating on a subset of the principal eigenvalues and eigenvectors of the residue pair covariance matrix calculated from MD. The trajectory of the ATP-Hsp90 NTD complex was thus projected onto the first eigenvector and the residue-based root mean fluctuations (rmsf spectrum) along this main direction were calculated (Fig. 2). The active site lid segment appears to concentrate most of the essential motions in the ATP-Hsp90 NTD complex. The rest of the rmsf spectrum appears to be quite flat, with the exception of the first 25 N-terminal residues. The ATP-lid and N-terminal region are, thus, the only two significantly flexible regions, suggesting once more a globally constrained state of the domain in the ATP-bound complex (Figs. 1 and 2 and Fig. S4). Analysis of the 3D structures obtained from the trajectory projection on the first eigenvector shows that the main motion swings the lid from the open conformation along the direction leading to the closed conformation, accompanied by extensive fluctuations of the N-terminal 1–25 region. The dynamic coupling of these two regions of the protein has important functional significance as shown by recent structural and biochemical data. Richter and coworkers (18), using NMR relaxation techniques, showed that removal of the first 24 residues of Hsp90 NTD perturbs the dynamical properties particularly of the ATP-lid region. Interestingly, the Δ1–24 deletion results in the complete loss of the ATPase activity, showing that reactivity of the chaperone depends on the intradomain dynamical coupling of the N-terminal and ATP-lid regions of Hsp90 (18). It is worth noting that ATP-induced conformational changes have been also observed in GRP94, the endoplasmatic reticulum paralog of Hsp90 (25). In this case, ATP binding was shown to induce dramatic conformational rearrangements in the helices 1, 4, and 5 (corresponding to the N-terminal helix and the ATP-lid), which in turn resulted in the assembly of a second NTD in a functional dimer (25). In contrast, we have found that deletion of the terminal phosphate group in the ADP complex and binding with the inhibitors can minimize repulsive interactions (Fig. 1 b and c), and thereby lead to profound differences in local and global conformational dynamics of Hsp90. The lid in the Hsp90 NTD complexes with ADP and studied inhibitors populates mainly α-helical conformations (Fig. 1 and Fig. S4). According to the results, Hsp90 NTD binding with the inhibitors can block the large fluctuations of the lid induced by ATP binding, necessary for the activation of Hsp90 dimerization and chaperone function. The conformational transition and switching of the lid segment, modulated by the inhibitor binding, locks the open or lid-up conformation (14), which impedes lid folding over the nucleotide binding site conformations (Fig. 1 and Fig. S4). The differences in the dynamics of the active site lid observed in the inhibitor and ADP-bound complexes with respect to the ATP-bound complex are also reflected at the level of global flexibility. Indeed, the analysis of residue-based rmsf after projection along the main ED eigenvector indicates that in the complexes with ADP and bound inhibitors the dynamic motions are more evenly distributed along the entire protein domain, with only several regions fluctuating appreciably along the principal direction of motion (Fig. 2). Hence, the unifying characteristic of the Hsp90 binding-coupled dynamics is the increasing global flexibility of the domain during the hydrolysis of ATP to ADP or binding with the active inhibitors, where the conformational equilibrium of Hsp90 NTD tends to be shifted toward more relaxed, nonfunctional states.
Fig. 2.
Flexibility of different complexes from MD simulations. The residue-based root mean square fluctuation (rmsf) was calculated after projecting each trajectory along the respective first principal component calculated through ED analysis.
Functional Analysis of the Conformational Transitions in the Hsp90 Complexes.
The results of simulations suggest the existence of important and functionally relevant differences between conformational dynamics of the Hsp90 NTD bound to ATP, compared with a dynamical signature of the protein in complexes with other ligands and the apo structure (Fig. S4). We find that the differences in the underlying protein dynamics as a function of the binding partner may be largely determined by conformational switching of the active site lid. Structural and biochemical experiments have suggested that the transitions from the lid-up conformation to the lid-down conformation, observed in the full-length dimer complex with AMPPNP and p23, are important in regulating ATPase-coupled chaperone activity (14, 18). The inhibitor-stabilized open conformation of the segment would sterically collide with its equivalent in a second Hsp90 NTD monomer and with the contact region of the middle domain during formation of the active full-length Hsp90 dimer, resulting in a loss of chaperone function. These data have suggested that the lid may act as an intrinsic kinetic inhibitor preventing Hsp90 dimerization (Fig. S3). Stabilization of the open conformation in the apo form or by ADP may have further functional implications in the client loading and client release steps. In the client loading step, the active full-length dimer is stabilized by contacts involving only the C-terminal domains favoring a rather extended conformation efficient at binding client proteins (17). In the release step, the compact state of the client-bound conformation must undergo relaxation after ATP hydrolysis to ADP to complete the release of the folded substrate. The dynamic signature of the Hsp90 NTD in complexes with ADP and the inhibitors, which favors open conformations of the lid segment, may have an important functional role in the regulation of these steps. In the client loading phase (apo protein), the destabilization of contacts with a second Hsp90 NTD would favor the extended conformation of the full-length dimer. In the release phase (ADP bound), the increase of steric hindrance in the putative compact dimer could spark the disruption of the compact client-bound complex.
Ligand Modulation of the Global Conformational Dynamics of Hsp90 N-Terminal Domain.
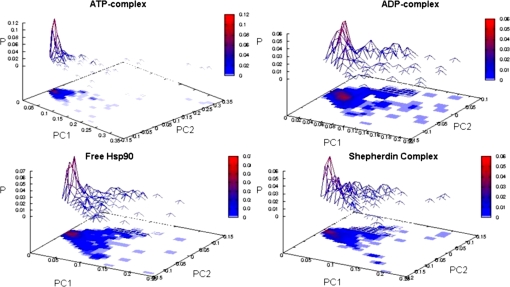
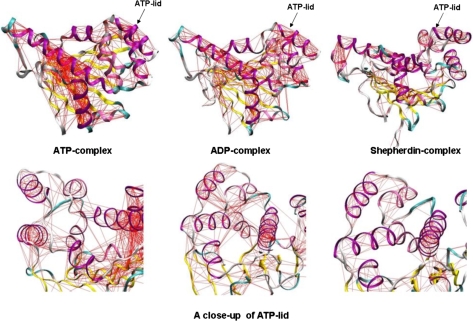
The results suggest that local conformational transitions of the lid segment, which is modulated by binding to different partners, may be intimately coupled to, and thus regulate, conformational dynamics of the whole Hsp90 molecular chaperone. The question to address is whether it is possible to not only analyze the variation of flexibility in other regions as we have reported above but also pinpoint protein regions whose dynamics is correlated with that of the active site lid and identify global dynamic differences among different complexes as a function of the ligand identity. A convenient framework for the identification of concerted nonrandom structural fluctuations in a protein is the analysis of the degree of covariance of pairs of residues. The covariance matrices for Hsp90 NTD are calculated from each simulation. First, we generated approximate free energy surfaces by projecting each trajectory on the “essential plane” defined by the two principal components with the highest eigenvalues calculated from the ED analysis (Fig. 3). This analysis reveals that the global dynamics of the ATP-bound complex is limited to a much narrower portion of this free energy surface compared with the other complexes and to the unliganded Hsp90 NTD. The full covariance matrix can be used to identify global dynamic properties of the protein and the specific regions that are involved in correlated motions (26). In this method, covariance web plots of the Hsp90 NTD complexes with binding partners are generated via drawing a line between pairs of atoms with a correlation coefficient higher than a given threshold of 0.5 (Fig. 4). This threshold typically guarantees that the majority of secondary structure elements are included in the analysis of correlated motions. The covariance web of the ATP-complex represents an extreme case, in which the connections extend well beyond secondary structure elements and involve most of the protein domain elements, particularly revealing the correlation between the N-terminal sequence (strand 1 and helix 1, residues 1–25) and the active site lid. The intramolecular couplings also involve the principal helix 2, helix 8, and the β-sheet motif, defining the basis of the ATP binding pocket. In contrast, for the Hsp90 complex with Shepherdin, only secondary structure elements are highly connected, and no correlation line can be seen between different elements of a secondary structure, in particular between the active site lid and the N-terminal region (Fig. 4). The corresponding covariance web plots for the ADP-complex and unliganded N-terminal Hsp90 reveal some extra connections between residues that belong to different secondary structure elements (Fig. 4). The covariance connections for the ATP-complex extend well beyond the secondary structure and local elements, which shows that the ATP-complex has a more diffuse and stronger interaction network in a sharp contrast with all other cases analyzed here. Despite a loss of secondary structure content in the ATP-lid segment, an overall increase in the structural rigidity and a globally more ordered state is a fundamental characteristic of the Hsp90 complex with ATP. These results suggest that structural plasticity of the Hsp90 NTD can be exploited by the molecular chaperone machinery to modulate enhanced structural rigidity during ATP binding and increased protein flexibility as a consequence of the inhibitor binding.
Fig. 3.
Energy landscape for different complexes from MD simulations. The free energy surfaces were obtained by projecting each trajectory on the essential plane defined by the two principal components with the highest eigenvalues calculated from the ED analysis of the respective simulation.
Fig. 4.
Ligand-modulation of protein dynamics. The pairwise covariance matrix accounting for coherent motions in the protein is represented as a web plot on the representative structure from each simulation. The matrix has been computed only considering the Cα atoms. The red lines connect pairs of atoms with a pairwise covariance >0.5.
Global Flexibility Analysis and Topological Similarity of the Hsp90 Dynamical Spaces.
The similarity and differences among the essential subspaces of the N-terminal Hsp90 bound to different ligands were calculated by using the RWSIP parameter. The similarity of the essential subspaces is reported in Tables S1 and S2 and Fig. S5a. According to this metric, the dynamics of ATP-Hsp90 complex is distant from all other complexes with the bound inhibitors and from the apo Hsp90 NTD. Within the limits of this approximation, the inhibitor bound complexes display similar essential subspaces. Interestingly, the two AICAR-bound complexes are the most distant from the ATP complex, despite spanning different subspaces with respect to each other. The Shepherdin complex yields a dynamical behavior that is very similar to that of the apo structure and is rather close to all complexes. Interestingly, the essential dynamical space of the uncomplexed Hsp90 NTD is shared in the “neighbor list” of Hsp90 NTD complexes with different binding partners. Indeed, the dynamics of the apo Hsp90 is characterized by a moderate α-helical content in the ATP-lid region compared with that of the ATP complex. According to the global flexibility analysis, the essential dynamical spaces of the ATP and ADP complexes are very different, although their respective structures from simulations may be highly similar. The emerging picture is that the activity of the molecular chaperone may be critically linked with the differences in the protein conformational dynamics specifically modulated by the binding partners. To shed light on this point, we have calculated the global flexibility of Hsp90 NTD bound to different ligands. The global flexibility parameter is defined as the sum of the average fluctuations of Cα atoms during the molecular dynamics trajectory. In the presence of ATP, the Hsp90 NTD shows a remarkable rigidity in comparison with the other complexes. In particular, small molecule inhibitors enhance the global flexibility of the domain also with respect to the apo and the ADP bound case (Table S3 and Fig. S5b). To further corroborate these findings, we calculated the approximate configurational entropy from the mass-weighted covariance matrix, using Schlitter's approximation (27). The configurational entropy is minimal for the ATP complex, whereas it strongly increases for the unbound protein and even more strongly increases in the presence of ADP or of the inhibitors. Altogether, these data suggest that, globally, the dynamics of the ATP-bound complex is limited to a much narrower portion of the available states on the free energy surface for Hsp90 compared with all of the other complexes and to the unliganded N-terminal Hsp90. This picture is consistent with the notion that functionally coupled ATPase cycle is determined by binding of ATP, stabilizing a “tense” conformational state of Hsp90, so that subsequent hydrolysis to ADP destabilizes that conformation and allows the Hsp90 dimer to relax to a default state. Hence, the ATP-bound state appears to be a constrained state, in which the relevant dynamics involves the structural remodeling motions of the active site lid and the N-terminal portion forming a coupled conformational switch.
Energy Landscape Model of Ligand-Based hsp90 NTD Modulation.
According to the energy landscape theory, coupling between folding and binding is often accompanied by conformational transitions associated with the biological functions of proteins and, as such, are intimately connected to the underlying energy landscape (28–33). A “conformational selection” model of biomolecular binding based on the energy landscape theory implies that conformational flexibility of the binding partners and statistical characterization of the binding mechanism may be determined by the dynamic equilibrium of the protein and the ligand conformational states, which may be shifted upon binding toward thermodynamically most stable complexes (34). The results of MD simulations suggest that energy landscape of the apo Hsp90 NTD may be populated by structurally different conformational states, featuring local conformational switching of the ATP-lid, which is accessible on longer time scales. According to the energy landscape model of binding, we conjecture that apo Hsp90 NTD may absorb conformational states with distinct lid conformations, and dynamic equilibrium between conformational states of the apo Hsp90 NTD can be modulated by ligand binding, leading to diversity of functionally relevant complexes. To test the feasibility of the energy landscape model assuming equilibrium between functionally important conformational states in the native Hsp90, we have also conducted longer MD simulations of the apo Hsp90 with implicit solvent. Importantly, we have observed the existence of spontaneous and reversible conformational transitions of the lid in which this segment can restructure from its initial position, effectively folding over the ATP-binding pocket (Fig. S6). Consequently, Hsp90 NTD may not have a single relaxed conformation but exists as a continuum of rather flexible conformations. By contrast, the ATP-bound state is a highly constrained structure whose formation involves coupled conformational switches in the N terminus and lid of the N-terminal domain. In this framework, binding of different ligands modulates the degree of local energetic frustration of Hsp90 NTD surface regions important for function, thus modulating the dynamics of protein–protein complex formation (35).
The results of this study suggest that flexible Hsp90 complexes with the inhibitors may have a more rugged bottom of the binding energy landscape with the low barriers between structurally similar conformers of the complex. In contrast, ligand-based modulation of the Hsp90 binding energy landscape can result in a dramatically different and a highly specific complex with ATP, which is considerably more rigid, featuring a steep funnel of conformations leading to the tense, native structure. According to the proposed model, the ligand-based modulation of the Hsp90 dynamics can induce functionally important conformational transitions between these tense and relaxed protein states, thereby providing a mechanism for allosteric regulation and inhibition.
Conclusions
The results of this study suggest that the activity of the Hsp90 molecular chaperone may be linked to (i) local conformational transitions and switching of the N-terminal lid upon binding and (ii) notable differences in the underlying protein dynamics as a function of the binding partner. The emergence of ligand-based modulation of the Hsp90 N-domain conformational dynamics supports the mechanism of the active site lid as a nucleotide sensitive conformational switch of the molecular chaperone activity. Hence, the local energetic frustration inherently present in a folded Hsp90 N-terminal may allow a considerable functional diversity of the protein dynamics, which is modulated by the presence of the specific binding partner. The present study agrees with the experimental structural data and provides a plausible molecular model for understanding mechanisms of modulation of molecular chaperone activities by binding partners.
Methods
Molecular Dynamics of Hsp90 Complexes.
All-atom molecular dynamics (MD) simulations in explicit water have been independently carried out on a long simulation time scale of at least 70 ns for (i) the unbound apo form of the Hsp90 NTD in solution, (ii) Hsp90 NTD complexes with the natural substrates ATP and ADP, and (iii) Hsp90 NTD complexes with the active inhibitors. The crystal structure of human Hsp90 in complex with ADP, solved by Obermann and coworkers (PDB entry 1BYQ) (15), was used directly in the simulations of the ADP-Hsp90 NTD complex. In the absence of crystal structures for the unliganded human Hsp90 NTD and cocrystals of the nucleotide bound with the isolated Hsp90 NTD, the ATP complex was initially modeled starting from the ADP complex by adding one phosphate group and minimizing the resulting complex with the Macromodel program (36). The apo Hsp90 NTD structure was respectively generated from the ADP-Hsp90 NTD complex by removing the ligand from the active site. The important rationale behind these choices was to consistently simulate and analyze dynamics of the Hsp90 conformational transitions and monitor the mechanism of lid refolding, because the system moves from ADP to ATP and apo forms. Indeed, the subsequent crystal structures of the ATP-bound full-length dimer have revealed a major conformational change, involving swapping of the N-terminal chain and lid refolding on the ATP-binding site. The comparison of our modeled Hsp90 NTD structures with the crystal structures of GRP94, the ER paralog of Hsp90, in the complex with ATP (PDB entry 1TC0) (25), the apo form (PDB entry 2YT2) (37), and the structure of yeast Hsp90 (PDB entry 1AH6) (38) has confirmed that in the isolated N-domain the lid is in the open conformation. The inhibitor bound complexes were obtained starting from the complex of human Hsp90 in complex with geldanamycin (PDB entry 1YET) (16), removing the inhibitor and docking Shepherdin, Shepherdin[79–83] or AICAR as described in refs. 19–21 and SI Text. MD simulations of the Hsp90 complexes have reproduced the salient molecular determinants of Hsp90 inhibition and assisted in the successful design of novel Hsp90 inhibitors (20, 21). All simulations and the analysis of the trajectories were performed by using the GROMACS software package (39), using the GROMOS96 force field (40, 41) and the SPC water model (42). MD simulations have always been initiated from the open or lid-up conformation of the lid segment as determined in the crystal structures of Hsp90 complexes with ADP and geldanamycin (15–18). The details of the simulation set up and analysis are described in SI Text.
Essential Dynamics Analysis and Dynamics Similarity Metrics.
The covariance matrix for each of the simulated complexes of the Hsp90 NTD was built by averaging motions of Cα atoms deviating from the mean structure. Details of ED analysis can be found in SI Text. The essential dynamical spaces of Hsp90 NTD interacting with different binding partners were compared by the rooted weighted square inner product (RWSIP) (43):
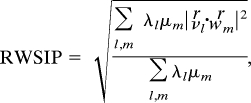 |
where λl and vl are the lth eigenvalue and lth eigenvector of the first system, and μm and wm are the mth eigenvalue and corresponding eigenvector for the second system. The similarities and differences among the essential subspaces spanned are analyzed by calculating RWSIP metric. By considering the RWSIP value as a distance parameter, it is possible to provide a quantitative measure of similarities and differences between dynamical subspaces, in particular by building a neighbor list describing the degree of similarity between dynamical spaces of the apo Hsp90 NTD and complexes with natural substrates and inhibitors.
Supplementary Material
Acknowledgments.
We thank Dr. G. Carrea for support. G.C. thanks Dr. F. Compostella for critical discussion and support. This work was supported by Sixth Research Framework Program of the European Union Grant LSHB-CT-2006-037325, Fondo per gli Investimenti della Ricerca di Base Program Grant RBNE03PX83, and the Ministero degli Esteri exchange program “Understanding the molecular determinants of amyloid fibril formation in human neurodegenerative diseases.”
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802879105/DCSupplemental.
References
- 1.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 3.Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- 4.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 5.Whitesell L, Lindquist SL. Hsp90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 6.Young JC, Moarefi I, Hartl FU. Hsp90: A specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neckers L. Heat shock protein 90: The cancer chaperone. J Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs JS, Xu WP, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 9.Janin YL. Heat Shock Protein 90 inhibitors. A text book example of medicinal chemistry? J Med Chem. 2005;48:7503–7512. doi: 10.1021/jm050759r. [DOI] [PubMed] [Google Scholar]
- 10.Vilenchik M, et al. Targeting wide-range oncogenic transformation via PU24FCL, a specific inhibitor of Tumor Hsp90. Chem Biol. 2004;11:787–797. doi: 10.1016/j.chembiol.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Powers MV, Workman P. Inhibitors of the heat shock response: Biology and pharmacology. FEBS Lett. 2007;581:3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 12.Workman P. Overview: Translating Hsp90 biology into Hsp90 drugs. Curr Cancer Drug Targets. 2003;3:297–300. doi: 10.2174/1568009033481868. [DOI] [PubMed] [Google Scholar]
- 13.Neckers L, Ivy SP. Heat Shock Protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Ali MMU, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stebbins CE, et al. Crystal structure of Hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 17.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E-coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 18.Richter K, et al. Intrinsic inhibition of the Hsp90 ATPase activity. J Biol Chem. 2006;281:11301–11311. doi: 10.1074/jbc.M510142200. [DOI] [PubMed] [Google Scholar]
- 19.Plescia J, et al. Rational design of Shepherdin, a novel anticancer agent. Cancer Cell. 2005;7:457–467. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 20.Gyurkocza B, et al. Antileukemic activity of shepherdin and molecular diversity of hsp90 inhibitors. J Natl Cancer Inst. 2006;98:1068–1077. doi: 10.1093/jnci/djj300. [DOI] [PubMed] [Google Scholar]
- 21.Meli M, et al. Small-molecule targeting of heat shock protein 90 chaperone function: Rational identification of a new anticancer lead. J Med Chem. 2006;49:7721–7730. doi: 10.1021/jm060836y. [DOI] [PubMed] [Google Scholar]
- 22.Gung BW, et al. Quantitative study of interactions between oxygen lone pair and aromatic rings: Substituent effect and the importance of closeness of contact. J Org Chem. 2008;73:689–693. doi: 10.1021/jo702170j. [DOI] [PubMed] [Google Scholar]
- 23.Dorn T, Janiak C, Abu-Shandi K. Hydrogen-bonding, pi-stacking and Cl-anion-π interactions of linear bipyridinium cations with phosphate, chloride and [CoCl4](2-) anions. CrystEngComm. 2005;7:633–641. [Google Scholar]
- 24.Amadei A, Linssen ABM, Berendsen HJC. Essential dynamics of proteins. Proteins Struct Funct Genet. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 25.Immormino RM, et al. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279:46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 26.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and hck underlies their inactivation by C-terminal tyrosine Phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 27.Schlitter J. Estimation of absolute and relative entropies of macromolecules using the covariance matrix. Chem Phys Lett. 1993;215:617–621. [Google Scholar]
- 28.Wolynes PG. Recent successes of the energy landscape theory of protein folding and function. Q Rev Biophys. 2005;38:405–410. doi: 10.1017/S0033583505004075. [DOI] [PubMed] [Google Scholar]
- 29.Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103:11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy Y, Cho SS, Onuchic JN, Wolynes PG. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J Mol Biol. 2005;346:1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 31.Levy Y, Wolynes PG, Onuchic JN. Protein topology determines binding mechanism. Proc Natl Acad Sci USA. 2004;101:511–516. doi: 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papoian GA, Wolynes PG. The physics and Bioinformatics of binding and folding - An energy landscape perspective. Biopolymers. 2003;68:333–349. doi: 10.1002/bip.10286. [DOI] [PubMed] [Google Scholar]
- 33.Miller DW, Dill KA. Ligand binding to proteins: The binding landscape model. Protein Sci. 1997;6:2166–2179. doi: 10.1002/pro.5560061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Verkhivker GM. Energy landscape theory, funnels, specificity, and optimal criterion of biomolecular binding. Phys Rev Lett. 2003;90:188181. doi: 10.1103/PhysRevLett.90.188101. [DOI] [PubMed] [Google Scholar]
- 35.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. Local frustration in native proteins and protein assemblies. Proc Natl Acad Sci USA. 2007;104:19819–19824. doi: 10.1073/pnas.0709915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrödinger . MacroModel, version 9.0. New York: Schrödinger; 2005. [Google Scholar]
- 37.Dollins DE, Immormino RM, Gewirth DT. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90: Basis for ligand induced conformational changes. J Biol Chem. 2005;280:30438–30447. doi: 10.1074/jbc.M503761200. [DOI] [PubMed] [Google Scholar]
- 38.Prodromou C, Roe SM, Pearl LH. Molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nat Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- 39.van der Spoel D, et al. Gromacs User Manual Version 3.2. 2004. Available at www.gromacs.org.
- 40.van Gunsteren WF, Berendsen HJC. GROMOS (GROningen MOlecular Simulation package) The Netherlands: Biomos BV; 1987. [Google Scholar]
- 41.van Gunsteren, et al. Biomolecular modeling: Goals, problems, perspectives. Angew Chem Int Ed. 2006;45:4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- 42.Berendsen HJC, Grigera JR, Straatsma PR. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 43.Carnevale V, Pontiggia F, Micheletti C. Structural and dynamical alignment of enzymes with partial structural similarity. J Phys Condens Matter. 2007;19:285206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.