Abstract
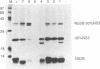
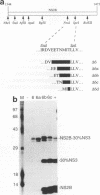
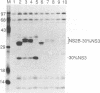
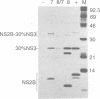
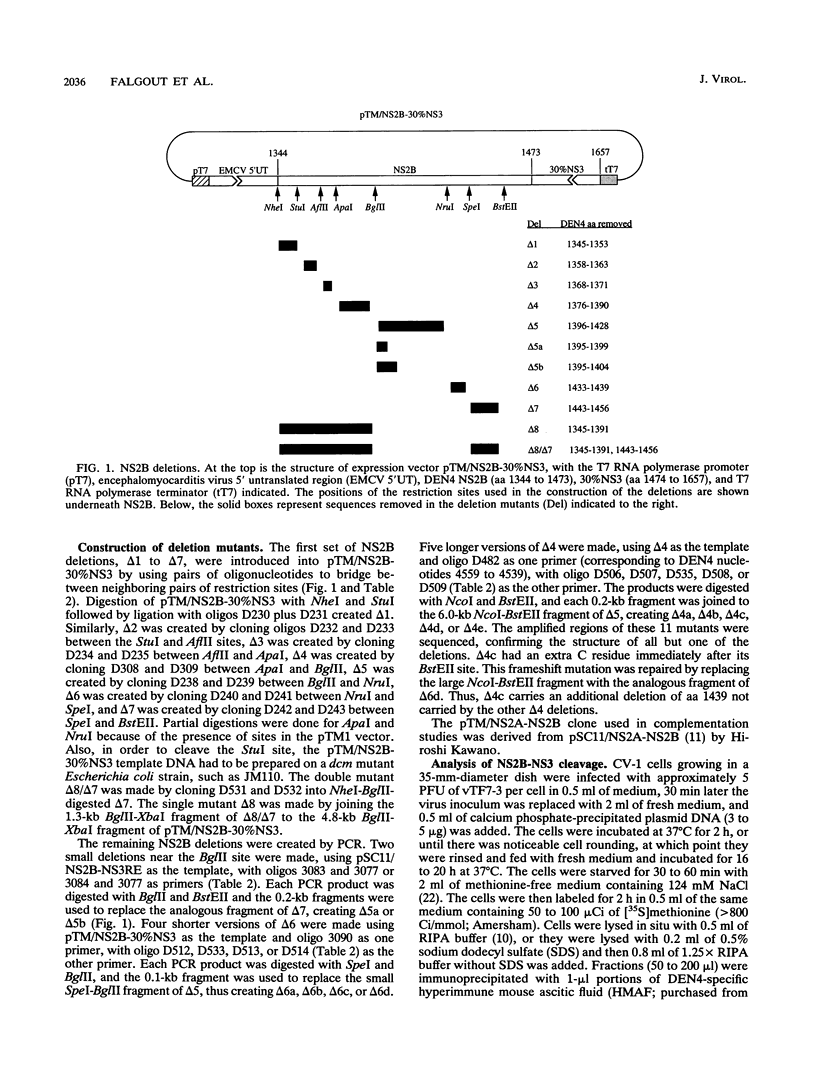
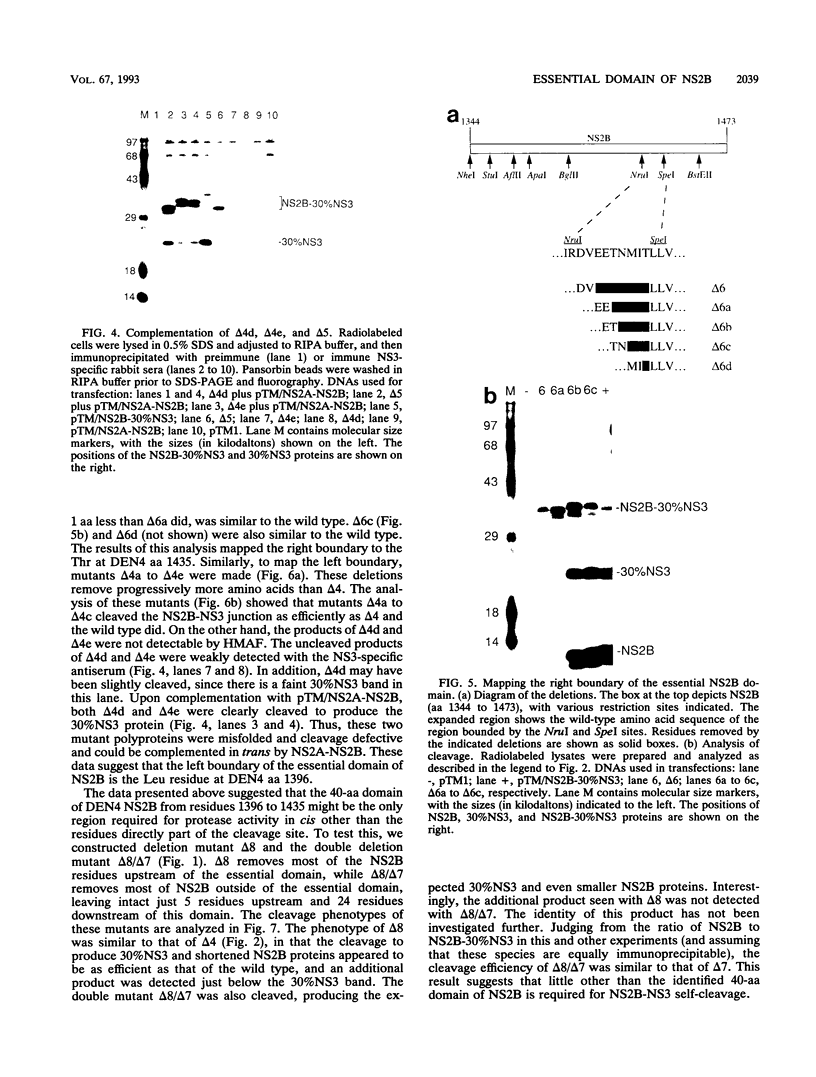
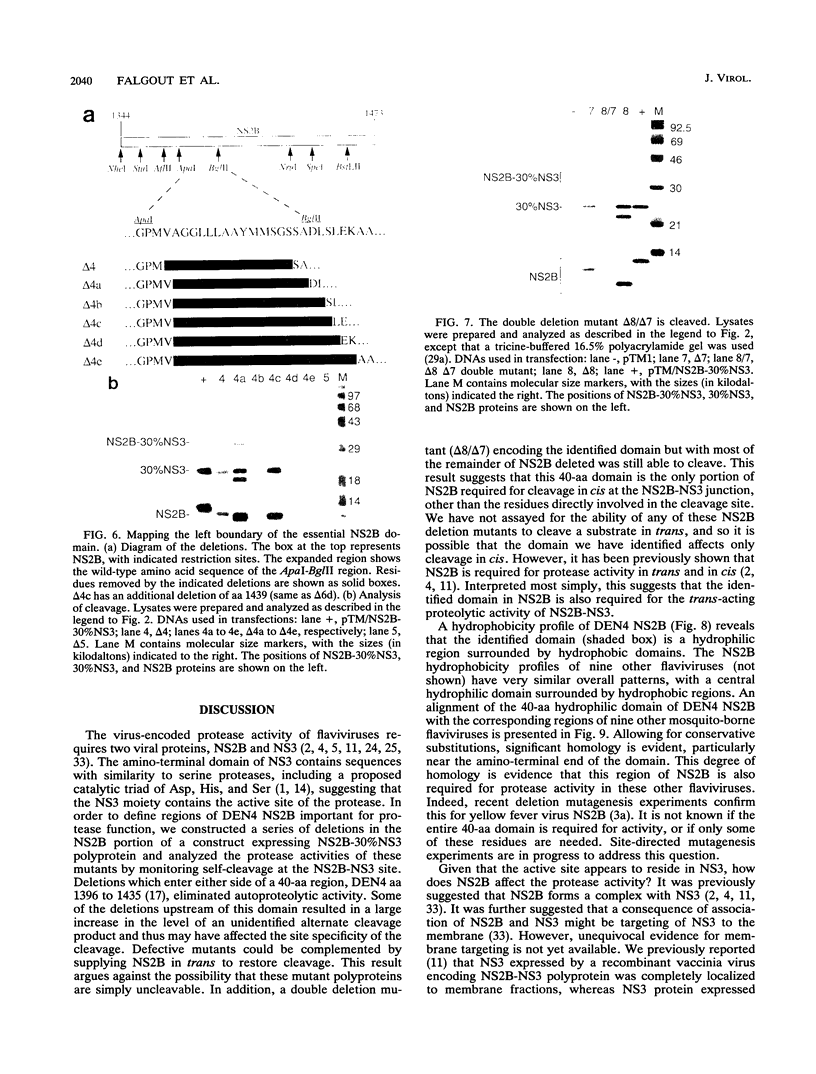
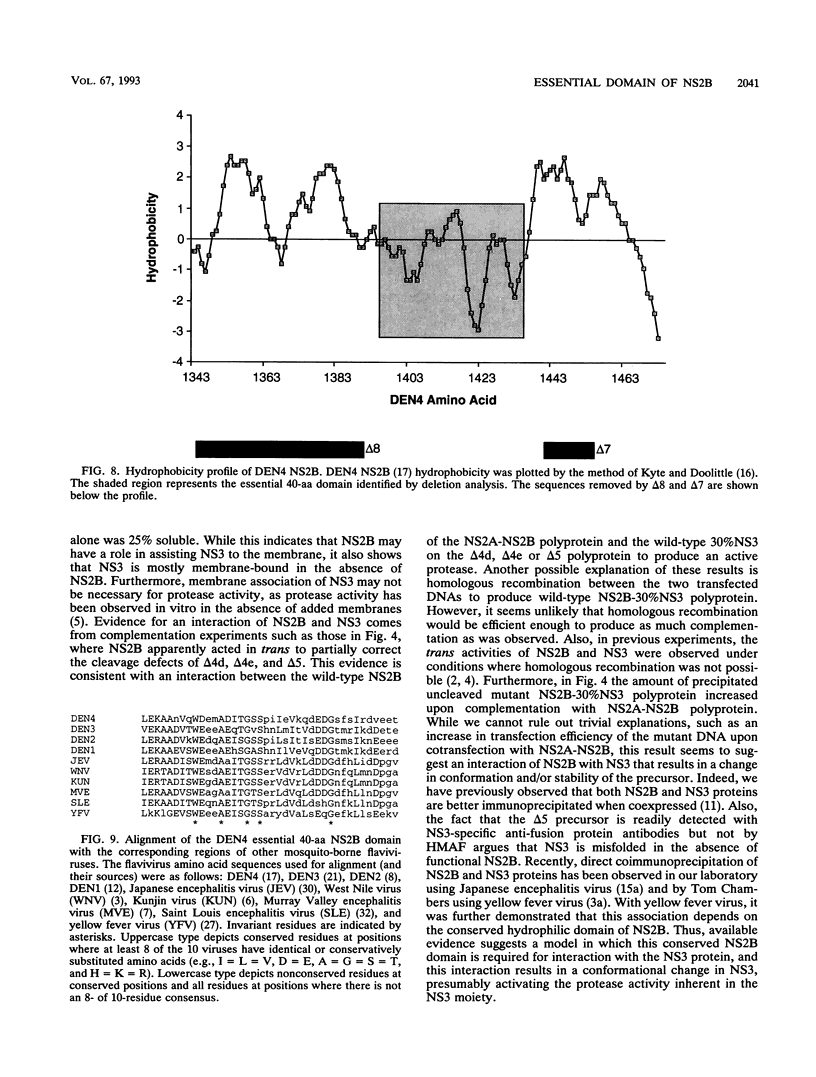
Most proteolytic cleavages in the nonstructural protein (NS) region of the flavivirus polyprotein are effected by a virus-encoded protease composed of two viral proteins, NS2B and NS3. The N-terminal 180-amino-acid-region of NS3 includes sequences with homology to the active sites of serine proteases, and there is evidence that this portion of NS3 can mediate proteolytic cleavages. In contrast, nothing is known about required sequences in NS2B. We constructed a series of deletion mutations in the NS2B portion of plasmid pTM/NS2B-30% NS3, which expresses dengue virus type 4 (DEN4) cDNA encoding NS2B and the N-terminal 184 residues of NS3 from the T7 RNA polymerase promoter. Mutant or wild-type plasmids were transfected into cells that had been infected with a recombinant vaccinia virus expressing T7 RNA polymerase, and the protease activities of the expressed polyproteins were assayed by examining the extent of self-cleavage at the NS2B-NS3 junction. The results identify a 40-amino-acid segment of NS2B (DEN4 amino acids 1396 to 1435) essential for protease activity. A hydrophobicity profile of DEN4 NS2B predicts this segment constitutes a hydrophilic domain surrounded by hydrophobic regions. Hydrophobicity profiles of the NS2B proteins of other flaviviruses show similar patterns. Amino acid sequence alignment of this domain of DEN4 NS2B with comparable regions of other proteins of flaviviruses indicates significant sequence conservation, especially at the N-terminal end. These observations suggest that the central hydrophilic domain of NS2B of these other flaviviruses will also prove to be essential for protease activity.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazan J. F., Fletterick R. J. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989 Aug;171(2):637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Cahour A., Falgout B., Lai C. J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992 Mar;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle E., Leidner U., Nowak T., Wengler G., Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986 Feb;149(1):10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia G., Parker M. D., Speight G., Byrne M. E., Westaway E. G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988 Jan;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- Dalgarno L., Trent D. W., Strauss J. H., Rice C. M. Partial nucleotide sequence of the Murray Valley encephalitis virus genome. Comparison of the encoded polypeptides with yellow fever virus structural and non-structural proteins. J Mol Biol. 1986 Feb 5;187(3):309–323. doi: 10.1016/0022-2836(86)90435-3. [DOI] [PubMed] [Google Scholar]
- Deubel V., Kinney R. M., Trent D. W. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology. 1988 Jul;165(1):234–244. doi: 10.1016/0042-6822(88)90677-0. [DOI] [PubMed] [Google Scholar]
- Elroy-Stein O., Fuerst T. R., Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5' sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C. J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J Virol. 1989 May;63(5):1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Tan B. H., Yap E. H., Chan Y. C., Tan Y. H. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90). Virology. 1992 Jun;188(2):953–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Donchenko A. P., Koonin E. V., Blinov V. M. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989 May 25;17(10):3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori H., Lai C. J. Cleavage of dengue virus NS1-NS2A requires an octapeptide sequence at the C terminus of NS1. J Virol. 1990 Sep;64(9):4573–4577. doi: 10.1128/jvi.64.9.4573-4577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Mackow E., Makino Y., Zhao B. T., Zhang Y. M., Markoff L., Buckler-White A., Guiler M., Chanock R., Lai C. J. The nucleotide sequence of dengue type 4 virus: analysis of genes coding for nonstructural proteins. Virology. 1987 Aug;159(2):217–228. doi: 10.1016/0042-6822(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Markoff L. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J Virol. 1989 Aug;63(8):3345–3352. doi: 10.1128/jvi.63.8.3345-3352.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Elroy-Stein O., Mizukami T., Alexander W. A., Fuerst T. R. Product review. New mammalian expression vectors. Nature. 1990 Nov 1;348(6296):91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Nowak T., Färber P. M., Wengler G., Wengler G. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989 Apr;169(2):365–376. doi: 10.1016/0042-6822(89)90162-1. [DOI] [PubMed] [Google Scholar]
- Osatomi K., Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990 Jun;176(2):643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- Pethel M., Falgout B., Lai C. J. Mutational analysis of the octapeptide sequence motif at the NS1-NS2A cleavage junction of dengue type 4 virus. J Virol. 1992 Dec;66(12):7225–7231. doi: 10.1128/jvi.66.12.7225-7231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat F., Lenches E. M., Strauss J. H. Flavivirus enzyme-substrate interactions studied with chimeric proteinases: identification of an intragenic locus important for substrate recognition. J Virol. 1991 Sep;65(9):4749–4758. doi: 10.1128/jvi.65.9.4749-4758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat F., Strauss J. H. Processing of nonstructural proteins NS4A and NS4B of dengue 2 virus in vitro and in vivo. Virology. 1991 Dec;185(2):689–697. doi: 10.1016/0042-6822(91)90540-r. [DOI] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph V. B., Winkler G., Stollar V. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology. 1990 Feb;174(2):450–458. doi: 10.1016/0042-6822(90)90099-d. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A., Cahour A., Després P., Girard M., Bouloy M. Processing of yellow fever virus polyprotein: role of cellular proteases in maturation of the structural proteins. J Virol. 1989 Oct;63(10):4199–4209. doi: 10.1128/jvi.63.10.4199-4209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi H., Mori C., Fuke I., Morita K., Kuhara S., Kondou J., Kikuchi Y., Nagamatu H., Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987 Dec;161(2):497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Ugarova T. Y., Chernovskaya T. V., Lyapustin V. N., Lashkevich V. A., Agol V. I. Translation of tick-borne encephalitis virus (flavivirus) genome in vitro: synthesis of two structural polypeptides. Virology. 1981 Apr 15;110(1):26–34. doi: 10.1016/0042-6822(81)90004-0. [DOI] [PubMed] [Google Scholar]
- Trent D. W., Kinney R. M., Johnson B. J., Vorndam A. V., Grant J. A., Deubel V., Rice C. M., Hahn C. Partial nucleotide sequence of St. Louis encephalitis virus RNA: structural proteins, NS1, ns2a, and ns2b. Virology. 1987 Feb;156(2):293–304. doi: 10.1016/0042-6822(87)90409-0. [DOI] [PubMed] [Google Scholar]
- Wengler G., Czaya G., Färber P. M., Hegemann J. H. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991 Apr;72(Pt 4):851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]