Abstract
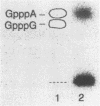
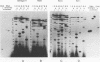
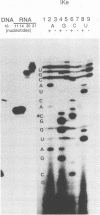
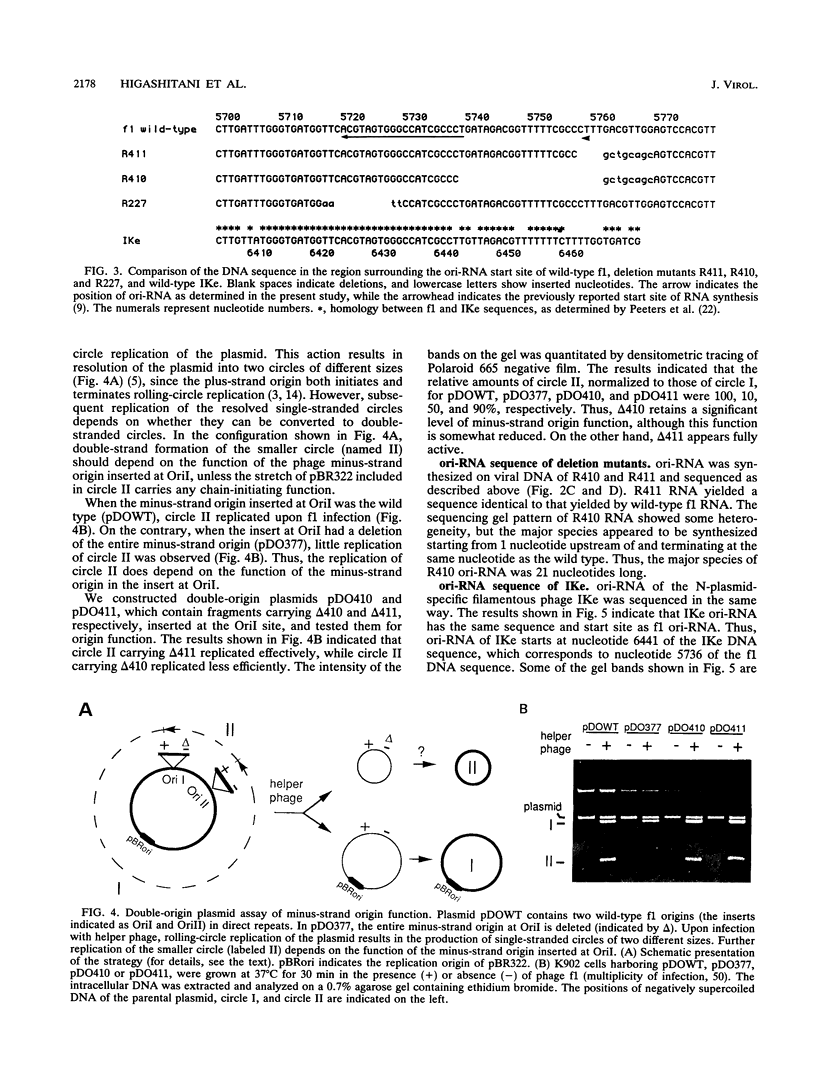
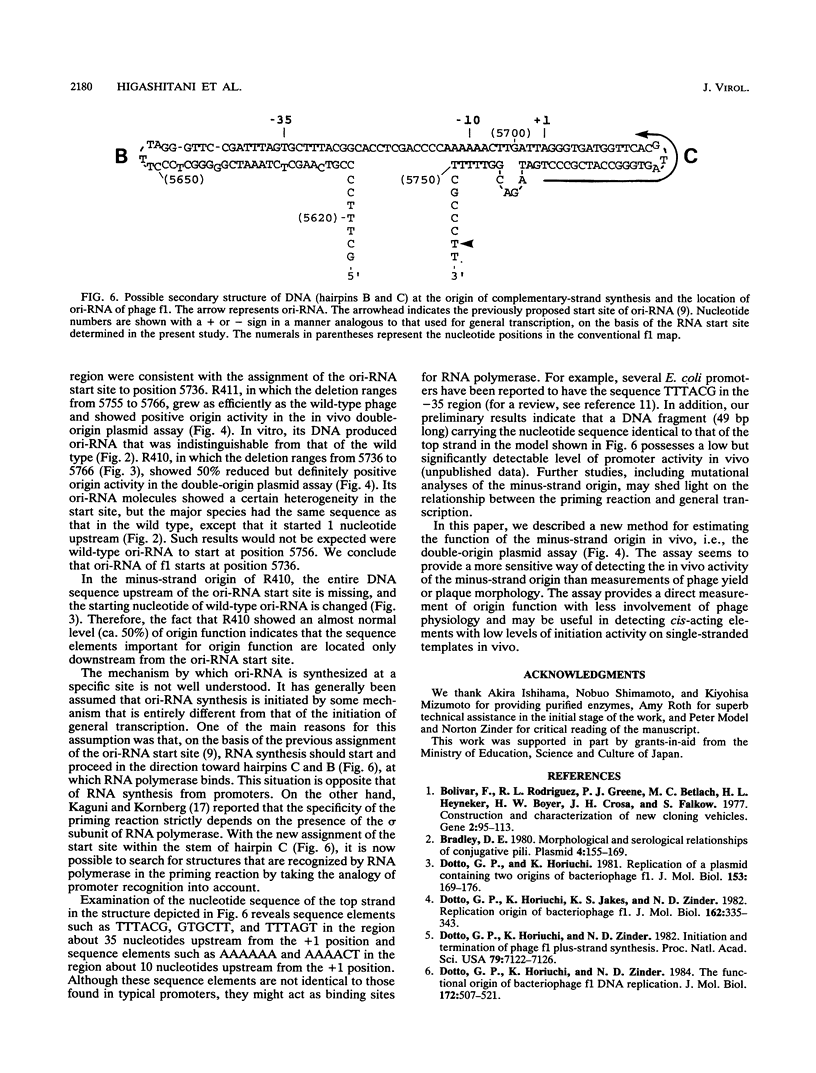
We determined the nucleotide sequence of RNA synthesized in vitro by Escherichia coli RNA polymerase at the complementary-strand replication origin on the single-stranded viral DNA of bacteriophages f1 and IKe (ori-RNA) by using chain-terminating ribonucleoside triphosphate analogs. The results indicated that the start site of f1 ori-RNA synthesis is 20 nucleotides downstream from the site previously reported (K. Geider, E. Beck, and H. Schaller, Proc. Natl. Acad. Sci. USA 75:645-649, 1978) and that the RNA sequence [(5')pppAGGGCGAUGGCCCACUACGU-OH(3')] is complementary to the f1 DNA sequence from nucleotides 5736 to 5717, with minor heterogeneity at the 3' end. IKe ori-RNA had a sequence identical to that of f1 ori-RNA, except for a single base substitution, and IKe RNA was complementary to a region of IKe DNA (from nucleotides 6441 to 6422) that was homologous to the f1 sequence. Phenotypes and ori-RNA sequences in the relevant region of the genome of f1 deletion mutants were consistent with the presently determined sequence of ori-RNA. A possibility that ori-RNA synthesis is initiated by a mechanism similar to that for general transcription is suggested as a result of the new assignment of the ori-RNA start site. The double-origin plasmid assay of minus-strand origin activity, a sensitive in vivo method for detecting cis-acting elements for the initiation of DNA replication on a single-stranded DNA template, is described.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bradley D. E. Morphological and serological relationships of conjugative pili. Plasmid. 1980 Sep;4(2):155–169. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Jakes K. S., Zinder N. D. Replication origin of bacteriophage f1. Two signals required for its function. J Mol Biol. 1982 Dec 5;162(2):335–343. doi: 10.1016/0022-2836(82)90530-7. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K. Replication of a plasmid containing two origins of bacteriophage. J Mol Biol. 1981 Nov 25;153(1):169–176. doi: 10.1016/0022-2836(81)90532-5. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. Initiation and termination of phage f1 plus-strand synthesis. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7122–7126. doi: 10.1073/pnas.79.23.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. The functional origin of bacteriophage f1 DNA replication. Its signals and domains. J Mol Biol. 1984 Feb 5;172(4):507–521. doi: 10.1016/s0022-2836(84)80020-0. [DOI] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford W., Model P. Gene X of bacteriophage f1 is required for phage DNA synthesis. Mutagenesis of in-frame overlapping genes. J Mol Biol. 1984 Sep 15;178(2):137–153. doi: 10.1016/0022-2836(84)90136-0. [DOI] [PubMed] [Google Scholar]
- Geider K., Beck E., Schaller H. An RNA transcribed from DNA at the origin of phage fd single strand to replicative form conversion. Proc Natl Acad Sci U S A. 1978 Feb;75(2):645–649. doi: 10.1073/pnas.75.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Kornberg A. Conversion of the M13 viral single strand to the double-stranded replicative forms by purified proteins. J Biol Chem. 1974 Jul 10;249(13):3999–4005. [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashitani N., Higashitani A., Roth A., Horiuchi K. SOS induction in Escherichia coli by infection with mutant filamentous phage that are defective in initiation of complementary-strand DNA synthesis. J Bacteriol. 1992 Mar;174(5):1612–1618. doi: 10.1128/jb.174.5.1612-1618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982 Oct;44(1):32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K. Origin of DNA replication of bacteriophage f1 as the signal for termination. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5226–5229. doi: 10.1073/pnas.77.9.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Azure mutants: a type of host-dependent mutant of the bacteriophage f2. Science. 1967 Jun 23;156(3782):1618–1623. doi: 10.1126/science.156.3782.1618. [DOI] [PubMed] [Google Scholar]
- Itoh N., Mizumoto K., Kaziro Y. Messenger RNA guanylyltransferase from Saccharomyces cerevisiae. I. Purification and subunit structure. J Biol Chem. 1984 Nov 25;259(22):13923–13929. [PubMed] [Google Scholar]
- Kaguni J. M., Kornberg A. The rho subunit of RNA polymerase holoenzyme confers specificity in priming M13 viral DNA replication. J Biol Chem. 1982 May 25;257(10):5437–5443. [PubMed] [Google Scholar]
- Kim M. H., Hines J. C., Ray D. S. Viable deletions of the M13 complementary strand origin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6784–6788. doi: 10.1073/pnas.78.11.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Lipmann F. Transmethylation and transguanylylation in 5'-RNA capping system isolated from rat liver nuclei. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4961–4965. doi: 10.1073/pnas.76.10.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte J. A., Kahn J. D., Hearst J. E. RNA folding during transcription by Escherichia coli RNA polymerase analyzed by RNA self-cleavage. Biochemistry. 1990 Aug 28;29(34):7882–7890. doi: 10.1021/bi00486a015. [DOI] [PubMed] [Google Scholar]
- Peeters B. P., Peters R. M., Schoenmakers J. G., Konings R. N. Nucleotide sequence and genetic organization of the genome of the N-specific filamentous bacteriophage IKe. Comparison with the genome of the F-specific filamentous phages M13, fd and f1. J Mol Biol. 1985 Jan 5;181(1):27–39. doi: 10.1016/0022-2836(85)90322-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H., Uhlmann A., Geider K. A DNA fragment from the origin of single-strand to double-strand DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1976 Jan;73(1):49–53. doi: 10.1073/pnas.73.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder N. D., Boeke J. D. The filamentous phage (Ff) as vectors for recombinant DNA--a review. Gene. 1982 Jul-Aug;19(1):1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]
- Zinder N. D., Horiuchi K. Multiregulatory element of filamentous bacteriophages. Microbiol Rev. 1985 Jun;49(2):101–106. doi: 10.1128/mr.49.2.101-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]