Abstract
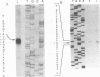
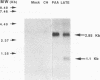
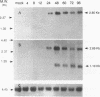
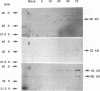
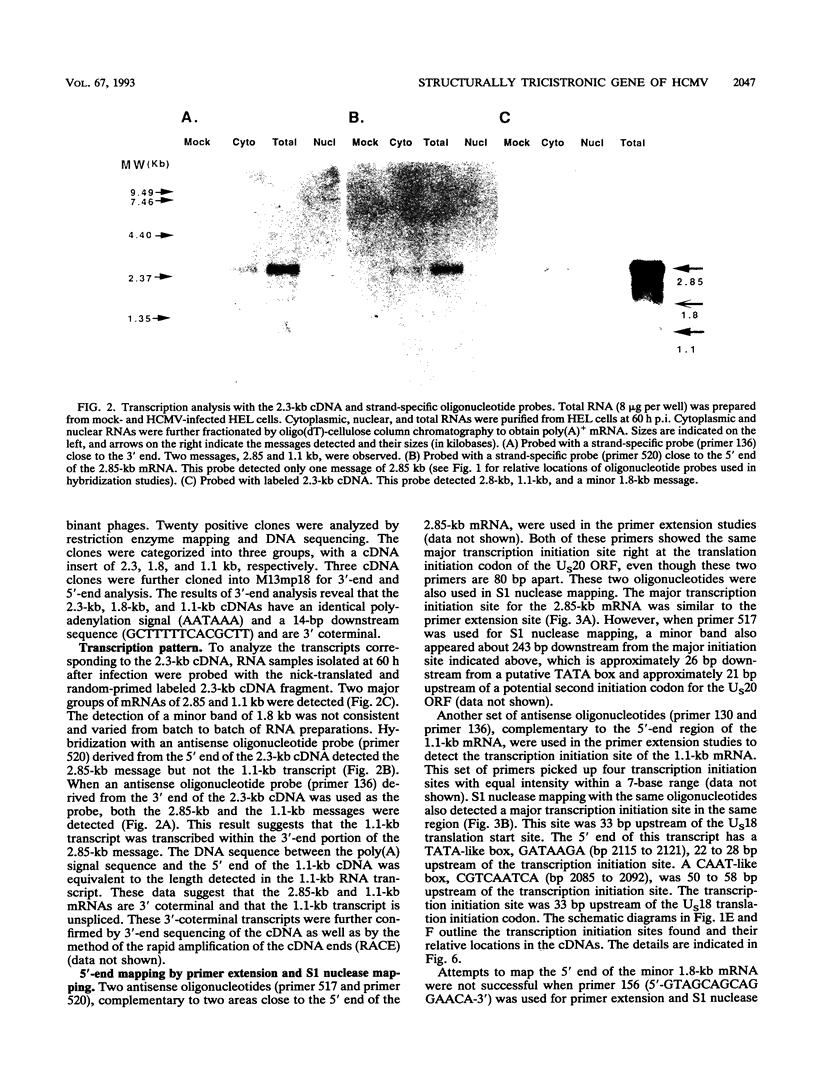
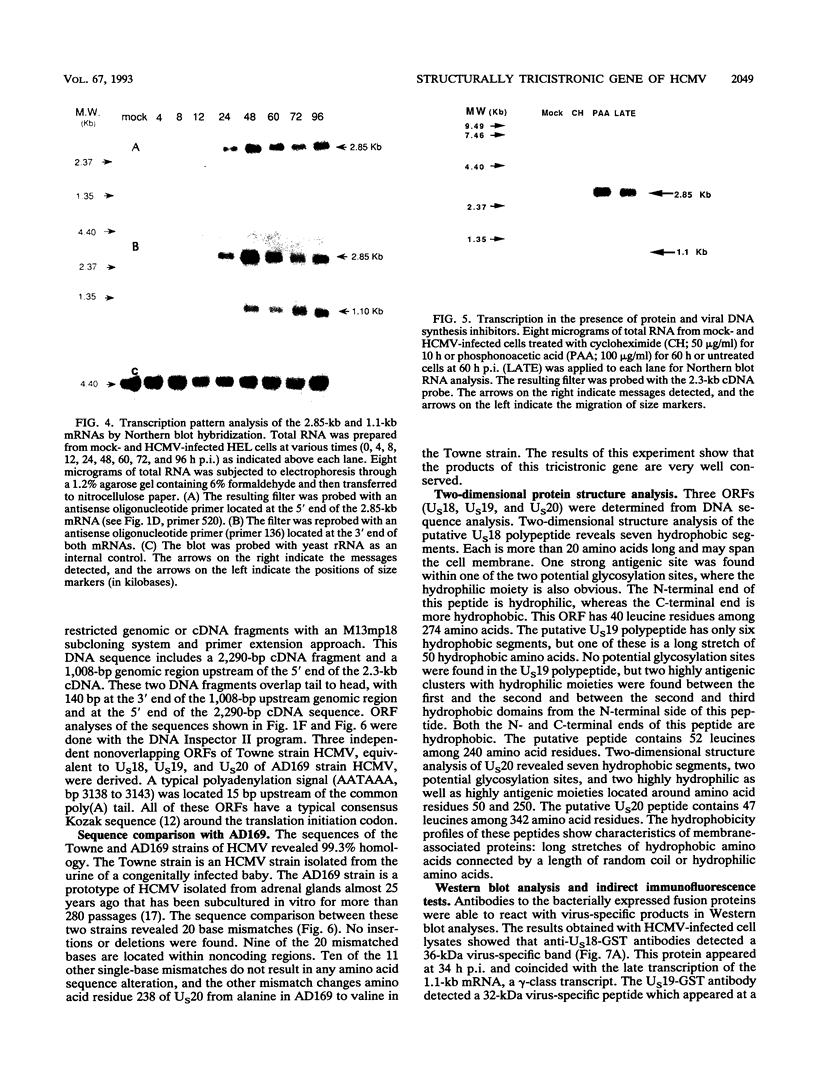
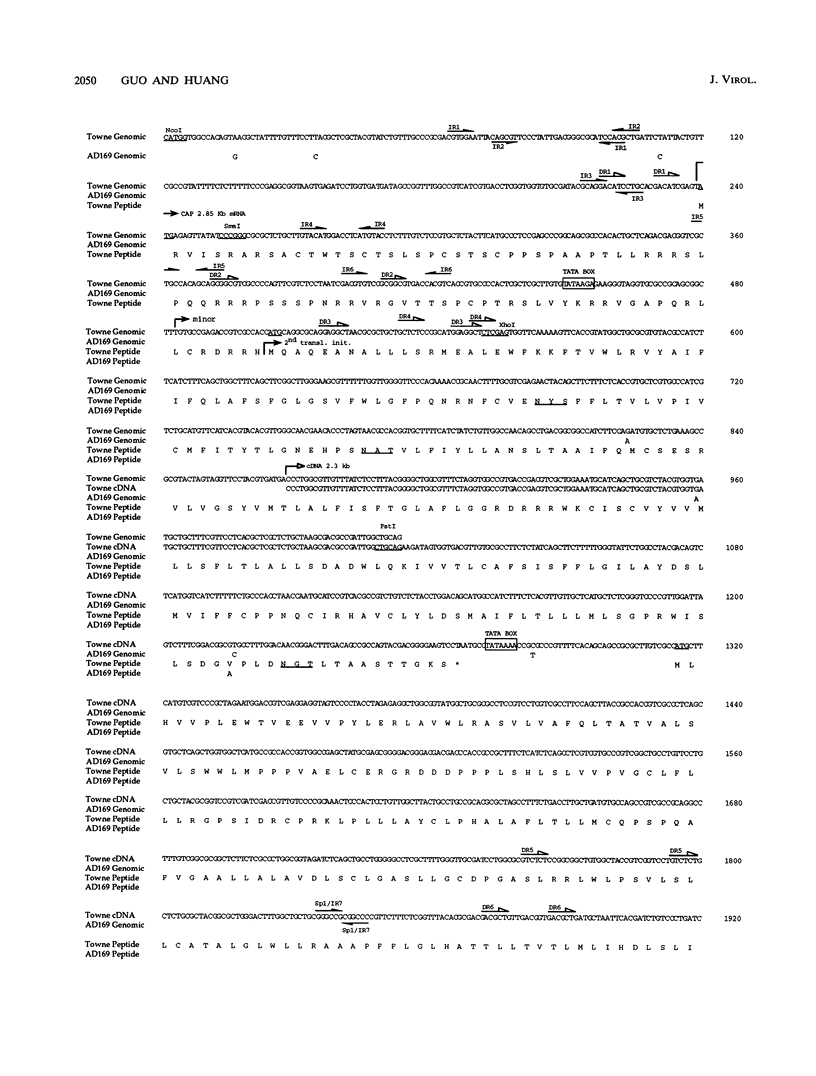
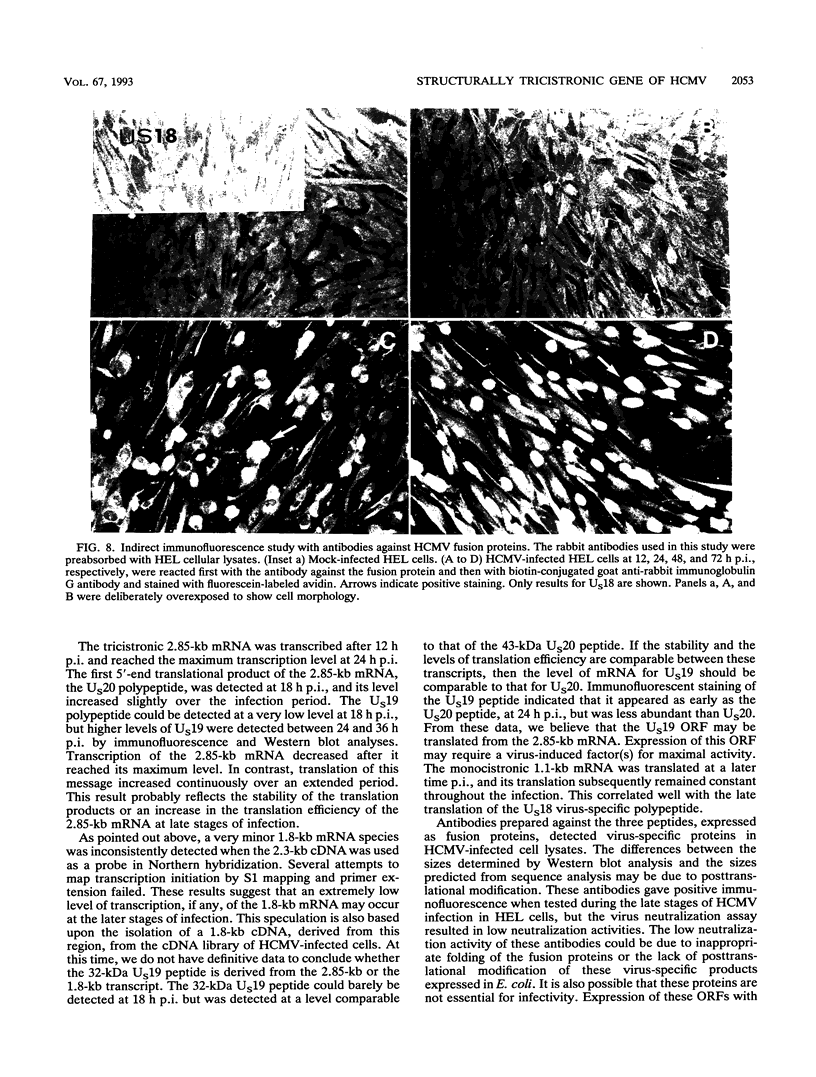
A tricistronic gene mapped between 0.91 and 0.93 map units within the EcoRI D fragment of the human cytomegalovirus unique short region (U(s)) has been cloned, sequenced, and expressed in vitro. Cloned cDNAs of 2.3, 1.8, and 1.1 kb derived from this region were isolated from a lambda gt11 cDNA library made from virus-infected fibroblasts and used for this study. Two major classes of 3'-coterminal mRNAs, 2.8 and 1.1 kb, were transcribed from this region. Sequence analysis of the cDNAs and the upstream genomic DNA revealed three open reading frames (ORFs), U(s)18, U(s)19, and U(s)20, and a common polyadenylation signal located 15 bases upstream of the poly(A) tail of both the 2.85- and 1.1-kb mRNAs. Protein structure analyses predicted the existence of multiple hydrophobic moieties, suggesting that the U(s)18, U(s)19, and U(s)20 polypeptides were transmembrane proteins. The major transcription initiation site, determined by primer extension and S1 nuclease mapping, for the 2.85-kb transcript was located right at the first initiation codon of the U(s)20 ORF. There was no typical TATA box or CAAT box upstream of the 2.85-kb mRNA cap site except for a TATAAGA sequence that was found about 210 bp downstream from the major cap site. The 1.1-kb transcript was initiated 33 bp upstream of the U(s)18 translation initiation site, and an atypical TATA box sequence (GATAAGA) was found 22 bp upstream of the transcription start site. Differences in transcription kinetics and sensitivities to metabolic inhibitors suggest that they were regulated by different mechanisms; the 2.85-kb mRNA belongs to the early (beta) class of transcripts, while the 1.1-kb mRNA is a late (gamma) message. Subgenomic DNA segments derived from the U(s)18, U(s)19, and U(s)20 ORFs were subcloned and overexpressed in Escherichia coli as fusion proteins with glutathione-s-transferase. Western immunoblot analysis with antibodies against the U(s)18, U(s)19, and U(s)20 fusion proteins detected virus-specific polypeptides with molecular sizes of 36, 32, and 43 kDa, respectively. All three antibodies also exhibited a positive immunofluorescence reaction with human cytomegalovirus-infected cells harvested at late stages of infection.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Chang C. P., Vesole D. H., Nelson J., Oldstone M. B., Stinski M. F. Identification and expression of a human cytomegalovirus early glycoprotein. J Virol. 1989 Aug;63(8):3330–3337. doi: 10.1128/jvi.63.8.3330-3337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Davis M. G., Huang E. S. Nucleotide sequence of a human cytomegalovirus DNA fragment encoding a 67-kilodalton phosphorylated viral protein. J Virol. 1985 Oct;56(1):7–11. doi: 10.1128/jvi.56.1.7-11.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretch D. R., Kari B., Gehrz R. C., Stinski M. F. A multigene family encodes the human cytomegalovirus glycoprotein complex gcII (gp47-52 complex). J Virol. 1988 Jun;62(6):1956–1962. doi: 10.1128/jvi.62.6.1956-1962.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. S., Xu L., Huang E. S. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J Virol. 1992 Feb;66(2):1098–1108. doi: 10.1128/jvi.66.2.1098-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick B. A., Huang E. S. Human cytomegalovirus genome: partial denaturation map and organization of genome sequences. J Virol. 1977 Oct;24(1):261–276. doi: 10.1128/jvi.24.1.261-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFemina R. L., Hayward G. S. Replicative forms of human cytomegalovirus DNA with joined termini are found in permissively infected human cells but not in non-permissive Balb/c-3T3 mouse cells. J Gen Virol. 1983 Feb;64(Pt 2):373–389. doi: 10.1099/0022-1317-64-2-373. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Pereira L., McCormick A. L. Human cytomegalovirus ICP22, the product of the HWLF1 reading frame, is an early nuclear protein that is released from cells. J Gen Virol. 1988 Oct;69(Pt 10):2613–2621. doi: 10.1099/0022-1317-69-10-2613. [DOI] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- ROWE W. P., HARTLEY J. W., WATERMAN S., TURNER H. C., HUEBNER R. J. Cytopathogenic agent resembling human salivary gland virus recovered from tissue cultures of human adenoids. Proc Soc Exp Biol Med. 1956 Jun;92(2):418–424. [PubMed] [Google Scholar]
- Rüger B., Klages S., Walla B., Albrecht J., Fleckenstein B., Tomlinson P., Barrell B. Primary structure and transcription of the genes coding for the two virion phosphoproteins pp65 and pp71 of human cytomegalovirus. J Virol. 1987 Feb;61(2):446–453. doi: 10.1128/jvi.61.2.446-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Stinski M. F. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J Virol. 1978 Jun;26(3):686–701. doi: 10.1128/jvi.26.3.686-701.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen M. W., Stinski M. F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982 Feb;41(2):462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K. An enhancer element in the short unique region of human cytomegalovirus regulates the production of a group of abundant immediate early transcripts. Virology. 1988 Feb;162(2):406–416. doi: 10.1016/0042-6822(88)90481-3. [DOI] [PubMed] [Google Scholar]
- Weston K., Barrell B. G. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J Mol Biol. 1986 Nov 20;192(2):177–208. doi: 10.1016/0022-2836(86)90359-1. [DOI] [PubMed] [Google Scholar]