Abstract
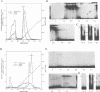
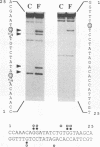
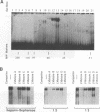
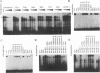
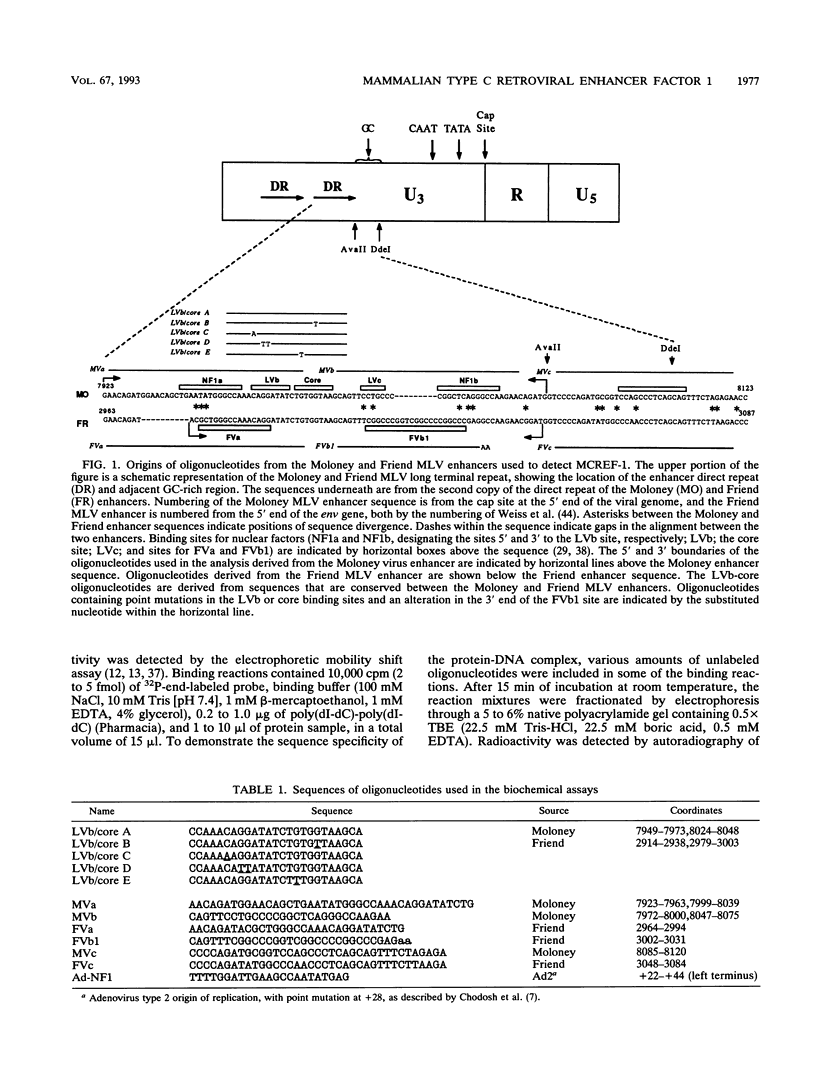
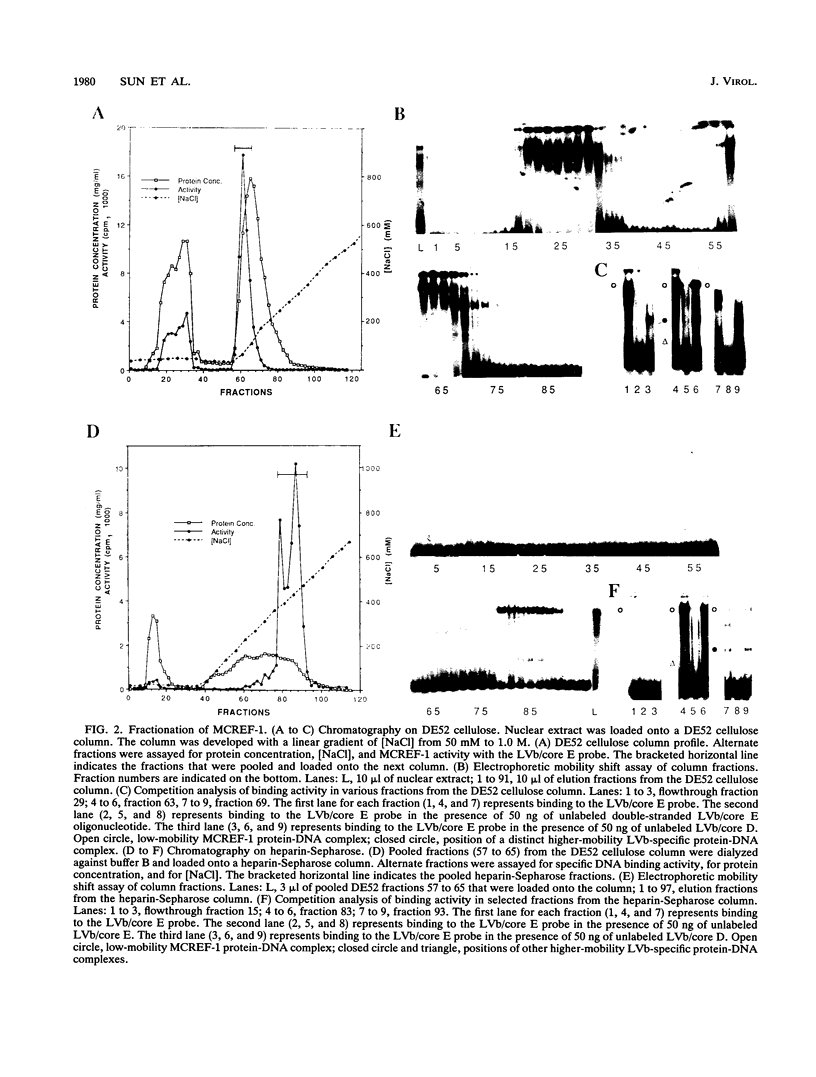
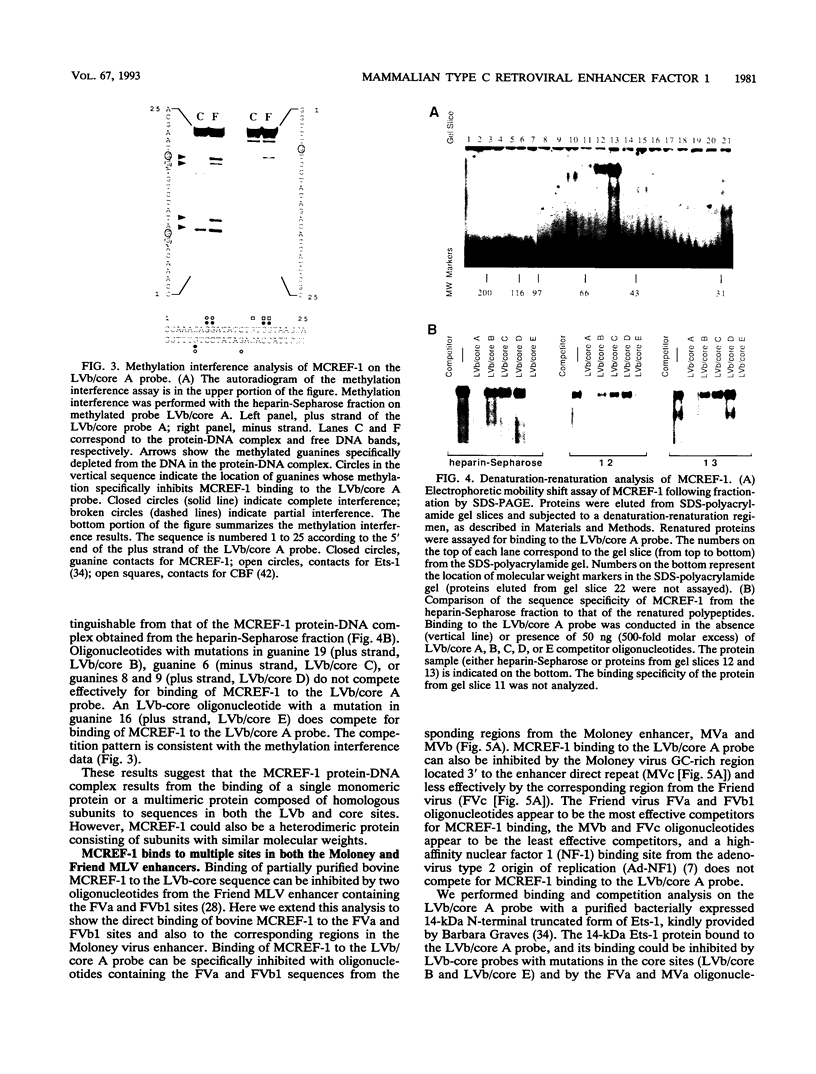
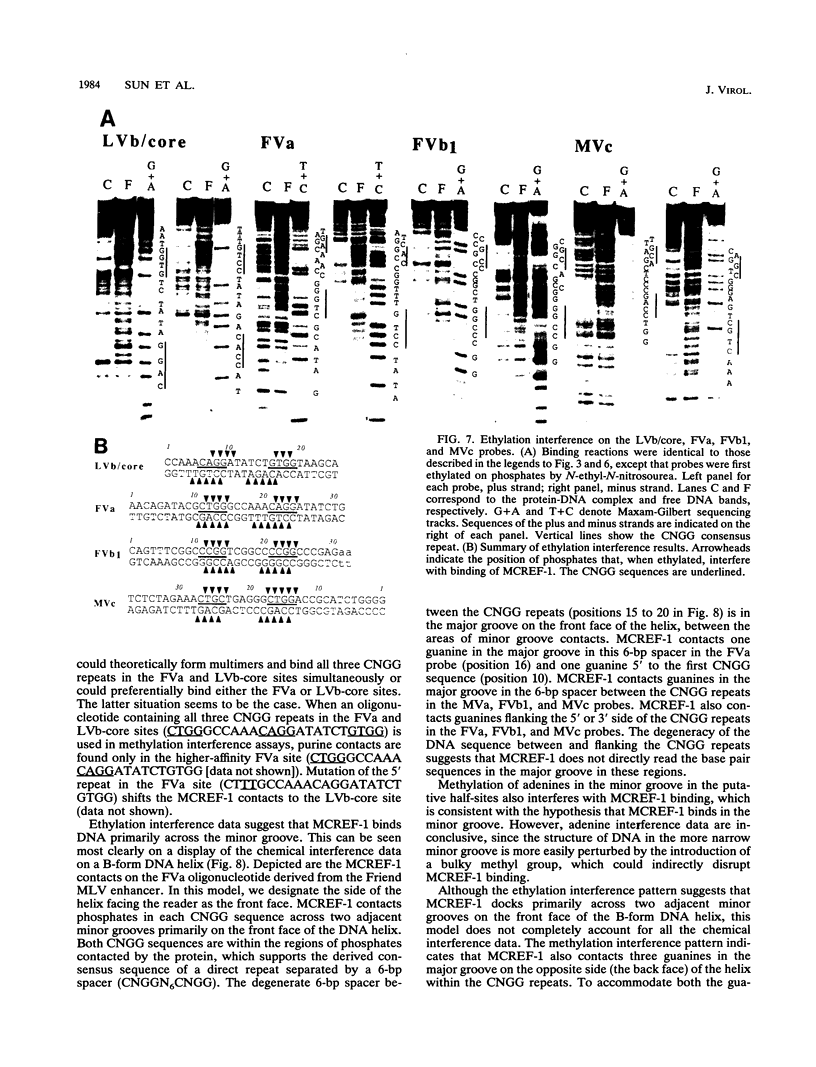
Mammalian type C retrovirus enhancer factor 1 (MCREF-1) is a nuclear protein that binds several directly repeated sequences (CNGGN6CNGG) in the Moloney and Friend murine leukemia virus (MLV) enhancers (N. R. Manley, M. O'Connell, W. Sun, N. A. Speck, and N. Hopkins, J. Virol. 67:1967-1975, 1993). In this paper, we describe the partial purification of MCREF-1 from calf thymus nuclei and further characterize the binding properties of MCREF-1. MCREF-1 binds four sites in the Moloney MLV enhancer and three sites in the Friend MLV enhancer. Ethylation interference analysis suggests that the MCREF-1 binding site spans two adjacent minor grooves of DNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. DNA binding properties of the purified Antennapedia homeodomain. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boral A. L., Okenquist S. A., Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989 Jan;63(1):76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Villemur R., Jolicoeur P. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3' end of env. J Virol. 1983 Jul;47(1):24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E. A., Speck N. A., Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990 Feb;64(2):534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hallberg B., Schmidt J., Luz A., Pedersen F. S., Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991 Aug;65(8):4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985 Feb;141(1):30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N. R., O'Connell M. A., Sharp P. A., Hopkins N. Nuclear factors that bind to the enhancer region of nondefective Friend murine leukemia virus. J Virol. 1989 Oct;63(10):4210–4223. doi: 10.1128/jvi.63.10.4210-4223.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N. R., O'Connell M., Sun W., Speck N. A., Hopkins N. Two factors that bind to highly conserved sequences in mammalian type C retroviral enhancers. J Virol. 1993 Apr;67(4):1967–1975. doi: 10.1128/jvi.67.4.1967-1975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mercurio F., Karin M. Transcription factors AP-3 and AP-2 interact with the SV40 enhancer in a mutually exclusive manner. EMBO J. 1989 May;8(5):1455–1460. doi: 10.1002/j.1460-2075.1989.tb03528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J. A., Graves B. J. Alkylation interference identifies essential DNA contacts for sequence-specific binding of the eukaryotic transcription factor C/EBP. Proc Natl Acad Sci U S A. 1990 May;87(10):3992–3996. doi: 10.1073/pnas.87.10.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Satake M., Inuzuka M., Shigesada K., Oikawa T., Ito Y. Differential expression of subspecies of polyomavirus and murine leukemia virus enhancer core binding protein, PEBP2, in various hematopoietic cells. Jpn J Cancer Res. 1992 Jul;83(7):714–722. doi: 10.1111/j.1349-7006.1992.tb01971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991 Jan;65(1):42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. W., Speck N. A. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992 Jan;12(1):89–102. doi: 10.1128/mcb.12.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]