Abstract
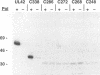
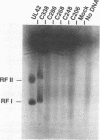
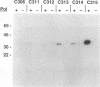
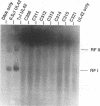
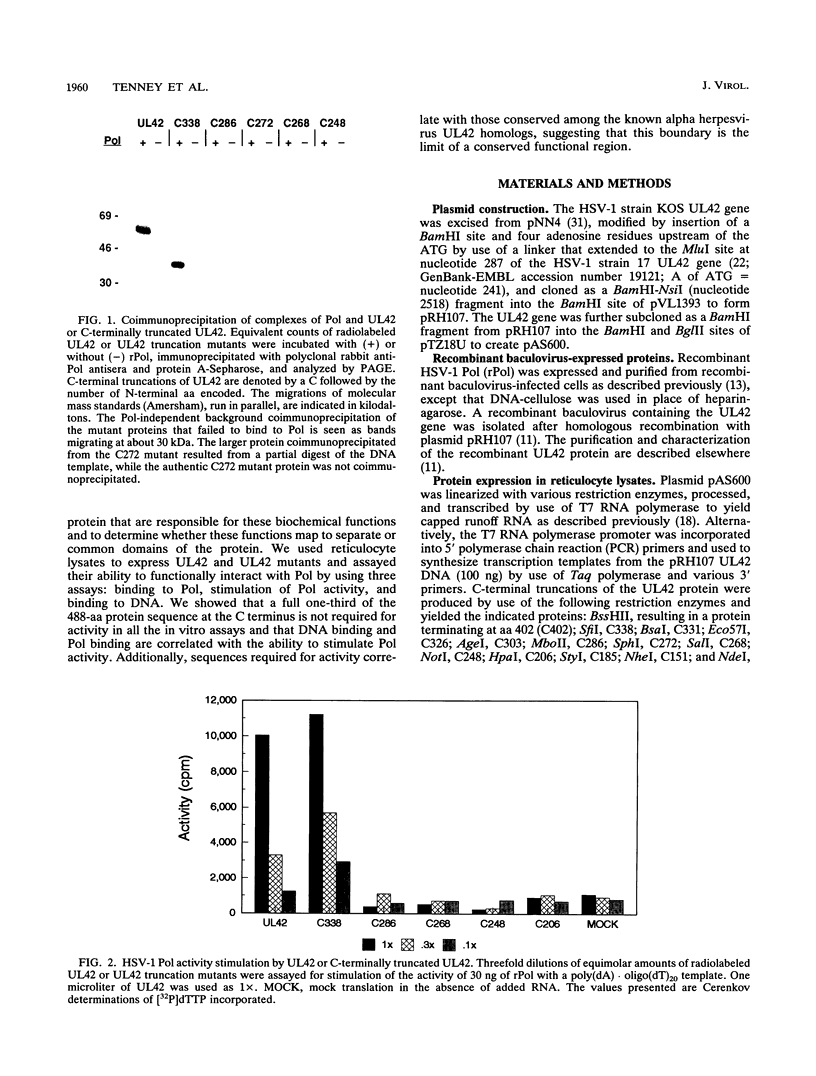
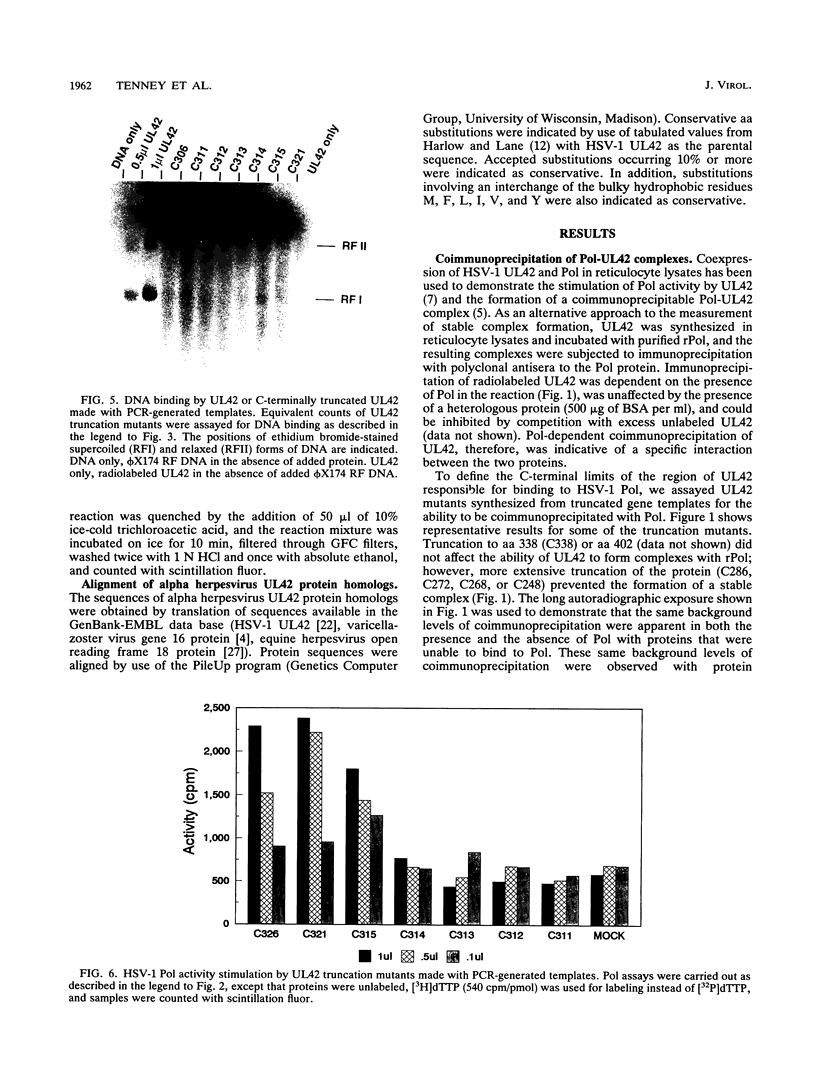
The herpes simplex virus type 1 UL42 protein was synthesized in reticulocyte lysates and assayed for activity in vitro. Three functional assays were used to examine the properties of in vitro-synthesized UL42: (i) coimmunoprecipitation to detect stable complex formation with purified herpes simplex virus type 1 DNA polymerase (Pol), (ii) a simple gel-based assay for DNA binding, and (iii) a sensitive assay for the stimulation of Pol activity. UL42 synthesized in reticulocyte lysates formed a stable coimmunoprecipitable complex with Pol, bound to double-stranded DNA, and stimulated the activity of Pol in vitro. Carboxy-terminal truncations of the UL42 protein were synthesized from restriction enzyme-digested UL42 gene templates and gene templates made by polymerase chain reaction and assayed for in vitro activity. Truncations of the 488-amino-acid (aa) UL42 protein to aa 315 did not abolish its ability to bind to Pol and DNA or to stimulate Pol activity. Proteins terminating at aas 314 and 313 showed reduced levels of binding to Pol, but these and shorter proteins were unable to bind to DNA or to stimulate Pol activity. These results suggest that all three of the biochemical functions of UL42 colocalize entirely within the N-terminal 315 aas of the UL42 protein. Amino acid sequence alignment of alpha herpesvirus UL42 homologs revealed that the N-terminal functional domain corresponds to the most highly conserved region of the protein, while the dispensable C terminus is not conserved. Conservative aa changes at the C terminus of the 315-aa truncated protein were used to show that conserved residues were important for activity. These results suggest that 173 aa of UL42 can be deleted without a loss of activity and that DNA-binding and Pol-binding activities are correlated with the ability of UL42 to stimulate Pol activity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Crute J. J., Lehman I. R. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5'----3' exonuclease with ribonuclease H activity. J Biol Chem. 1989 Nov 15;264(32):19266–19270. [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Digard P., Bebrin W. R., Weisshart K., Coen D. M. The extreme C terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J Virol. 1993 Jan;67(1):398–406. doi: 10.1128/jvi.67.1.398-406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digard P., Coen D. M. A novel functional domain of an alpha-like DNA polymerase. The binding site on the herpes simplex virus polymerase for the viral UL42 protein. J Biol Chem. 1990 Oct 15;265(29):17393–17396. [PubMed] [Google Scholar]
- Ertl P. F., Powell K. L. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J Virol. 1992 Jul;66(7):4126–4133. doi: 10.1128/jvi.66.7.4126-4133.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M. L., Dorsky D. I., Crumpacker C. S., Parris D. S. The essential 65-kilodalton DNA-binding protein of herpes simplex virus stimulates the virus-encoded DNA polymerase. J Virol. 1989 Dec;63(12):5023–5029. doi: 10.1128/jvi.63.12.5023-5029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M. L., Jackwood D. H., Murphy M., Marsden H. S., Parris D. S. Purification of the herpes simplex virus type 1 65-kilodalton DNA-binding protein: properties of the protein and evidence of its association with the virus-encoded DNA polymerase. J Virol. 1988 Aug;62(8):2874–2883. doi: 10.1128/jvi.62.8.2874-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Knipe D. M. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J Virol. 1991 May;65(5):2666–2675. doi: 10.1128/jvi.65.5.2666-2675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb J., Marcy A. I., Coen D. M., Challberg M. D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990 Dec;64(12):5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez T. R., Lehman I. R. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J Biol Chem. 1990 Jul 5;265(19):11227–11232. [PubMed] [Google Scholar]
- Johnson P. A., Best M. G., Friedmann T., Parris D. S. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J Virol. 1991 Feb;65(2):700–710. doi: 10.1128/jvi.65.2.700-710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl A., Dorsky D. I. Cooperation of EBV DNA polymerase and EA-D(BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991 Sep;184(1):330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Marchetti M. E., Smith C. A., Schaffer P. A. A temperature-sensitive mutation in a herpes simplex virus type 1 gene required for viral DNA synthesis maps to coordinates 0.609 through 0.614 in UL. J Virol. 1988 Mar;62(3):715–721. doi: 10.1128/jvi.62.3.715-721.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H. S., Campbell M. E., Haarr L., Frame M. C., Parris D. S., Murphy M., Hope R. G., Muller M. T., Preston C. M. The 65,000-Mr DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987 Aug;61(8):2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Dolan A., McNab D., Perry L. J., Taylor P., Challberg M. D. Structures of herpes simplex virus type 1 genes required for replication of virus DNA. J Virol. 1988 Feb;62(2):444–453. doi: 10.1128/jvi.62.2.444-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orberg P. K., Schaffer P. A. Expression of herpes simplex virus type 1 major DNA-binding protein, ICP8, in transformed cell lines: complementation of deletion mutants and inhibition of wild-type virus. J Virol. 1987 Apr;61(4):1136–1146. doi: 10.1128/jvi.61.4.1136-1146.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K. L., Purifoy D. J. Nonstructural proteins of herpes simplex virus. I. Purification of the induced DNA polymerase. J Virol. 1977 Nov;24(2):618–626. doi: 10.1128/jvi.24.2.618-626.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Sommer S. S. Access to a messenger RNA sequence or its protein product is not limited by tissue or species specificity. Science. 1989 Apr 21;244(4902):331–334. doi: 10.1126/science.2565599. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Herpes simplex virus type 1 origin-dependent DNA replication in insect cells using recombinant baculoviruses. J Gen Virol. 1992 Feb;73(Pt 2):313–321. doi: 10.1099/0022-1317-73-2-313. [DOI] [PubMed] [Google Scholar]
- Tenney D. J., Micheletti P. A., Stevens J. T., Hamatake R. K., Matthews J. T., Sanchez A. R., Hurlburt W. W., Bifano M., Cordingley M. G. Mutations in the C terminus of herpes simplex virus type 1 DNA polymerase can affect binding and stimulation by its accessory protein UL42 without affecting basal polymerase activity. J Virol. 1993 Jan;67(1):543–547. doi: 10.1128/jvi.67.1.543-547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan P. J., Purifoy D. J., Powell K. L. DNA-binding protein associated with herpes simplex virus DNA polymerase. J Virol. 1985 Feb;53(2):501–508. doi: 10.1128/jvi.53.2.501-508.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Nelson N. J., McGeoch D. J., Challberg M. D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988 Feb;62(2):435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]